To understand the mechanisms of the formation and action of human fusion gene breakpoints (FGBPs) in diseases, we performed classifying between human FGBP positive and negative sequences context using a convolutional neural network approach. Using FusionAI, we can study the features of fusion gene breakpoints, infer the potential breakage of the user's interested genomic regions.
 FusionAI architecture and training using deep learning
FusionAI architecture and training using deep learning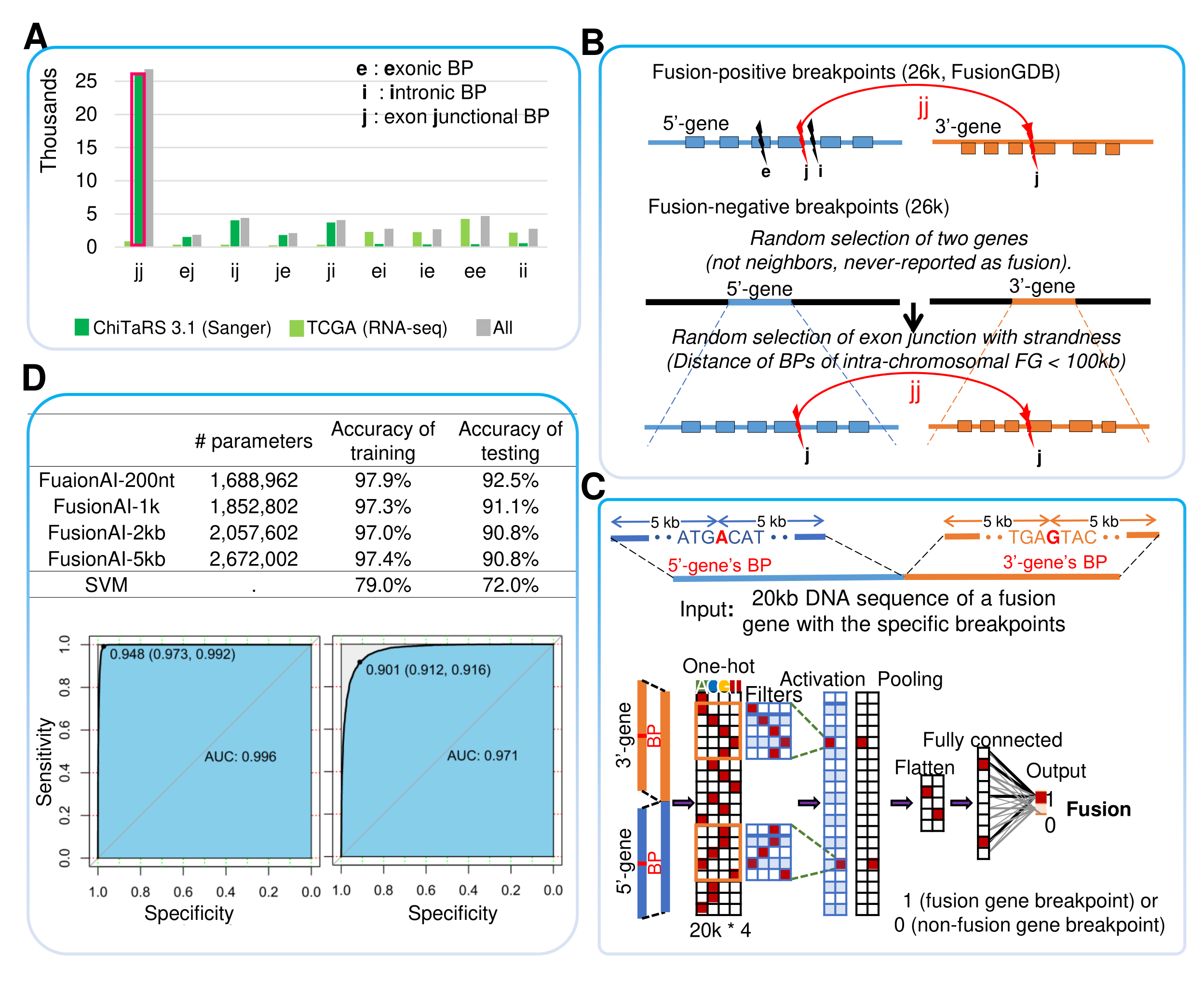
Figure 1. Overview of FusionAI. (a) The investigation of fusion gene breakpoints of 48K FGs from FusionGDB identified the BP location across the human genome. (b) Making training and test datasets of fusion-positive and -negative breakpoints. (c) Diagram of fusion gene breakpoints classification by FusionAI. (d) Effect of the size of the input sequence context on the accuracy.
 Tutorial for the FusionAI
Tutorial for the FusionAISoftware and algorithms to run FusionAI
- python (>=3)
- python modules of tensorflow and keras
:conda install -c bioconda tensorflow
:conda install -c conda-forge keras
Software and algorithms to draw feature landscape image (if you do not need to draw this image, then don't need to install these software)
nibFrag: http://hgdownload.soe.ucsc.edu/admin/jksrc.zip or here
R (>=3.5): https://www.r-project.org/
bedtools (>=2.26.0): https://bedtools.readthedocs.io/en/latest/content/installation.html
bedtoolsr (2.30.0.1): http://phanstiel-lab.med.unc.edu/bedtoolsr-install.html
optparse (>=1.6.0): https://cran.r-project.org/web/packages/optparse/index.html
doParallel (1.0.16): https://cran.r-project.org/web/packages/doParallel/index.html
Iterators (1.0.13): https://cran.r-project.org/web/packages/iterators/index.html
magrittr (2.0.1): https://cran.r-project.org/web/packages/magrittr/index.html
foreach (1.5.1): https://cran.r-project.org/web/packages/foreach/index.html
ggplot2 (3.3.5): https://cran.r-project.org/web/packages/ggplot2/index.html
gridExtra (2.3): https://cran.r-project.org/web/packages/gridExtra/index.html
scales (1.1.1): https://cran.r-project.org/web/packages/scales/index.html
cowplot (1.1.1): https://cran.r-project.org/web/packages/cowplot/index.html
ggpubr (>=0.1.7): https://cran.r-project.org/web/packages/ggpubr/index.html
- gencode_hg19v19_.txt: GENCODE version 19 gene structure information
- nib_files_hg19.tar.gz: hg19 nib files
- chromosome_size.txt: chromosome size information
- features_info.txt: feature information
- features.tar.gz: 44 genomic features of human genome
- newdat_newmod_jj.h5: The FusionAI CNN model
- pre_processing_for_FusionAI_from_tab_delim.py: A python script to make input data for FusionAI from fusion gene information
- FusionAI_pred.py: A python script to predict the ouput scores FusionAI model
- FusionAI_FIS.py: A python script to calculate the feature importance scores (FIS) across 20Kb fusion DNA sequence
- FusionAI_genomic_features.R: An R script to draw a figure for the distribution of 44 human genomic feautures across 20Kb fusion DNA sequence
- FusionAI_genomic_features2.R: An R script to draw a figure for the distribution of 44 human genomic feautures across 20Kb fusion DNA sequence focusing on the top 10% FIS regions only
- k562_starfusion.txt: fusion genes that were predicted using STAR-fusion for K562 cell.
1. Data preparation
: Read fusion gene information as the input and make 20k DNA sequence by combining of +/- 5kb flanking sequence from two breakpoints as the input of FusionAI.
: $ python pre_processing_for_FusionAI_from_tab_delim.py [INPUT_FILE]
: $ python pre_processing_for_FusionAI_from_tab_delim.py k562_starfusion.txt
2. Run FusionAI
: Predict fusion breakpoint tendency using FusionAI model. Here the $INPUT_FILE is the output file after making 20Kb DNA sequence in step 2. $COLA and $COLB are the DNA sequences of 5' and 3' fusion partner genes that were created from step 2. If the user wants to run for one specific fusion gene, then set $INDEX_OF_FUSION as the number of raw in the fusion gene file from the previous step.
: $ python FusionAI_FIS.py [-h] -f FILENAME [-m MODEL, default: newdat_newmod_jj.h5] [-o OUTPUT] [-A COLA] [-B COLB] [-I ROWI] [-N NGPUS]
: $ python FusionAI_pred.py -f k562_starfusion.FusionAI.input -o k562_starfusion.FusionAI.output -m newdat_newmod_jj.h5
3. Select high FusionAI prediction scored fusions or interested fusions (using Excel program).
: The users can cut at 0.5 or 0.9 or 0.95 based on the numbers and overalpping with fusion prediction results by other tools.
4. Calculate the feature importance scores across 20Kb fusion DNA sequence.
: Check the feature importance across 20Kb fusion DNA sequence of the selected fusion genes. If the user wants to run for one specific fusion gene, then set $INDEX_OF_FUSION as the number of raw in the fusion gene file from the previous step.
: $ python FusionAI_FIS.py [-h] -f [INPUT_FILE] -m [MODEL] -o [OUTPUT] -A [COLA] -B [COLB] -I [INDEX_OF_FUSION]
: $ python FusionAI_FIS.py -f k562_starfusion.FusionAI.output -o k562_starfusion.FusionAI.output.FIS
5. Visualize 44 human genomic features across 20Kb DNA sequence.
5-1. Draw the distribution of 44 human genomic feautures across 20Kb fusion DNA sequence.
: $ Rscript FusionAI_genomic_features.R -g [FUSION_GENE_FILE] -f [FEATURE_PATH] -s [CHROMOSOME_SIZE_FILE] -i [FEATURE_INFO_FILE] -o [OUTPUT_FILE_PATH]
: $ Rscript FusionAI_genomic_features.R -g K562_STARfusion.FusionAI.output.FIS -f ./features/ -s chromosome_size.txt -i features_info.txt -o ./K562/whole_features/
5-2. Draw the distribution of 44 human genomic feautures across 20Kb fusion DNA sequence focusing on the top 10% FIS regions only.
: $ Rscript FusionAI_genomic_features2.R -g [FUSION_GENE_FILE] -f [FEATURE_PATH] -s [CHROMOSOME_SIZE_FILE] -i [FEATURE_INFO_FILE] -o [OUTPUT_FILE_PATH]
: $ Rscript FusionAI_genomic_features2.R -g K562_STARfusion.FusionAI.output.FIS -f ./features/ -s chromosome_size.txt -i features_info.txt -o ./K562/top1pct_features/
- Fusion data
: We used exon junction-junction breakpoint fusions from TCGA fusion data (26K=18K training + 8K test)
- Training data set
: ~ 18K fusion-positive breakpoint sequences + ~ 18K negative fusion-negative breakpoint sequences
: Hgene\tHchr\tHbp\tHstrand\tTgene\tTchr\tTbp\tTstrand\tSequence
- Training data information
: Hgene\tHchr\tHbp\tHstrand\tTgene\tTchr\tTbp\tTstrand\tT or F(1 or 0)
- Simulation RNA-seq split reads of training data
: Simulation RNA-seq split reads aligned at the exon junction-junction breakpoints of fusion events in training data set
- Test data set
: ~ 8K fusion-positive breakpoint sequences + ~ 8K negative fusion-negative breakpoint sequences
: Hgene\tHchr\tHbp\tHstrand\tTgene\tTchr\tTbp\tTstrand\tSequence
- Test data information
: Hgene\tHchr\tHbp\tHstrand\tTgene\tTchr\tTbp\tTstrand\tT or F(1 or 0)
- Simulation RNA-seq split reads of teset data
: Simulation RNA-seq split reads aligned at the exon junction-junction breakpoints of fusion events in test data set
- Sanger sequece based fusion data for validation
: Sanger sequece based exon junction-junction breakpoints of fusion events from ChiTaRs-3.1
: Input for FusionAI (20Kbp sequence)
: Simulation RNA-seq split reads
- 2200 fusion events from ~600 cancer cell-lines for validation
: Sanger sequece based exon junction-junction breakpoints of fusion events from 600 cancer cell-lines
: Input for FusionAI (20Kbp sequence)
: Simulation RNA-seq split reads
-Input by the user (Input in step 1)
:Hgene\tHchr\tHbp\tHstrand\tTgene\tTchr\tTbp\tTstrand
-Input of FusionAI after pro_processing (Output in step 1-2/Input in step 2)
:Hgene\tHchr\tHbp\tHstrand\tTgene\tTchr\tTbp\tTstrand\t5'-gene sequence (10Kb)\t3'-gene sequence (10Kb)
-output
:Fusion-negative probability\tFusion-positive probability (Output in step 3)
 FusionGDB2: fusion gene annotation update aided by deep learning
FusionGDB2: fusion gene annotation update aided by deep learning About us
About usSchool of Biomedical Informatics
The University of Texas Health Science Center at Houston
7000 Fannin Street, Houston, TX 77030
 Citation
CitationTo cite the FusionGDB website in a publication, please quote the following:
- Kim P*, Tan H, Liu J, Lee H, Jung H, Kumar H, and Zhou X. FusionGDB 2.0: fusion gene annotation update aided by deep learning. Nucleic Acids Res. 2021 Nov 10; doi: 10.1093/nar/gkab1056
- Kim P*, Tan H, Liu J, Yang M, and Zhou X*, FusionAI: Predicting fusion breakpoint from DNA sequence with deep learning. iScience. 2021 Sep 25; 24(10):103164. doi: 10.1016/j.isci.2021.103164.
- Kim P*, Yiya K*, and Zhou X*. FGviewer: an online visualization tool for functional features of human fusion genes. Nucleic Acids Res. 2020 Jul 2;48(W1):W313-W320.
- Kim P and Zhou X*. FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res. 2019 Jan 8;47(D1):D994-D1004
