
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:YWHAZ-EIF3A (FusionGDB2 ID:100213) |
Fusion Gene Summary for YWHAZ-EIF3A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: YWHAZ-EIF3A | Fusion gene ID: 100213 | Hgene | Tgene | Gene symbol | YWHAZ | EIF3A | Gene ID | 7534 | 8661 |
| Gene name | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | eukaryotic translation initiation factor 3 subunit A | |
| Synonyms | 14-3-3-zeta|HEL-S-3|HEL-S-93|HEL4|KCIP-1|POPCHAS|YWHAD | EIF3|EIF3S10|P167|TIF32|eIF3-p170|eIF3-theta|p180|p185 | |
| Cytomap | 8q22.3 | 10q26.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | 14-3-3 protein zeta/delta14-3-3 delta14-3-3 protein/cytosolic phospholipase A214-3-3 zetaepididymis luminal protein 4epididymis secretory protein Li 3epididymis secretory protein Li 93phospholipase A2protein kinase C inhibitor protein-1tyrosine 3 | eukaryotic translation initiation factor 3 subunit AEIF3, p180 subunitcentrosomin homologcytoplasmic protein p167eIF-3-thetaeIF3 p167eIF3 p180eIF3 p185eukaryotic translation initiation factor 3 subunit 10eukaryotic translation initiation factor 3 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | Q14152 | |
| Ensembl transtripts involved in fusion gene | ENST00000353245, ENST00000395948, ENST00000395956, ENST00000395958, ENST00000457309, ENST00000521309, ENST00000522542, ENST00000395951, ENST00000395953, ENST00000395957, ENST00000419477, ENST00000522819, ENST00000492736, | ENST00000369144, ENST00000541549, | |
| Fusion gene scores | * DoF score | 22 X 20 X 10=4400 | 8 X 9 X 3=216 |
| # samples | 29 | 9 | |
| ** MAII score | log2(29/4400*10)=-3.92337871839709 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/216*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: YWHAZ [Title/Abstract] AND EIF3A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | YWHAZ(101937143)-EIF3A(120796821), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | YWHAZ-EIF3A seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. YWHAZ-EIF3A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. YWHAZ-EIF3A seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. YWHAZ-EIF3A seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. YWHAZ-EIF3A seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | YWHAZ | GO:0070372 | regulation of ERK1 and ERK2 cascade | 31024343 |
| Hgene | YWHAZ | GO:0090128 | regulation of synapse maturation | 16959763 |
| Tgene | EIF3A | GO:0001732 | formation of cytoplasmic translation initiation complex | 17581632 |
| Tgene | EIF3A | GO:0075522 | IRES-dependent viral translational initiation | 9573242|23766293|24357634 |
| Tgene | EIF3A | GO:0075525 | viral translational termination-reinitiation | 21347434 |
 Fusion gene breakpoints across YWHAZ (5'-gene) Fusion gene breakpoints across YWHAZ (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
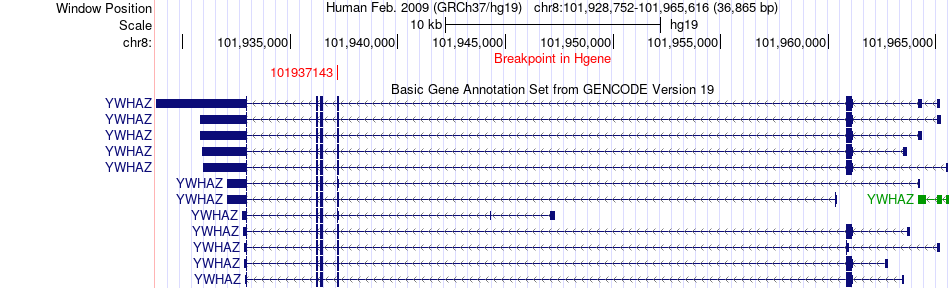 |
 Fusion gene breakpoints across EIF3A (3'-gene) Fusion gene breakpoints across EIF3A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
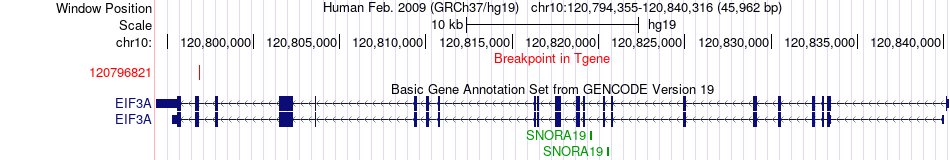 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-FD-A5BY | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
Top |
Fusion Gene ORF analysis for YWHAZ-EIF3A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000353245 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000353245 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000395948 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000395948 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000395956 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000395956 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000395958 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000395958 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000457309 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000457309 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000521309 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000521309 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000522542 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| Frame-shift | ENST00000522542 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000395951 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000395951 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000395953 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000395953 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000395957 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000395957 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000419477 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000419477 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000522819 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| In-frame | ENST00000522819 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| intron-3CDS | ENST00000492736 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
| intron-3CDS | ENST00000492736 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000395957 | YWHAZ | chr8 | 101937143 | - | ENST00000369144 | EIF3A | chr10 | 120796821 | - | 2376 | 760 | 978 | 340 | 212 |
| ENST00000395957 | YWHAZ | chr8 | 101937143 | - | ENST00000541549 | EIF3A | chr10 | 120796821 | - | 1427 | 760 | 978 | 340 | 212 |
| ENST00000522819 | YWHAZ | chr8 | 101937143 | - | ENST00000369144 | EIF3A | chr10 | 120796821 | - | 2004 | 388 | 597 | 842 | 81 |
| ENST00000522819 | YWHAZ | chr8 | 101937143 | - | ENST00000541549 | EIF3A | chr10 | 120796821 | - | 1055 | 388 | 606 | 103 | 167 |
| ENST00000395953 | YWHAZ | chr8 | 101937143 | - | ENST00000369144 | EIF3A | chr10 | 120796821 | - | 2184 | 568 | 786 | 148 | 212 |
| ENST00000395953 | YWHAZ | chr8 | 101937143 | - | ENST00000541549 | EIF3A | chr10 | 120796821 | - | 1235 | 568 | 786 | 148 | 212 |
| ENST00000395951 | YWHAZ | chr8 | 101937143 | - | ENST00000369144 | EIF3A | chr10 | 120796821 | - | 2197 | 581 | 25 | 702 | 225 |
| ENST00000395951 | YWHAZ | chr8 | 101937143 | - | ENST00000541549 | EIF3A | chr10 | 120796821 | - | 1248 | 581 | 25 | 702 | 225 |
| ENST00000419477 | YWHAZ | chr8 | 101937143 | - | ENST00000369144 | EIF3A | chr10 | 120796821 | - | 2136 | 520 | 738 | 100 | 212 |
| ENST00000419477 | YWHAZ | chr8 | 101937143 | - | ENST00000541549 | EIF3A | chr10 | 120796821 | - | 1187 | 520 | 738 | 100 | 212 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000395957 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.001620949 | 0.998379 |
| ENST00000395957 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.003400997 | 0.996599 |
| ENST00000522819 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.45217726 | 0.5478227 |
| ENST00000522819 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.35373938 | 0.6462607 |
| ENST00000395953 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.000717964 | 0.99928206 |
| ENST00000395953 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.001562406 | 0.9984376 |
| ENST00000395951 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.001034001 | 0.998966 |
| ENST00000395951 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.001697163 | 0.9983028 |
| ENST00000419477 | ENST00000369144 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.000641226 | 0.99935883 |
| ENST00000419477 | ENST00000541549 | YWHAZ | chr8 | 101937143 | - | EIF3A | chr10 | 120796821 | - | 0.001621743 | 0.9983783 |
Top |
Fusion Genomic Features for YWHAZ-EIF3A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
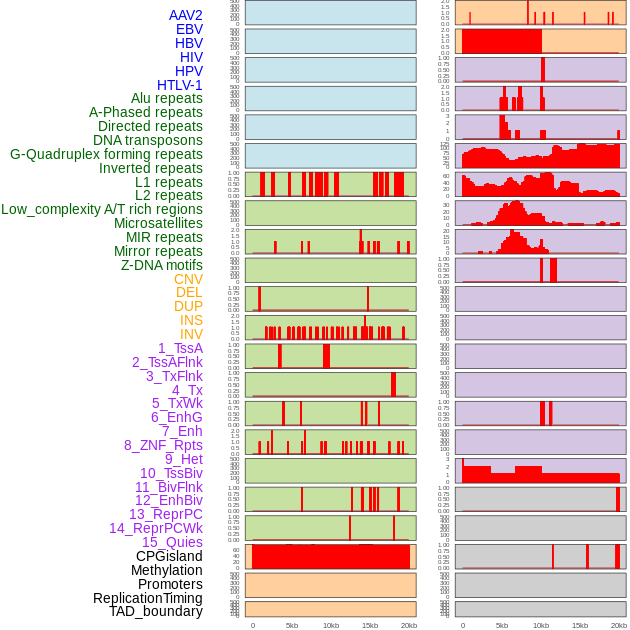 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for YWHAZ-EIF3A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:101937143/chr10:120796821) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | EIF3A |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: RNA-binding component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis (PubMed:17581632, PubMed:25849773). The eIF-3 complex associates with the 40S ribosome and facilitates the recruitment of eIF-1, eIF-1A, eIF-2:GTP:methionyl-tRNAi and eIF-5 to form the 43S pre-initiation complex (43S PIC). The eIF-3 complex stimulates mRNA recruitment to the 43S PIC and scanning of the mRNA for AUG recognition. The eIF-3 complex is also required for disassembly and recycling of post-termination ribosomal complexes and subsequently prevents premature joining of the 40S and 60S ribosomal subunits prior to initiation (PubMed:17581632, PubMed:11169732). The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation, including cell cycling, differentiation and apoptosis, and uses different modes of RNA stem-loop binding to exert either translational activation or repression (PubMed:25849773, PubMed:27462815). {ECO:0000255|HAMAP-Rule:MF_03000, ECO:0000269|PubMed:11169732, ECO:0000269|PubMed:17581632, ECO:0000269|PubMed:25849773, ECO:0000269|PubMed:27462815}.; FUNCTION: (Microbial infection) Essential for the initiation of translation on type-1 viral ribosomal entry sites (IRESs), like for HCV, PV, EV71 or BEV translation (PubMed:23766293, PubMed:24357634). {ECO:0000269|PubMed:23766293, ECO:0000269|PubMed:24357634}.; FUNCTION: (Microbial infection) In case of FCV infection, plays a role in the ribosomal termination-reinitiation event leading to the translation of VP2 (PubMed:18056426). {ECO:0000269|PubMed:18056426}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 82_120 | 1242 | 1383.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 576_875 | 1242 | 1383.0 | Compositional bias | Note=Glu-rich | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 925_1294 | 1242 | 1383.0 | Compositional bias | Note=Asp-rich | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 315_498 | 1242 | 1383.0 | Domain | PCI | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 925_1172 | 1242 | 1383.0 | Region | Note=25 X 10 AA approximate tandem repeats of [DE]-[DE]-[DE]-R-[SEVGFPILV]-[HPSN]-[RSW]-[RL]-[DRGTIHN]-[EPMANLGDT] | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1003_1012 | 1242 | 1383.0 | Repeat | Note=9 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1013_1022 | 1242 | 1383.0 | Repeat | Note=10 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1023_1032 | 1242 | 1383.0 | Repeat | Note=11 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1033_1042 | 1242 | 1383.0 | Repeat | Note=12 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1043_1052 | 1242 | 1383.0 | Repeat | Note=13 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1054_1063 | 1242 | 1383.0 | Repeat | Note=14 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1064_1073 | 1242 | 1383.0 | Repeat | Note=15 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1074_1083 | 1242 | 1383.0 | Repeat | Note=16 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1084_1093 | 1242 | 1383.0 | Repeat | Note=17 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1094_1103 | 1242 | 1383.0 | Repeat | Note=18 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1104_1113 | 1242 | 1383.0 | Repeat | Note=19 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1114_1123 | 1242 | 1383.0 | Repeat | Note=20 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1124_1133 | 1242 | 1383.0 | Repeat | Note=21 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1134_1143 | 1242 | 1383.0 | Repeat | Note=22 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1144_1152 | 1242 | 1383.0 | Repeat | Note=23%3B truncated | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1153_1162 | 1242 | 1383.0 | Repeat | Note=24 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 1163_1172 | 1242 | 1383.0 | Repeat | Note=25%3B approximate | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 925_934 | 1242 | 1383.0 | Repeat | Note=1 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 935_942 | 1242 | 1383.0 | Repeat | Note=2%3B truncated | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 943_952 | 1242 | 1383.0 | Repeat | Note=3 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 953_962 | 1242 | 1383.0 | Repeat | Note=4 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 963_972 | 1242 | 1383.0 | Repeat | Note=5 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 973_982 | 1242 | 1383.0 | Repeat | Note=6 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 983_992 | 1242 | 1383.0 | Repeat | Note=7 | |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 993_1002 | 1242 | 1383.0 | Repeat | Note=8 |
Top |
Fusion Gene Sequence for YWHAZ-EIF3A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >100213_100213_1_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395951_EIF3A_chr10_120796821_ENST00000369144_length(transcript)=2197nt_BP=581nt ATGACAAAGGAGGAGAGTCGGGGACTTGAGCCGTGGCTCCGACTTGGGCGGAGCCTGGAGGGGGGTGGTTGCGATACGCGGACCGGAGAA TTTGCACTTTAAAGTCCGGGTCTGCCCGTTTTCGTTTCACAGTAACCGACGTCTCAAGTCAGAACATCCAGTCATGGATAAAAATGAGCT GGTTCAGAAGGCCAAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATGAAGTCTGTAACTGAGCAAGGAGCTGAATT ATCCAATGAGGAGAGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGGTCATCTTGGAGGGTCGTCTCAAGTATTGA ACAAAAGACGGAAGGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATTGAGACGGAGCTAAGAGATATCTGCAATGA TGTACTGTCTCTTTTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTA CCGTTACTTGGCTGAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGG AGAGACTCAAGTCGCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTA AGAGACGACAGGGACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGAC CGGGTGGAAGAGCGGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGT GAAAAAGAGAAGGCCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACA GTACGACGTTAAGTCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAAC CAATCTAAATTGGATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACC ATATCCACAGGAGGTTGGAAAAACCATGCCATTTTCTGGAATTTAAGGGTGTTGCATTATTTCATCAATCATTTGTTGACAAAAAAGAAA AACTAAAAAATAAATTTAAAATGTGACCCTTCAGGTATTGAGTAACACCTTTATCTTGGTATAGAACTGATACTTTTTTTTTGATTTTGA AATATCTGATAATAATTTGGAATGAAGTAAGGTTCTGTTAAAATATATTTGAAGACCCTTTAAAGCAGTGAATCTGAAACAATTTTCACA CCCTTAAGTGGTTGATACGTACCTATTTTAGGTATTTTGAGGTATTTACCATAAACTAAATTTAGAAATTTTTTAGATTCACTTGAAGTA AACATTACAAACATTGGATACGGTGGGGTTTTCTTTAGATTTTACTTGAGAGAAGGTGAGTACAAAGCAATTTGCAGTTGTTGTAATGAC AAGATTACTGCGCAAGTGTGAATCCAAACAGTATAGCTTTTAAATTTTAAAGCATTTGGTAAATTATCGCTGAGTTTTTTTCTGTTGCCA ATAGCAAACTGCTTTTCCATTAATGGAGAATTCATGCCTTTCAAGCATTTTAAATATGACAATATTTATAAATGTATGGTTTGGAGGAAT CGTTTAAATTCTCTTTCCTAATTTTCTTTCTTTTGAAGATAGATTCTTTCAACAAGTAATTTGTAGTAATGACTGTGTTGACTTCAATTT TGGAGCGCAGTAGCTATGTTAAAGATGAACTATTTGGTCTCATTGAAGCCAACACAGAACTTGCTGCTGTGTTTTTTCTTCAGTGATAAA TAAAATACTTACAGAATTTGTTTTAGTGTTGATTTGTGGTTATAGTATTTTGTTTATAATGGTAAGTTTGCATATTCAGTTGAGTTTTTT TTACTTGAATTTTTATCAGTGCCATTAAATGTCTGTGTTTAGTATCAATGAAAATGAACCTAAATATAACAAAGAAAGCATATGTGGCTA >100213_100213_1_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395951_EIF3A_chr10_120796821_ENST00000369144_length(amino acids)=225AA_BP=185 MSRGSDLGGAWRGVVAIRGPENLHFKVRVCPFSFHSNRRLKSEHPVMDKNELVQKAKLAEQAERYDDMAACMKSVTEQGAELSNEERNLL SVAYKNVVGARRSSWRVVSSIEQKTEGAEKKQQMAREYREKIETELRDICNDVLSLLEKFLIPNASQAESKVFYLKMKGDYYRYLAEVAA -------------------------------------------------------------- >100213_100213_2_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395951_EIF3A_chr10_120796821_ENST00000541549_length(transcript)=1248nt_BP=581nt ATGACAAAGGAGGAGAGTCGGGGACTTGAGCCGTGGCTCCGACTTGGGCGGAGCCTGGAGGGGGGTGGTTGCGATACGCGGACCGGAGAA TTTGCACTTTAAAGTCCGGGTCTGCCCGTTTTCGTTTCACAGTAACCGACGTCTCAAGTCAGAACATCCAGTCATGGATAAAAATGAGCT GGTTCAGAAGGCCAAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATGAAGTCTGTAACTGAGCAAGGAGCTGAATT ATCCAATGAGGAGAGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGGTCATCTTGGAGGGTCGTCTCAAGTATTGA ACAAAAGACGGAAGGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATTGAGACGGAGCTAAGAGATATCTGCAATGA TGTACTGTCTCTTTTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTA CCGTTACTTGGCTGAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGG AGAGACTCAAGTCGCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTA AGAGACGACAGGGACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGAC CGGGTGGAAGAGCGGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGT GAAAAAGAGAAGGCCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACA GTACGACGTTAAGTCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAAC CAATCTAAATTGGATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACC >100213_100213_2_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395951_EIF3A_chr10_120796821_ENST00000541549_length(amino acids)=225AA_BP=185 MSRGSDLGGAWRGVVAIRGPENLHFKVRVCPFSFHSNRRLKSEHPVMDKNELVQKAKLAEQAERYDDMAACMKSVTEQGAELSNEERNLL SVAYKNVVGARRSSWRVVSSIEQKTEGAEKKQQMAREYREKIETELRDICNDVLSLLEKFLIPNASQAESKVFYLKMKGDYYRYLAEVAA -------------------------------------------------------------- >100213_100213_3_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395953_EIF3A_chr10_120796821_ENST00000369144_length(transcript)=2184nt_BP=568nt GCATGGCCGGGGCGCCGCCGGCCGCGATCCATCCCCACCCTTGTCTCTGCGGGCTTCCACCCCCTCCCTCGCCGCCCTTCTCGCTTCCGC CCGAGATAATGACTCGGCGGCGGCGGCGGCGGGCATCGCAGTGGCGCAGAACATCCAGTCATGGATAAAAATGAGCTGGTTCAGAAGGCC AAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATGAAGTCTGTAACTGAGCAAGGAGCTGAATTATCCAATGAGGAG AGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGGTCATCTTGGAGGGTCGTCTCAAGTATTGAACAAAAGACGGAA GGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATTGAGACGGAGCTAAGAGATATCTGCAATGATGTACTGTCTCTT TTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTACCGTTACTTGGCT GAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGGAGAGACTCAAGTC GCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTAAGAGACGACAGGG ACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGACCGGGTGGAAGAGC GGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGTGAAAAAGAGAAGG CCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACAGTACGACGTTAAG TCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAACCAATCTAAATTGG ATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACCATATCCACAGGAG GTTGGAAAAACCATGCCATTTTCTGGAATTTAAGGGTGTTGCATTATTTCATCAATCATTTGTTGACAAAAAAGAAAAACTAAAAAATAA ATTTAAAATGTGACCCTTCAGGTATTGAGTAACACCTTTATCTTGGTATAGAACTGATACTTTTTTTTTGATTTTGAAATATCTGATAAT AATTTGGAATGAAGTAAGGTTCTGTTAAAATATATTTGAAGACCCTTTAAAGCAGTGAATCTGAAACAATTTTCACACCCTTAAGTGGTT GATACGTACCTATTTTAGGTATTTTGAGGTATTTACCATAAACTAAATTTAGAAATTTTTTAGATTCACTTGAAGTAAACATTACAAACA TTGGATACGGTGGGGTTTTCTTTAGATTTTACTTGAGAGAAGGTGAGTACAAAGCAATTTGCAGTTGTTGTAATGACAAGATTACTGCGC AAGTGTGAATCCAAACAGTATAGCTTTTAAATTTTAAAGCATTTGGTAAATTATCGCTGAGTTTTTTTCTGTTGCCAATAGCAAACTGCT TTTCCATTAATGGAGAATTCATGCCTTTCAAGCATTTTAAATATGACAATATTTATAAATGTATGGTTTGGAGGAATCGTTTAAATTCTC TTTCCTAATTTTCTTTCTTTTGAAGATAGATTCTTTCAACAAGTAATTTGTAGTAATGACTGTGTTGACTTCAATTTTGGAGCGCAGTAG CTATGTTAAAGATGAACTATTTGGTCTCATTGAAGCCAACACAGAACTTGCTGCTGTGTTTTTTCTTCAGTGATAAATAAAATACTTACA GAATTTGTTTTAGTGTTGATTTGTGGTTATAGTATTTTGTTTATAATGGTAAGTTTGCATATTCAGTTGAGTTTTTTTTACTTGAATTTT TATCAGTGCCATTAAATGTCTGTGTTTAGTATCAATGAAAATGAACCTAAATATAACAAAGAAAGCATATGTGGCTAGGATGTCCCCCTA >100213_100213_3_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395953_EIF3A_chr10_120796821_ENST00000369144_length(amino acids)=212AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL LSFSNRRLCSLLVKHWGSRTFPKETVHHCRYLLAPSQFSLCILEPSAVFSQHLPSFVQYLRRPSKMTYGLLQHFYKQLREDSSPHWIIQL -------------------------------------------------------------- >100213_100213_4_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395953_EIF3A_chr10_120796821_ENST00000541549_length(transcript)=1235nt_BP=568nt GCATGGCCGGGGCGCCGCCGGCCGCGATCCATCCCCACCCTTGTCTCTGCGGGCTTCCACCCCCTCCCTCGCCGCCCTTCTCGCTTCCGC CCGAGATAATGACTCGGCGGCGGCGGCGGCGGGCATCGCAGTGGCGCAGAACATCCAGTCATGGATAAAAATGAGCTGGTTCAGAAGGCC AAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATGAAGTCTGTAACTGAGCAAGGAGCTGAATTATCCAATGAGGAG AGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGGTCATCTTGGAGGGTCGTCTCAAGTATTGAACAAAAGACGGAA GGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATTGAGACGGAGCTAAGAGATATCTGCAATGATGTACTGTCTCTT TTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTACCGTTACTTGGCT GAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGGAGAGACTCAAGTC GCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTAAGAGACGACAGGG ACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGACCGGGTGGAAGAGC GGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGTGAAAAAGAGAAGG CCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACAGTACGACGTTAAG TCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAACCAATCTAAATTGG ATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACCATATCCACAGGAG >100213_100213_4_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395953_EIF3A_chr10_120796821_ENST00000541549_length(amino acids)=212AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL LSFSNRRLCSLLVKHWGSRTFPKETVHHCRYLLAPSQFSLCILEPSAVFSQHLPSFVQYLRRPSKMTYGLLQHFYKQLREDSSPHWIIQL -------------------------------------------------------------- >100213_100213_5_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395957_EIF3A_chr10_120796821_ENST00000369144_length(transcript)=2376nt_BP=760nt CGCAGTCACCGAGCGCAGATCCCGCGCTCTCGATTGGAACGCCTCCCCATCACTCAGCCACACTCAGGGGCTGGACGCCTCACTCCCGTT TCCGAGCCATAAAAGGTCTAGGACCGCTTCCCCCGAGCCAGCAGCGTTTGACGTCATCGTGCGTGTGGTGCCCCTGCTGCCGGGGCTGGT GATTGGAGGAAACCCCGTGTCTGCGGAGCGGCTGTAGCCTGTGAGCAGCGAGATCCAGGGACAGAGTCTCAGCCTCGCCGCTGCTGCCGC CGCCGCCGCCCAGAGACTGCTGAGCCCGTCCGTCCGCCGCCACCACCCACTCCGGACACAGAACATCCAGTCATGGATAAAAATGAGCTG GTTCAGAAGGCCAAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATGAAGTCTGTAACTGAGCAAGGAGCTGAATTA TCCAATGAGGAGAGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGGTCATCTTGGAGGGTCGTCTCAAGTATTGAA CAAAAGACGGAAGGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATTGAGACGGAGCTAAGAGATATCTGCAATGAT GTACTGTCTCTTTTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTAC CGTTACTTGGCTGAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGGA GAGACTCAAGTCGCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTAA GAGACGACAGGGACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGACC GGGTGGAAGAGCGGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGTG AAAAAGAGAAGGCCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACAG TACGACGTTAAGTCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAACC AATCTAAATTGGATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACCA TATCCACAGGAGGTTGGAAAAACCATGCCATTTTCTGGAATTTAAGGGTGTTGCATTATTTCATCAATCATTTGTTGACAAAAAAGAAAA ACTAAAAAATAAATTTAAAATGTGACCCTTCAGGTATTGAGTAACACCTTTATCTTGGTATAGAACTGATACTTTTTTTTTGATTTTGAA ATATCTGATAATAATTTGGAATGAAGTAAGGTTCTGTTAAAATATATTTGAAGACCCTTTAAAGCAGTGAATCTGAAACAATTTTCACAC CCTTAAGTGGTTGATACGTACCTATTTTAGGTATTTTGAGGTATTTACCATAAACTAAATTTAGAAATTTTTTAGATTCACTTGAAGTAA ACATTACAAACATTGGATACGGTGGGGTTTTCTTTAGATTTTACTTGAGAGAAGGTGAGTACAAAGCAATTTGCAGTTGTTGTAATGACA AGATTACTGCGCAAGTGTGAATCCAAACAGTATAGCTTTTAAATTTTAAAGCATTTGGTAAATTATCGCTGAGTTTTTTTCTGTTGCCAA TAGCAAACTGCTTTTCCATTAATGGAGAATTCATGCCTTTCAAGCATTTTAAATATGACAATATTTATAAATGTATGGTTTGGAGGAATC GTTTAAATTCTCTTTCCTAATTTTCTTTCTTTTGAAGATAGATTCTTTCAACAAGTAATTTGTAGTAATGACTGTGTTGACTTCAATTTT GGAGCGCAGTAGCTATGTTAAAGATGAACTATTTGGTCTCATTGAAGCCAACACAGAACTTGCTGCTGTGTTTTTTCTTCAGTGATAAAT AAAATACTTACAGAATTTGTTTTAGTGTTGATTTGTGGTTATAGTATTTTGTTTATAATGGTAAGTTTGCATATTCAGTTGAGTTTTTTT TACTTGAATTTTTATCAGTGCCATTAAATGTCTGTGTTTAGTATCAATGAAAATGAACCTAAATATAACAAAGAAAGCATATGTGGCTAG >100213_100213_5_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395957_EIF3A_chr10_120796821_ENST00000369144_length(amino acids)=212AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL LSFSNRRLCSLLVKHWGSRTFPKETVHHCRYLLAPSQFSLCILEPSAVFSQHLPSFVQYLRRPSKMTYGLLQHFYKQLREDSSPHWIIQL -------------------------------------------------------------- >100213_100213_6_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395957_EIF3A_chr10_120796821_ENST00000541549_length(transcript)=1427nt_BP=760nt CGCAGTCACCGAGCGCAGATCCCGCGCTCTCGATTGGAACGCCTCCCCATCACTCAGCCACACTCAGGGGCTGGACGCCTCACTCCCGTT TCCGAGCCATAAAAGGTCTAGGACCGCTTCCCCCGAGCCAGCAGCGTTTGACGTCATCGTGCGTGTGGTGCCCCTGCTGCCGGGGCTGGT GATTGGAGGAAACCCCGTGTCTGCGGAGCGGCTGTAGCCTGTGAGCAGCGAGATCCAGGGACAGAGTCTCAGCCTCGCCGCTGCTGCCGC CGCCGCCGCCCAGAGACTGCTGAGCCCGTCCGTCCGCCGCCACCACCCACTCCGGACACAGAACATCCAGTCATGGATAAAAATGAGCTG GTTCAGAAGGCCAAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATGAAGTCTGTAACTGAGCAAGGAGCTGAATTA TCCAATGAGGAGAGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGGTCATCTTGGAGGGTCGTCTCAAGTATTGAA CAAAAGACGGAAGGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATTGAGACGGAGCTAAGAGATATCTGCAATGAT GTACTGTCTCTTTTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTAC CGTTACTTGGCTGAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGGA GAGACTCAAGTCGCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTAA GAGACGACAGGGACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGACC GGGTGGAAGAGCGGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGTG AAAAAGAGAAGGCCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACAG TACGACGTTAAGTCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAACC AATCTAAATTGGATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACCA >100213_100213_6_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000395957_EIF3A_chr10_120796821_ENST00000541549_length(amino acids)=212AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL LSFSNRRLCSLLVKHWGSRTFPKETVHHCRYLLAPSQFSLCILEPSAVFSQHLPSFVQYLRRPSKMTYGLLQHFYKQLREDSSPHWIIQL -------------------------------------------------------------- >100213_100213_7_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000419477_EIF3A_chr10_120796821_ENST00000369144_length(transcript)=2136nt_BP=520nt GAGCGGGCCGGAGCCCAGGCCTGGCGCGGCGGCGCCCGCCAGGTAGACCGGGGCGAGGCTGCGCGCCGGCGGGAGGGGAGTCCACTGCGG GAACATCCAGTCATGGATAAAAATGAGCTGGTTCAGAAGGCCAAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATG AAGTCTGTAACTGAGCAAGGAGCTGAATTATCCAATGAGGAGAGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGG TCATCTTGGAGGGTCGTCTCAAGTATTGAACAAAAGACGGAAGGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATT GAGACGGAGCTAAGAGATATCTGCAATGATGTACTGTCTCTTTTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTC TTCTATTTGAAAATGAAAGGAGATTACTACCGTTACTTGGCTGAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAA GAGGACCAGCTGAGGAATCTTCAAGCTGGAGAGACTCAAGTCGCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACC GGCGTGATCTAAGAGAAAGACGAGATCTAAGAGACGACAGGGACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTT GGAGACGTGCTGATGACAGGAAAGATGACCGGGTGGAAGAGCGGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAG AAAGAGACCGAGACCGAGAAAGAGAAGGTGAAAAAGAGAAGGCCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAA ATGAGACTGATGAAGATGGATGGACCACAGTACGACGTTAAGTCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACA TTCAAGGATTATTATACTTGTGCTTCAACCAATCTAAATTGGATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATT AATCCTATATAATAGATTGATCATGCACCATATCCACAGGAGGTTGGAAAAACCATGCCATTTTCTGGAATTTAAGGGTGTTGCATTATT TCATCAATCATTTGTTGACAAAAAAGAAAAACTAAAAAATAAATTTAAAATGTGACCCTTCAGGTATTGAGTAACACCTTTATCTTGGTA TAGAACTGATACTTTTTTTTTGATTTTGAAATATCTGATAATAATTTGGAATGAAGTAAGGTTCTGTTAAAATATATTTGAAGACCCTTT AAAGCAGTGAATCTGAAACAATTTTCACACCCTTAAGTGGTTGATACGTACCTATTTTAGGTATTTTGAGGTATTTACCATAAACTAAAT TTAGAAATTTTTTAGATTCACTTGAAGTAAACATTACAAACATTGGATACGGTGGGGTTTTCTTTAGATTTTACTTGAGAGAAGGTGAGT ACAAAGCAATTTGCAGTTGTTGTAATGACAAGATTACTGCGCAAGTGTGAATCCAAACAGTATAGCTTTTAAATTTTAAAGCATTTGGTA AATTATCGCTGAGTTTTTTTCTGTTGCCAATAGCAAACTGCTTTTCCATTAATGGAGAATTCATGCCTTTCAAGCATTTTAAATATGACA ATATTTATAAATGTATGGTTTGGAGGAATCGTTTAAATTCTCTTTCCTAATTTTCTTTCTTTTGAAGATAGATTCTTTCAACAAGTAATT TGTAGTAATGACTGTGTTGACTTCAATTTTGGAGCGCAGTAGCTATGTTAAAGATGAACTATTTGGTCTCATTGAAGCCAACACAGAACT TGCTGCTGTGTTTTTTCTTCAGTGATAAATAAAATACTTACAGAATTTGTTTTAGTGTTGATTTGTGGTTATAGTATTTTGTTTATAATG GTAAGTTTGCATATTCAGTTGAGTTTTTTTTACTTGAATTTTTATCAGTGCCATTAAATGTCTGTGTTTAGTATCAATGAAAATGAACCT >100213_100213_7_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000419477_EIF3A_chr10_120796821_ENST00000369144_length(amino acids)=212AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL LSFSNRRLCSLLVKHWGSRTFPKETVHHCRYLLAPSQFSLCILEPSAVFSQHLPSFVQYLRRPSKMTYGLLQHFYKQLREDSSPHWIIQL -------------------------------------------------------------- >100213_100213_8_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000419477_EIF3A_chr10_120796821_ENST00000541549_length(transcript)=1187nt_BP=520nt GAGCGGGCCGGAGCCCAGGCCTGGCGCGGCGGCGCCCGCCAGGTAGACCGGGGCGAGGCTGCGCGCCGGCGGGAGGGGAGTCCACTGCGG GAACATCCAGTCATGGATAAAAATGAGCTGGTTCAGAAGGCCAAACTGGCCGAGCAGGCTGAGCGATATGATGACATGGCAGCCTGCATG AAGTCTGTAACTGAGCAAGGAGCTGAATTATCCAATGAGGAGAGGAATCTTCTCTCAGTTGCTTATAAAAATGTTGTAGGAGCCCGTAGG TCATCTTGGAGGGTCGTCTCAAGTATTGAACAAAAGACGGAAGGTGCTGAGAAAAAACAGCAGATGGCTCGAGAATACAGAGAGAAAATT GAGACGGAGCTAAGAGATATCTGCAATGATGTACTGTCTCTTTTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTC TTCTATTTGAAAATGAAAGGAGATTACTACCGTTACTTGGCTGAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAA GAGGACCAGCTGAGGAATCTTCAAGCTGGAGAGACTCAAGTCGCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACC GGCGTGATCTAAGAGAAAGACGAGATCTAAGAGACGACAGGGACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTT GGAGACGTGCTGATGACAGGAAAGATGACCGGGTGGAAGAGCGGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAG AAAGAGACCGAGACCGAGAAAGAGAAGGTGAAAAAGAGAAGGCCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAA ATGAGACTGATGAAGATGGATGGACCACAGTACGACGTTAAGTCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACA TTCAAGGATTATTATACTTGTGCTTCAACCAATCTAAATTGGATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATT AATCCTATATAATAGATTGATCATGCACCATATCCACAGGAGGTTGGAAAAACCATGCCATTTTCTGGAATTTAAGGGTGTTGCATTATT >100213_100213_8_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000419477_EIF3A_chr10_120796821_ENST00000541549_length(amino acids)=212AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL LSFSNRRLCSLLVKHWGSRTFPKETVHHCRYLLAPSQFSLCILEPSAVFSQHLPSFVQYLRRPSKMTYGLLQHFYKQLREDSSPHWIIQL -------------------------------------------------------------- >100213_100213_9_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000522819_EIF3A_chr10_120796821_ENST00000369144_length(transcript)=2004nt_BP=388nt TCAGCCTCCCGAGTAGCTGGGATTACAGTCGTGTGCCACCACATCCAATTTTTTTTTTTTTCCCAGTAGAGACGGGGTGTCACCCAGGCT GGTAATCTTTATTTTAAATTGTACTTGCTCCTAAGAATTTAGGTGTTAAGAGCATAGCATGTAGAAGGAAAAAATAAAGCTGCTGTAACC TCATCCTGCCTTCTTGGAGATGAGAATCTGAGACCCAGGAAGATTTCAGAGTTTTGTCAAGGTGACAGAGTTTGAATTTAGGCATCTCTT TTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTACCGTTACTTGGCT GAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGGAGAGACTCAAGTC GCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTAAGAGACGACAGGG ACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGACCGGGTGGAAGAGC GGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGTGAAAAAGAGAAGG CCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACAGTACGACGTTAAG TCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAACCAATCTAAATTGG ATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACCATATCCACAGGAG GTTGGAAAAACCATGCCATTTTCTGGAATTTAAGGGTGTTGCATTATTTCATCAATCATTTGTTGACAAAAAAGAAAAACTAAAAAATAA ATTTAAAATGTGACCCTTCAGGTATTGAGTAACACCTTTATCTTGGTATAGAACTGATACTTTTTTTTTGATTTTGAAATATCTGATAAT AATTTGGAATGAAGTAAGGTTCTGTTAAAATATATTTGAAGACCCTTTAAAGCAGTGAATCTGAAACAATTTTCACACCCTTAAGTGGTT GATACGTACCTATTTTAGGTATTTTGAGGTATTTACCATAAACTAAATTTAGAAATTTTTTAGATTCACTTGAAGTAAACATTACAAACA TTGGATACGGTGGGGTTTTCTTTAGATTTTACTTGAGAGAAGGTGAGTACAAAGCAATTTGCAGTTGTTGTAATGACAAGATTACTGCGC AAGTGTGAATCCAAACAGTATAGCTTTTAAATTTTAAAGCATTTGGTAAATTATCGCTGAGTTTTTTTCTGTTGCCAATAGCAAACTGCT TTTCCATTAATGGAGAATTCATGCCTTTCAAGCATTTTAAATATGACAATATTTATAAATGTATGGTTTGGAGGAATCGTTTAAATTCTC TTTCCTAATTTTCTTTCTTTTGAAGATAGATTCTTTCAACAAGTAATTTGTAGTAATGACTGTGTTGACTTCAATTTTGGAGCGCAGTAG CTATGTTAAAGATGAACTATTTGGTCTCATTGAAGCCAACACAGAACTTGCTGCTGTGTTTTTTCTTCAGTGATAAATAAAATACTTACA GAATTTGTTTTAGTGTTGATTTGTGGTTATAGTATTTTGTTTATAATGGTAAGTTTGCATATTCAGTTGAGTTTTTTTTACTTGAATTTT TATCAGTGCCATTAAATGTCTGTGTTTAGTATCAATGAAAATGAACCTAAATATAACAAAGAAAGCATATGTGGCTAGGATGTCCCCCTA >100213_100213_9_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000522819_EIF3A_chr10_120796821_ENST00000369144_length(amino acids)=81AA_BP= -------------------------------------------------------------- >100213_100213_10_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000522819_EIF3A_chr10_120796821_ENST00000541549_length(transcript)=1055nt_BP=388nt TCAGCCTCCCGAGTAGCTGGGATTACAGTCGTGTGCCACCACATCCAATTTTTTTTTTTTTCCCAGTAGAGACGGGGTGTCACCCAGGCT GGTAATCTTTATTTTAAATTGTACTTGCTCCTAAGAATTTAGGTGTTAAGAGCATAGCATGTAGAAGGAAAAAATAAAGCTGCTGTAACC TCATCCTGCCTTCTTGGAGATGAGAATCTGAGACCCAGGAAGATTTCAGAGTTTTGTCAAGGTGACAGAGTTTGAATTTAGGCATCTCTT TTGGAAAAGTTCTTGATCCCCAATGCTTCACAAGCAGAGAGCAAAGTCTTCTATTTGAAAATGAAAGGAGATTACTACCGTTACTTGGCT GAGGTTGCCGCTGGTGATGACAAGAAAGGGATGAAGGTGGCTGGAGAAGAGGACCAGCTGAGGAATCTTCAAGCTGGAGAGACTCAAGTC GCCGGGACGATAGGGATAGGGATGACCGTCGCCGTGAGAGGGATGACCGGCGTGATCTAAGAGAAAGACGAGATCTAAGAGACGACAGGG ACCGAAGAGGACCTCCACTCAGATCAGAACGTGAAGAAGTAAGTTCTTGGAGACGTGCTGATGACAGGAAAGATGACCGGGTGGAAGAGC GGGACCCTCCTCGTCGAGTTCCTCCCCCAGCTCTTTCAAGAGACCGAGAAAGAGACCGAGACCGAGAAAGAGAAGGTGAAAAAGAGAAGG CCTCATGGAGAGCTGAGAAAGATAGGGAATCTCTCCGTCGTACTAAAAATGAGACTGATGAAGATGGATGGACCACAGTACGACGTTAAG TCTCAAGATAATGGATTTAAACTGGTGTCTTAAATAGGTTTGATCACATTCAAGGATTATTATACTTGTGCTTCAACCAATCTAAATTGG ATTCTTTAATGTTGTTTCACCATAACACAAAAAGCATGAACTTGTATTAATCCTATATAATAGATTGATCATGCACCATATCCACAGGAG >100213_100213_10_YWHAZ-EIF3A_YWHAZ_chr8_101937143_ENST00000522819_EIF3A_chr10_120796821_ENST00000541549_length(amino acids)=167AA_BP=1 MSSARLQELTSSRSDLSGGPLRSLSSLRSRLSLRSRRSSLSRRRSSLSLSSRRLESLQLEDSSAGPLLQPPSSLSCHHQRQPQPSNGSNL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for YWHAZ-EIF3A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | EIF3A | chr8:101937143 | chr10:120796821 | ENST00000369144 | 19 | 22 | 664_835 | 1242.6666666666667 | 1383.0 | EIF3B |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for YWHAZ-EIF3A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for YWHAZ-EIF3A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
