
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZC3H7B-L3MBTL2 (FusionGDB2 ID:100591) |
Fusion Gene Summary for ZC3H7B-L3MBTL2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZC3H7B-L3MBTL2 | Fusion gene ID: 100591 | Hgene | Tgene | Gene symbol | ZC3H7B | L3MBTL2 | Gene ID | 23264 | 83746 |
| Gene name | zinc finger CCCH-type containing 7B | L3MBTL histone methyl-lysine binding protein 2 | |
| Synonyms | ROXAN1|RoXaN | H-l(3)mbt-l|L3MBT | |
| Cytomap | 22q13.2 | 22q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger CCCH domain-containing protein 7BRotavirus 'X' associated non-structural proteinrotavirus 'X'-associated non-structural proteinrotavirus X protein associated with NSP3ubiquitous tetratricopeptide containing protein RoXaN | lethal(3)malignant brain tumor-like protein 2H-l(3)mbt-like protein 2L(3)mbt-like protein 2L3MBTL2, polycomb repressive complex 1 subunitl(3)mbt-like 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q969R5 | |
| Ensembl transtripts involved in fusion gene | ENST00000351589, ENST00000352645, | ENST00000489136, ENST00000216237, | |
| Fusion gene scores | * DoF score | 15 X 12 X 9=1620 | 3 X 8 X 4=96 |
| # samples | 18 | 7 | |
| ** MAII score | log2(18/1620*10)=-3.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/96*10)=-0.45567948377619 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZC3H7B [Title/Abstract] AND L3MBTL2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZC3H7B(41697776)-L3MBTL2(41615423), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ZC3H7B (5'-gene) Fusion gene breakpoints across ZC3H7B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
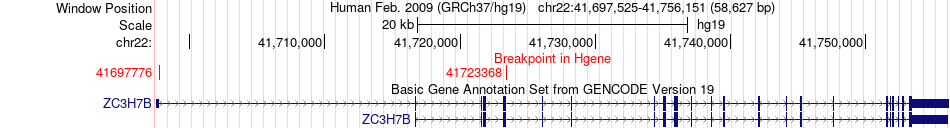 |
 Fusion gene breakpoints across L3MBTL2 (3'-gene) Fusion gene breakpoints across L3MBTL2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
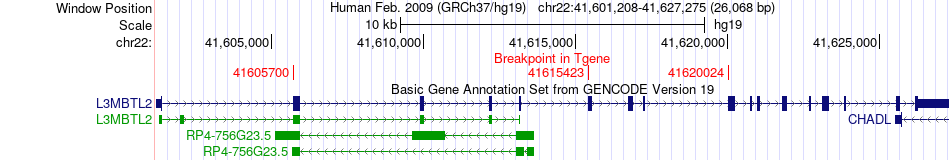 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-EW-A6SB-01A | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41605700 | + |
| ChimerDB4 | BRCA | TCGA-EW-A6SB-01A | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + |
| ChimerDB4 | LIHC | TCGA-2Y-A9H3-01A | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + |
| ChimerDB4 | LIHC | TCGA-CC-A9FU-01A | ZC3H7B | chr22 | 41697776 | - | L3MBTL2 | chr22 | 41615423 | + |
| ChimerDB4 | LIHC | TCGA-CC-A9FU-01A | ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615423 | + |
Top |
Fusion Gene ORF analysis for ZC3H7B-L3MBTL2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000351589 | ENST00000489136 | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + |
| 5CDS-3UTR | ENST00000352645 | ENST00000489136 | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + |
| 5CDS-intron | ENST00000351589 | ENST00000489136 | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + |
| 5CDS-intron | ENST00000352645 | ENST00000489136 | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + |
| 5UTR-3CDS | ENST00000352645 | ENST00000216237 | ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615423 | + |
| 5UTR-intron | ENST00000352645 | ENST00000489136 | ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615423 | + |
| In-frame | ENST00000351589 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + |
| In-frame | ENST00000351589 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + |
| In-frame | ENST00000352645 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + |
| In-frame | ENST00000352645 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + |
| intron-3CDS | ENST00000351589 | ENST00000216237 | ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615423 | + |
| intron-intron | ENST00000351589 | ENST00000489136 | ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615423 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000352645 | ZC3H7B | chr22 | 41723368 | + | ENST00000216237 | L3MBTL2 | chr22 | 41605700 | + | 3815 | 701 | 254 | 2794 | 846 |
| ENST00000351589 | ZC3H7B | chr22 | 41723368 | + | ENST00000216237 | L3MBTL2 | chr22 | 41605700 | + | 3558 | 444 | 0 | 2537 | 845 |
| ENST00000352645 | ZC3H7B | chr22 | 41723368 | - | ENST00000216237 | L3MBTL2 | chr22 | 41620024 | + | 2897 | 701 | 254 | 1876 | 540 |
| ENST00000351589 | ZC3H7B | chr22 | 41723368 | - | ENST00000216237 | L3MBTL2 | chr22 | 41620024 | + | 2640 | 444 | 0 | 1619 | 539 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000352645 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + | 0.002754472 | 0.99724555 |
| ENST00000351589 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605700 | + | 0.002823552 | 0.9971764 |
| ENST00000352645 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + | 0.004507 | 0.99549294 |
| ENST00000351589 | ENST00000216237 | ZC3H7B | chr22 | 41723368 | - | L3MBTL2 | chr22 | 41620024 | + | 0.004626217 | 0.9953738 |
Top |
Fusion Genomic Features for ZC3H7B-L3MBTL2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605699 | + | 3.40E-10 | 1 |
| ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615422 | + | 2.66E-09 | 1 |
| ZC3H7B | chr22 | 41723368 | + | L3MBTL2 | chr22 | 41605699 | + | 3.40E-10 | 1 |
| ZC3H7B | chr22 | 41697776 | + | L3MBTL2 | chr22 | 41615422 | + | 2.66E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
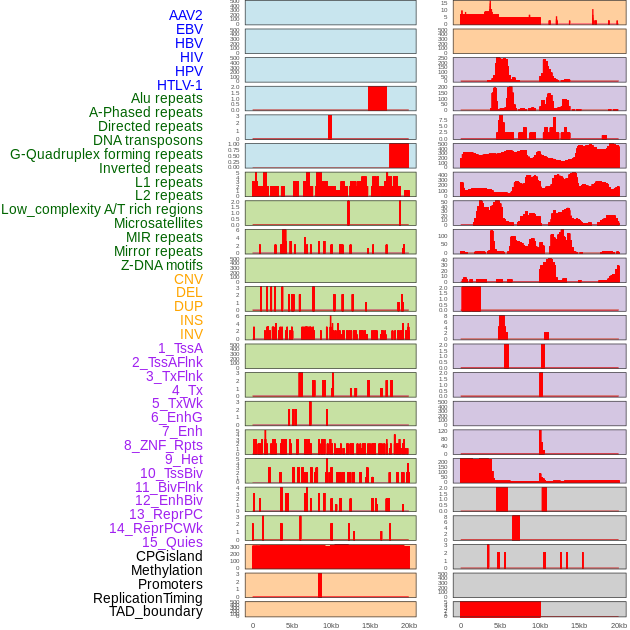 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
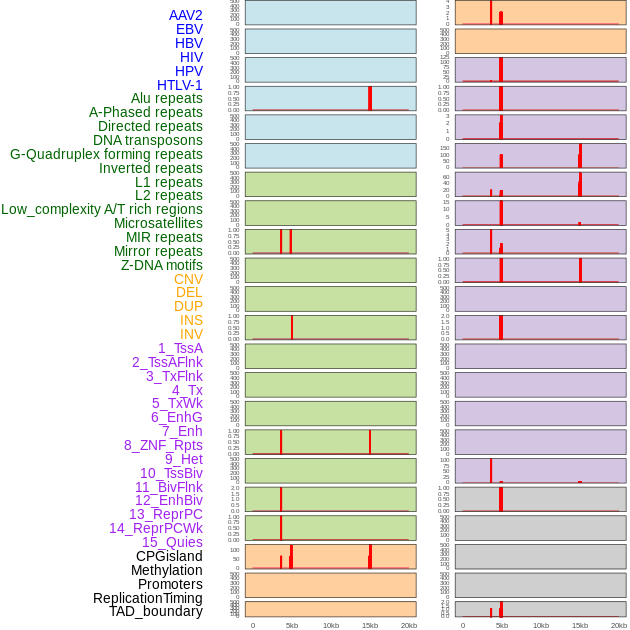 |
Top |
Fusion Protein Features for ZC3H7B-L3MBTL2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:41697776/chr22:41615423) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | L3MBTL2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Putative Polycomb group (PcG) protein. PcG proteins maintain the transcriptionally repressive state of genes, probably via a modification of chromatin, rendering it heritably changed in its expressibility. Its association with a chromatin-remodeling complex suggests that it may contribute to prevent expression of genes that trigger the cell into mitosis. Binds to monomethylated and dimethylated 'Lys-20' on histone H4. Binds histone H3 peptides that are monomethylated or dimethylated on 'Lys-4', 'Lys-9' or 'Lys-27'. {ECO:0000269|PubMed:19233876}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 1_27 | 148 | 978.0 | Repeat | TPR 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 36_69 | 148 | 978.0 | Repeat | TPR 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 82_115 | 148 | 978.0 | Repeat | TPR 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 1_27 | 148 | 978.0 | Repeat | TPR 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 36_69 | 148 | 978.0 | Repeat | TPR 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 82_115 | 148 | 978.0 | Repeat | TPR 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 1_27 | 148 | 978.0 | Repeat | TPR 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 36_69 | 148 | 978.0 | Repeat | TPR 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 82_115 | 148 | 978.0 | Repeat | TPR 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 1_27 | 148 | 978.0 | Repeat | TPR 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 36_69 | 148 | 978.0 | Repeat | TPR 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 82_115 | 148 | 978.0 | Repeat | TPR 3 |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 17_20 | 8 | 706.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 620_624 | 8 | 706.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 620_624 | 314 | 706.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 179_283 | 8 | 706.0 | Repeat | Note=MBT 1 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 291_391 | 8 | 706.0 | Repeat | Note=MBT 2 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 397_500 | 8 | 706.0 | Repeat | Note=MBT 3 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 508_604 | 8 | 706.0 | Repeat | Note=MBT 4 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 397_500 | 314 | 706.0 | Repeat | Note=MBT 3 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 508_604 | 314 | 706.0 | Repeat | Note=MBT 4 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41605700 | ENST00000216237 | 0 | 17 | 81_116 | 8 | 706.0 | Zinc finger | FCS-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 484_508 | 148 | 978.0 | Zinc finger | C3H1-type 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 616_638 | 148 | 978.0 | Zinc finger | C3H1-type 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 754_782 | 148 | 978.0 | Zinc finger | C3H1-type 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 842_866 | 148 | 978.0 | Zinc finger | C2H2-type |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000351589 | + | 4 | 22 | 886_914 | 148 | 978.0 | Zinc finger | C3H1-type 4 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 484_508 | 148 | 978.0 | Zinc finger | C3H1-type 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 616_638 | 148 | 978.0 | Zinc finger | C3H1-type 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 754_782 | 148 | 978.0 | Zinc finger | C3H1-type 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 842_866 | 148 | 978.0 | Zinc finger | C2H2-type |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41605700 | ENST00000352645 | + | 5 | 23 | 886_914 | 148 | 978.0 | Zinc finger | C3H1-type 4 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 484_508 | 148 | 978.0 | Zinc finger | C3H1-type 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 616_638 | 148 | 978.0 | Zinc finger | C3H1-type 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 754_782 | 148 | 978.0 | Zinc finger | C3H1-type 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 842_866 | 148 | 978.0 | Zinc finger | C2H2-type |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000351589 | - | 4 | 22 | 886_914 | 148 | 978.0 | Zinc finger | C3H1-type 4 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 484_508 | 148 | 978.0 | Zinc finger | C3H1-type 1 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 616_638 | 148 | 978.0 | Zinc finger | C3H1-type 2 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 754_782 | 148 | 978.0 | Zinc finger | C3H1-type 3 |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 842_866 | 148 | 978.0 | Zinc finger | C2H2-type |
| Hgene | ZC3H7B | chr22:41723368 | chr22:41620024 | ENST00000352645 | - | 5 | 23 | 886_914 | 148 | 978.0 | Zinc finger | C3H1-type 4 |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 17_20 | 314 | 706.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 179_283 | 314 | 706.0 | Repeat | Note=MBT 1 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 291_391 | 314 | 706.0 | Repeat | Note=MBT 2 | |
| Tgene | L3MBTL2 | chr22:41723368 | chr22:41620024 | ENST00000216237 | 7 | 17 | 81_116 | 314 | 706.0 | Zinc finger | FCS-type |
Top |
Fusion Gene Sequence for ZC3H7B-L3MBTL2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >100591_100591_1_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000351589_L3MBTL2_chr22_41605700_ENST00000216237_length(transcript)=3558nt_BP=444nt ATGGAGAGGCAGAAACGGAAGGCGGACATCGAGAAAGGGCTGCAGTTCATTCAGTCGACACTACCCCTAAAGCAAGAAGAATATGAGGCC TTTCTGCTCAAGCTGGTGCAGAATCTGTTTGCTGAGGGCAATGATCTGTTCCGGGAGAAGGACTATAAGCAGGCTCTGGTGCAGTACATG GAAGGGCTGAACGTGGCCGACTACGCTGCCTCTGACCAGGTGGCCCTGCCCCGGGAGCTGCTGTGCAAGCTGCATGTCAATAGGGCCGCC TGCTACTTCACCATGGGCCTGTATGAGAAGGCGCTGGAGGACAGCGAGAAGGCGCTGGGCCTGGACAGTGAGAGTATCCGGGCGTTGTTC CGCAAGGCACGCGCTCTCAATGAACTGGGACGCCACAAGGAGGCCTACGAGTGCAGCAGCCGGTGTTCCCTCGCCCTGCCCCACGAGACC CCATCTTCAGAACCAATGGAGGAAGAGGAAGATGACGACTTGGAGCTGTTTGGTGGCTATGATAGTTTCCGGAGTTATAACAGCAGTGTG GGCAGTGAGAGCAGCTCCTATCTGGAGGAGTCAAGTGAAGCAGAAAATGAGGATCGGGAAGCAGGGGAACTGCCGACCTCCCCGCTGCAT TTGCTCAGCCCTGGGACTCCTCGCTCCTTGGATGGCAGTGGTTCTGAGCCAGCTGTCTGTGAGATGTGTGGTATCGTGGGTACAAGGGAA GCCTTCTTCTCCAAGACCAAGAGGTTCTGCAGCGTCTCCTGCTCCAGGAGCTACTCCTCCAACTCCAAGAAAGCCAGTATCTTGGCTAGG TTACAGGGAAAACCACCGACCAAAAAAGCCAAAGTCCTGCACAAGGCTGCCTGGTCTGCCAAAATTGGAGCCTTCCTCCACTCTCAAGGG ACAGGACAGCTGGCAGATGGGACACCAACAGGACAAGACGCTCTGGTCTTGGGCTTCGACTGGGGGAAGTTCCTGAAGGATCACAGTTAC AAGGCTGCTCCCGTCAGCTGTTTCAAGCACGTCCCACTCTATGACCAGTGGGAGGATGTGATGAAAGGGATGAAGGTGGAGGTGCTCAAC AGTGATGCTGTGCTCCCCAGCCGGGTGTACTGGATCGCCTCTGTCATCCAGACAGCAGGGTATCGGGTGCTGCTTCGGTATGAAGGCTTT GAAAATGACGCCAGCCATGACTTCTGGTGCAACCTGGGAACAGTGGATGTCCACCCCATTGGCTGGTGTGCCATCAACAGCAAGATCCTA GTGCCCCCACGGACCATCCATGCCAAGTTCACCGACTGGAAGGGCTACCTCATGAAACGGCTGGTGGGCTCCAGGACGCTTCCCGTGGAT TTCCACATCAAGATGGTGGAGAGCATGAAGTACCCCTTTAGGCAGGGCATGCGGCTGGAAGTGGTGGACAAGTCCCAGGTGTCACGCACT CGCATGGCTGTGGTGGACACAGTAATCGGGGGTCGCCTACGGCTCCTCTACGAGGATGGTGACAGTGACGACGACTTCTGGTGCCACATG TGGAGCCCCCTGATCCACCCAGTGGGTTGGTCACGACGTGTGGGCCACGGCATCAAGATGTCAGAGAGGCGAAGTGACATGGCCCATCAC CCCACCTTCCGGAAGATCTACTGTGATGCCGTTCCTTACCTCTTCAAGAAGGTACGAGCAGTCTACACAGAAGGCGGTTGGTTTGAGGAA GGGATGAAGCTGGAGGCCATTGACCCCCTGAATCTGGGCAACATCTGCGTGGCAACTGTCTGTAAGGTTCTCCTGGATGGATACCTGATG ATCTGTGTGGACGGGGGGCCCTCCACAGATGGCTTGGACTGGTTCTGCTACCATGCCTCTTCCCACGCCATCTTCCCGGCCACCTTCTGT CAGAAGAATGACATTGAGCTCACACCGCCAAAAGGTTATGAGGCACAGACTTTCAACTGGGAGAACTACTTGGAGAAGACCAAGTCGAAA GCCGCTCCATCGAGACTCTTTAACATGGATTGCCCAAACCATGGCTTCAAGGTGGGCATGAAGCTGGAGGCCGTGGACCTGATGGAGCCC CGGCTCATCTGTGTGGCCACGGTGAAACGAGTGGTGCATCGGCTCCTCAGCATCCACTTTGACGGCTGGGACAGCGAGTACGACCAGTGG GTGGACTGCGAGTCCCCAGACATCTACCCCGTCGGCTGGTGTGAGCTCACCGGCTACCAGCTCCAGCCTCCTGTGGCCGCAGAACCGGCC ACACCGCTGAAGGCCAAAGAGGCCACAAAGAAGAAAAAGAAACAGTTTGGGAAGAAAAGGAAAAGAATCCCGCCCACTAAGACGCGACCC CTCAGACAGGGGTCCAAGAAGCCCCTGCTGGAGGACGACCCTCAGGGTGCCAGGAAGATCTCGTCGGAGCCTGTTCCTGGCGAGATCATT GCTGTGCGTGTGAAGGAAGAGCATCTAGACGTGGCCTCGCCCGACAAGGCTTCAAGTCCAGAGCTGCCTGTCTCCGTCGAGAACATCAAG CAGGAAACAGACGACTGAGCCTTCCTGCCTCCAGCCTGGCTTCTAGCTGGAAGCCAGCCCAGCGTTTCTCTACCACCACCACCATGCCTC CACCTGACTTTGGCTTGGAGACTGATCCTCTCTGTGTAAATTCTGCCCGGTGCTGTGAAGGCTGGACGGTGGAGGACCTGCTGGGGTCTC CTGGGACCCGCCTGTTGCTTCTGCCCTCCCCTGTGGAAAGGTCTATATGACGGGCCGCCTGAGGCCCCAGAACTCGTCTGTGAACCACCT TTTCCAGCCAGAGTTCCCAAAGCTGGAACGCTAGCTGCCTGCTCTTCCTTAAGATGGCCTCCCCCCGACCCGCCACGGCCCTCAGTTGCC AGGGATGGGGCCACCACTGTCACACTGTGGAATACAAGACAGTGAACTCTGTCTGCCTGAACGAGCCATGTAAATTAAGTTCTAGAGCAG CTCTCTGAGCAGGATAAGGTCCCCTGACAGTGAGTTGTGTGGTGGGGGCAGCCTCTGCCTCAAAAATTCACCAAGCAGAATGCCTCTCAG CCTCATGTGTTGGTCCTCTGCTCCTCCTAGCTCCCCAGGGATGTTGGGGACCCAGCTTGTCTCGGCAGCTAAGAAGCAGTGACCAGGATG TGGATTTTGGCGACCTGTGTGGTGGCCTTGAGCTGCTTTCTGTGTTTGTGAGGACTGACTCCCATTTCCTAAAGGAAATGCCCCCGGGGA GGACATTGGGAGGAAGATGGCCTGAGTGTGCACTTTGGCTCTGCTACCTGCTCCTGAAGCCCCGCTAAAAATAATTCATCCAAGATTCCT TTGTAGTTAAAGGGTCCAGTTCTGACTGGAGCCTCTAGAGAGCTGGGCTTGTATGTTCTTTTGGCCTTTTGTTCCTACCTAAATGAAGAA ACCATGCCTGGAGGGGCCGTGAACACAGAACCCTCAAGACAAGGATGACAGAGCTGGAGGACACATCTAGCTGCCATTGCAACCTCACTG >100591_100591_1_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000351589_L3MBTL2_chr22_41605700_ENST00000216237_length(amino acids)=845AA_BP=148 MERQKRKADIEKGLQFIQSTLPLKQEEYEAFLLKLVQNLFAEGNDLFREKDYKQALVQYMEGLNVADYAASDQVALPRELLCKLHVNRAA CYFTMGLYEKALEDSEKALGLDSESIRALFRKARALNELGRHKEAYECSSRCSLALPHETPSSEPMEEEEDDDLELFGGYDSFRSYNSSV GSESSSYLEESSEAENEDREAGELPTSPLHLLSPGTPRSLDGSGSEPAVCEMCGIVGTREAFFSKTKRFCSVSCSRSYSSNSKKASILAR LQGKPPTKKAKVLHKAAWSAKIGAFLHSQGTGQLADGTPTGQDALVLGFDWGKFLKDHSYKAAPVSCFKHVPLYDQWEDVMKGMKVEVLN SDAVLPSRVYWIASVIQTAGYRVLLRYEGFENDASHDFWCNLGTVDVHPIGWCAINSKILVPPRTIHAKFTDWKGYLMKRLVGSRTLPVD FHIKMVESMKYPFRQGMRLEVVDKSQVSRTRMAVVDTVIGGRLRLLYEDGDSDDDFWCHMWSPLIHPVGWSRRVGHGIKMSERRSDMAHH PTFRKIYCDAVPYLFKKVRAVYTEGGWFEEGMKLEAIDPLNLGNICVATVCKVLLDGYLMICVDGGPSTDGLDWFCYHASSHAIFPATFC QKNDIELTPPKGYEAQTFNWENYLEKTKSKAAPSRLFNMDCPNHGFKVGMKLEAVDLMEPRLICVATVKRVVHRLLSIHFDGWDSEYDQW VDCESPDIYPVGWCELTGYQLQPPVAAEPATPLKAKEATKKKKKQFGKKRKRIPPTKTRPLRQGSKKPLLEDDPQGARKISSEPVPGEII -------------------------------------------------------------- >100591_100591_2_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000351589_L3MBTL2_chr22_41620024_ENST00000216237_length(transcript)=2640nt_BP=444nt ATGGAGAGGCAGAAACGGAAGGCGGACATCGAGAAAGGGCTGCAGTTCATTCAGTCGACACTACCCCTAAAGCAAGAAGAATATGAGGCC TTTCTGCTCAAGCTGGTGCAGAATCTGTTTGCTGAGGGCAATGATCTGTTCCGGGAGAAGGACTATAAGCAGGCTCTGGTGCAGTACATG GAAGGGCTGAACGTGGCCGACTACGCTGCCTCTGACCAGGTGGCCCTGCCCCGGGAGCTGCTGTGCAAGCTGCATGTCAATAGGGCCGCC TGCTACTTCACCATGGGCCTGTATGAGAAGGCGCTGGAGGACAGCGAGAAGGCGCTGGGCCTGGACAGTGAGAGTATCCGGGCGTTGTTC CGCAAGGCACGCGCTCTCAATGAACTGGGACGCCACAAGGAGGCCTACGAGTGCAGCAGCCGGTGTTCCCTCGCCCTGCCCCACATGGTG GAGAGCATGAAGTACCCCTTTAGGCAGGGCATGCGGCTGGAAGTGGTGGACAAGTCCCAGGTGTCACGCACTCGCATGGCTGTGGTGGAC ACAGTAATCGGGGGTCGCCTACGGCTCCTCTACGAGGATGGTGACAGTGACGACGACTTCTGGTGCCACATGTGGAGCCCCCTGATCCAC CCAGTGGGTTGGTCACGACGTGTGGGCCACGGCATCAAGATGTCAGAGAGGCGAAGTGACATGGCCCATCACCCCACCTTCCGGAAGATC TACTGTGATGCCGTTCCTTACCTCTTCAAGAAGGTACGAGCAGTCTACACAGAAGGCGGTTGGTTTGAGGAAGGGATGAAGCTGGAGGCC ATTGACCCCCTGAATCTGGGCAACATCTGCGTGGCAACTGTCTGTAAGGTTCTCCTGGATGGATACCTGATGATCTGTGTGGACGGGGGG CCCTCCACAGATGGCTTGGACTGGTTCTGCTACCATGCCTCTTCCCACGCCATCTTCCCGGCCACCTTCTGTCAGAAGAATGACATTGAG CTCACACCGCCAAAAGGTTATGAGGCACAGACTTTCAACTGGGAGAACTACTTGGAGAAGACCAAGTCGAAAGCCGCTCCATCGAGACTC TTTAACATGGATTGCCCAAACCATGGCTTCAAGGTGGGCATGAAGCTGGAGGCCGTGGACCTGATGGAGCCCCGGCTCATCTGTGTGGCC ACGGTGAAACGAGTGGTGCATCGGCTCCTCAGCATCCACTTTGACGGCTGGGACAGCGAGTACGACCAGTGGGTGGACTGCGAGTCCCCA GACATCTACCCCGTCGGCTGGTGTGAGCTCACCGGCTACCAGCTCCAGCCTCCTGTGGCCGCAGAACCGGCCACACCGCTGAAGGCCAAA GAGGCCACAAAGAAGAAAAAGAAACAGTTTGGGAAGAAAAGGAAAAGAATCCCGCCCACTAAGACGCGACCCCTCAGACAGGGGTCCAAG AAGCCCCTGCTGGAGGACGACCCTCAGGGTGCCAGGAAGATCTCGTCGGAGCCTGTTCCTGGCGAGATCATTGCTGTGCGTGTGAAGGAA GAGCATCTAGACGTGGCCTCGCCCGACAAGGCTTCAAGTCCAGAGCTGCCTGTCTCCGTCGAGAACATCAAGCAGGAAACAGACGACTGA GCCTTCCTGCCTCCAGCCTGGCTTCTAGCTGGAAGCCAGCCCAGCGTTTCTCTACCACCACCACCATGCCTCCACCTGACTTTGGCTTGG AGACTGATCCTCTCTGTGTAAATTCTGCCCGGTGCTGTGAAGGCTGGACGGTGGAGGACCTGCTGGGGTCTCCTGGGACCCGCCTGTTGC TTCTGCCCTCCCCTGTGGAAAGGTCTATATGACGGGCCGCCTGAGGCCCCAGAACTCGTCTGTGAACCACCTTTTCCAGCCAGAGTTCCC AAAGCTGGAACGCTAGCTGCCTGCTCTTCCTTAAGATGGCCTCCCCCCGACCCGCCACGGCCCTCAGTTGCCAGGGATGGGGCCACCACT GTCACACTGTGGAATACAAGACAGTGAACTCTGTCTGCCTGAACGAGCCATGTAAATTAAGTTCTAGAGCAGCTCTCTGAGCAGGATAAG GTCCCCTGACAGTGAGTTGTGTGGTGGGGGCAGCCTCTGCCTCAAAAATTCACCAAGCAGAATGCCTCTCAGCCTCATGTGTTGGTCCTC TGCTCCTCCTAGCTCCCCAGGGATGTTGGGGACCCAGCTTGTCTCGGCAGCTAAGAAGCAGTGACCAGGATGTGGATTTTGGCGACCTGT GTGGTGGCCTTGAGCTGCTTTCTGTGTTTGTGAGGACTGACTCCCATTTCCTAAAGGAAATGCCCCCGGGGAGGACATTGGGAGGAAGAT GGCCTGAGTGTGCACTTTGGCTCTGCTACCTGCTCCTGAAGCCCCGCTAAAAATAATTCATCCAAGATTCCTTTGTAGTTAAAGGGTCCA GTTCTGACTGGAGCCTCTAGAGAGCTGGGCTTGTATGTTCTTTTGGCCTTTTGTTCCTACCTAAATGAAGAAACCATGCCTGGAGGGGCC GTGAACACAGAACCCTCAAGACAAGGATGACAGAGCTGGAGGACACATCTAGCTGCCATTGCAACCTCACTGGGCTCCCCAGACTCTGTG >100591_100591_2_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000351589_L3MBTL2_chr22_41620024_ENST00000216237_length(amino acids)=539AA_BP=148 MERQKRKADIEKGLQFIQSTLPLKQEEYEAFLLKLVQNLFAEGNDLFREKDYKQALVQYMEGLNVADYAASDQVALPRELLCKLHVNRAA CYFTMGLYEKALEDSEKALGLDSESIRALFRKARALNELGRHKEAYECSSRCSLALPHMVESMKYPFRQGMRLEVVDKSQVSRTRMAVVD TVIGGRLRLLYEDGDSDDDFWCHMWSPLIHPVGWSRRVGHGIKMSERRSDMAHHPTFRKIYCDAVPYLFKKVRAVYTEGGWFEEGMKLEA IDPLNLGNICVATVCKVLLDGYLMICVDGGPSTDGLDWFCYHASSHAIFPATFCQKNDIELTPPKGYEAQTFNWENYLEKTKSKAAPSRL FNMDCPNHGFKVGMKLEAVDLMEPRLICVATVKRVVHRLLSIHFDGWDSEYDQWVDCESPDIYPVGWCELTGYQLQPPVAAEPATPLKAK -------------------------------------------------------------- >100591_100591_3_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000352645_L3MBTL2_chr22_41605700_ENST00000216237_length(transcript)=3815nt_BP=701nt AGGGAGACAGTGCCTCCAGCGGGTGCTGCCGCGAGCGGCCAGCCGAGGGGCTGGAAATGAAAGTAAAGCGCTCCAGAGCCACATGGACGG AGCTGCCGGGGCGGCGGCGCCGGGAGCAGGATGCGGCCGCCCGTAATTAAATAGCATTTACTCTTATTATTACTAATAATAATAACGTAA TCATACCTCTAGTCATAGCATACCATTTATCGGGCTCGGCGCAGGCCCGCGGGGAGCGCAGCCCGGCGGAGAGACTGATGGAGAGGCAGA AACGGAAGGCGGACATCGAGAAAGGGCTGCAGTTCATTCAGTCGACACTACCCCTAAAGCAAGAAGAATATGAGGCCTTTCTGCTCAAGC TGGTGCAGAATCTGTTTGCTGAGGGCAATGATCTGTTCCGGGAGAAGGACTATAAGCAGGCTCTGGTGCAGTACATGGAAGGGCTGAACG TGGCCGACTACGCTGCCTCTGACCAGGTGGCCCTGCCCCGGGAGCTGCTGTGCAAGCTGCATGTCAATAGGGCCGCCTGCTACTTCACCA TGGGCCTGTATGAGAAGGCGCTGGAGGACAGCGAGAAGGCGCTGGGCCTGGACAGTGAGAGTATCCGGGCGTTGTTCCGCAAGGCACGCG CTCTCAATGAACTGGGACGCCACAAGGAGGCCTACGAGTGCAGCAGCCGGTGTTCCCTCGCCCTGCCCCACGAGACCCCATCTTCAGAAC CAATGGAGGAAGAGGAAGATGACGACTTGGAGCTGTTTGGTGGCTATGATAGTTTCCGGAGTTATAACAGCAGTGTGGGCAGTGAGAGCA GCTCCTATCTGGAGGAGTCAAGTGAAGCAGAAAATGAGGATCGGGAAGCAGGGGAACTGCCGACCTCCCCGCTGCATTTGCTCAGCCCTG GGACTCCTCGCTCCTTGGATGGCAGTGGTTCTGAGCCAGCTGTCTGTGAGATGTGTGGTATCGTGGGTACAAGGGAAGCCTTCTTCTCCA AGACCAAGAGGTTCTGCAGCGTCTCCTGCTCCAGGAGCTACTCCTCCAACTCCAAGAAAGCCAGTATCTTGGCTAGGTTACAGGGAAAAC CACCGACCAAAAAAGCCAAAGTCCTGCACAAGGCTGCCTGGTCTGCCAAAATTGGAGCCTTCCTCCACTCTCAAGGGACAGGACAGCTGG CAGATGGGACACCAACAGGACAAGACGCTCTGGTCTTGGGCTTCGACTGGGGGAAGTTCCTGAAGGATCACAGTTACAAGGCTGCTCCCG TCAGCTGTTTCAAGCACGTCCCACTCTATGACCAGTGGGAGGATGTGATGAAAGGGATGAAGGTGGAGGTGCTCAACAGTGATGCTGTGC TCCCCAGCCGGGTGTACTGGATCGCCTCTGTCATCCAGACAGCAGGGTATCGGGTGCTGCTTCGGTATGAAGGCTTTGAAAATGACGCCA GCCATGACTTCTGGTGCAACCTGGGAACAGTGGATGTCCACCCCATTGGCTGGTGTGCCATCAACAGCAAGATCCTAGTGCCCCCACGGA CCATCCATGCCAAGTTCACCGACTGGAAGGGCTACCTCATGAAACGGCTGGTGGGCTCCAGGACGCTTCCCGTGGATTTCCACATCAAGA TGGTGGAGAGCATGAAGTACCCCTTTAGGCAGGGCATGCGGCTGGAAGTGGTGGACAAGTCCCAGGTGTCACGCACTCGCATGGCTGTGG TGGACACAGTAATCGGGGGTCGCCTACGGCTCCTCTACGAGGATGGTGACAGTGACGACGACTTCTGGTGCCACATGTGGAGCCCCCTGA TCCACCCAGTGGGTTGGTCACGACGTGTGGGCCACGGCATCAAGATGTCAGAGAGGCGAAGTGACATGGCCCATCACCCCACCTTCCGGA AGATCTACTGTGATGCCGTTCCTTACCTCTTCAAGAAGGTACGAGCAGTCTACACAGAAGGCGGTTGGTTTGAGGAAGGGATGAAGCTGG AGGCCATTGACCCCCTGAATCTGGGCAACATCTGCGTGGCAACTGTCTGTAAGGTTCTCCTGGATGGATACCTGATGATCTGTGTGGACG GGGGGCCCTCCACAGATGGCTTGGACTGGTTCTGCTACCATGCCTCTTCCCACGCCATCTTCCCGGCCACCTTCTGTCAGAAGAATGACA TTGAGCTCACACCGCCAAAAGGTTATGAGGCACAGACTTTCAACTGGGAGAACTACTTGGAGAAGACCAAGTCGAAAGCCGCTCCATCGA GACTCTTTAACATGGATTGCCCAAACCATGGCTTCAAGGTGGGCATGAAGCTGGAGGCCGTGGACCTGATGGAGCCCCGGCTCATCTGTG TGGCCACGGTGAAACGAGTGGTGCATCGGCTCCTCAGCATCCACTTTGACGGCTGGGACAGCGAGTACGACCAGTGGGTGGACTGCGAGT CCCCAGACATCTACCCCGTCGGCTGGTGTGAGCTCACCGGCTACCAGCTCCAGCCTCCTGTGGCCGCAGAACCGGCCACACCGCTGAAGG CCAAAGAGGCCACAAAGAAGAAAAAGAAACAGTTTGGGAAGAAAAGGAAAAGAATCCCGCCCACTAAGACGCGACCCCTCAGACAGGGGT CCAAGAAGCCCCTGCTGGAGGACGACCCTCAGGGTGCCAGGAAGATCTCGTCGGAGCCTGTTCCTGGCGAGATCATTGCTGTGCGTGTGA AGGAAGAGCATCTAGACGTGGCCTCGCCCGACAAGGCTTCAAGTCCAGAGCTGCCTGTCTCCGTCGAGAACATCAAGCAGGAAACAGACG ACTGAGCCTTCCTGCCTCCAGCCTGGCTTCTAGCTGGAAGCCAGCCCAGCGTTTCTCTACCACCACCACCATGCCTCCACCTGACTTTGG CTTGGAGACTGATCCTCTCTGTGTAAATTCTGCCCGGTGCTGTGAAGGCTGGACGGTGGAGGACCTGCTGGGGTCTCCTGGGACCCGCCT GTTGCTTCTGCCCTCCCCTGTGGAAAGGTCTATATGACGGGCCGCCTGAGGCCCCAGAACTCGTCTGTGAACCACCTTTTCCAGCCAGAG TTCCCAAAGCTGGAACGCTAGCTGCCTGCTCTTCCTTAAGATGGCCTCCCCCCGACCCGCCACGGCCCTCAGTTGCCAGGGATGGGGCCA CCACTGTCACACTGTGGAATACAAGACAGTGAACTCTGTCTGCCTGAACGAGCCATGTAAATTAAGTTCTAGAGCAGCTCTCTGAGCAGG ATAAGGTCCCCTGACAGTGAGTTGTGTGGTGGGGGCAGCCTCTGCCTCAAAAATTCACCAAGCAGAATGCCTCTCAGCCTCATGTGTTGG TCCTCTGCTCCTCCTAGCTCCCCAGGGATGTTGGGGACCCAGCTTGTCTCGGCAGCTAAGAAGCAGTGACCAGGATGTGGATTTTGGCGA CCTGTGTGGTGGCCTTGAGCTGCTTTCTGTGTTTGTGAGGACTGACTCCCATTTCCTAAAGGAAATGCCCCCGGGGAGGACATTGGGAGG AAGATGGCCTGAGTGTGCACTTTGGCTCTGCTACCTGCTCCTGAAGCCCCGCTAAAAATAATTCATCCAAGATTCCTTTGTAGTTAAAGG GTCCAGTTCTGACTGGAGCCTCTAGAGAGCTGGGCTTGTATGTTCTTTTGGCCTTTTGTTCCTACCTAAATGAAGAAACCATGCCTGGAG GGGCCGTGAACACAGAACCCTCAAGACAAGGATGACAGAGCTGGAGGACACATCTAGCTGCCATTGCAACCTCACTGGGCTCCCCAGACT >100591_100591_3_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000352645_L3MBTL2_chr22_41605700_ENST00000216237_length(amino acids)=846AA_BP=149 MMERQKRKADIEKGLQFIQSTLPLKQEEYEAFLLKLVQNLFAEGNDLFREKDYKQALVQYMEGLNVADYAASDQVALPRELLCKLHVNRA ACYFTMGLYEKALEDSEKALGLDSESIRALFRKARALNELGRHKEAYECSSRCSLALPHETPSSEPMEEEEDDDLELFGGYDSFRSYNSS VGSESSSYLEESSEAENEDREAGELPTSPLHLLSPGTPRSLDGSGSEPAVCEMCGIVGTREAFFSKTKRFCSVSCSRSYSSNSKKASILA RLQGKPPTKKAKVLHKAAWSAKIGAFLHSQGTGQLADGTPTGQDALVLGFDWGKFLKDHSYKAAPVSCFKHVPLYDQWEDVMKGMKVEVL NSDAVLPSRVYWIASVIQTAGYRVLLRYEGFENDASHDFWCNLGTVDVHPIGWCAINSKILVPPRTIHAKFTDWKGYLMKRLVGSRTLPV DFHIKMVESMKYPFRQGMRLEVVDKSQVSRTRMAVVDTVIGGRLRLLYEDGDSDDDFWCHMWSPLIHPVGWSRRVGHGIKMSERRSDMAH HPTFRKIYCDAVPYLFKKVRAVYTEGGWFEEGMKLEAIDPLNLGNICVATVCKVLLDGYLMICVDGGPSTDGLDWFCYHASSHAIFPATF CQKNDIELTPPKGYEAQTFNWENYLEKTKSKAAPSRLFNMDCPNHGFKVGMKLEAVDLMEPRLICVATVKRVVHRLLSIHFDGWDSEYDQ WVDCESPDIYPVGWCELTGYQLQPPVAAEPATPLKAKEATKKKKKQFGKKRKRIPPTKTRPLRQGSKKPLLEDDPQGARKISSEPVPGEI -------------------------------------------------------------- >100591_100591_4_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000352645_L3MBTL2_chr22_41620024_ENST00000216237_length(transcript)=2897nt_BP=701nt AGGGAGACAGTGCCTCCAGCGGGTGCTGCCGCGAGCGGCCAGCCGAGGGGCTGGAAATGAAAGTAAAGCGCTCCAGAGCCACATGGACGG AGCTGCCGGGGCGGCGGCGCCGGGAGCAGGATGCGGCCGCCCGTAATTAAATAGCATTTACTCTTATTATTACTAATAATAATAACGTAA TCATACCTCTAGTCATAGCATACCATTTATCGGGCTCGGCGCAGGCCCGCGGGGAGCGCAGCCCGGCGGAGAGACTGATGGAGAGGCAGA AACGGAAGGCGGACATCGAGAAAGGGCTGCAGTTCATTCAGTCGACACTACCCCTAAAGCAAGAAGAATATGAGGCCTTTCTGCTCAAGC TGGTGCAGAATCTGTTTGCTGAGGGCAATGATCTGTTCCGGGAGAAGGACTATAAGCAGGCTCTGGTGCAGTACATGGAAGGGCTGAACG TGGCCGACTACGCTGCCTCTGACCAGGTGGCCCTGCCCCGGGAGCTGCTGTGCAAGCTGCATGTCAATAGGGCCGCCTGCTACTTCACCA TGGGCCTGTATGAGAAGGCGCTGGAGGACAGCGAGAAGGCGCTGGGCCTGGACAGTGAGAGTATCCGGGCGTTGTTCCGCAAGGCACGCG CTCTCAATGAACTGGGACGCCACAAGGAGGCCTACGAGTGCAGCAGCCGGTGTTCCCTCGCCCTGCCCCACATGGTGGAGAGCATGAAGT ACCCCTTTAGGCAGGGCATGCGGCTGGAAGTGGTGGACAAGTCCCAGGTGTCACGCACTCGCATGGCTGTGGTGGACACAGTAATCGGGG GTCGCCTACGGCTCCTCTACGAGGATGGTGACAGTGACGACGACTTCTGGTGCCACATGTGGAGCCCCCTGATCCACCCAGTGGGTTGGT CACGACGTGTGGGCCACGGCATCAAGATGTCAGAGAGGCGAAGTGACATGGCCCATCACCCCACCTTCCGGAAGATCTACTGTGATGCCG TTCCTTACCTCTTCAAGAAGGTACGAGCAGTCTACACAGAAGGCGGTTGGTTTGAGGAAGGGATGAAGCTGGAGGCCATTGACCCCCTGA ATCTGGGCAACATCTGCGTGGCAACTGTCTGTAAGGTTCTCCTGGATGGATACCTGATGATCTGTGTGGACGGGGGGCCCTCCACAGATG GCTTGGACTGGTTCTGCTACCATGCCTCTTCCCACGCCATCTTCCCGGCCACCTTCTGTCAGAAGAATGACATTGAGCTCACACCGCCAA AAGGTTATGAGGCACAGACTTTCAACTGGGAGAACTACTTGGAGAAGACCAAGTCGAAAGCCGCTCCATCGAGACTCTTTAACATGGATT GCCCAAACCATGGCTTCAAGGTGGGCATGAAGCTGGAGGCCGTGGACCTGATGGAGCCCCGGCTCATCTGTGTGGCCACGGTGAAACGAG TGGTGCATCGGCTCCTCAGCATCCACTTTGACGGCTGGGACAGCGAGTACGACCAGTGGGTGGACTGCGAGTCCCCAGACATCTACCCCG TCGGCTGGTGTGAGCTCACCGGCTACCAGCTCCAGCCTCCTGTGGCCGCAGAACCGGCCACACCGCTGAAGGCCAAAGAGGCCACAAAGA AGAAAAAGAAACAGTTTGGGAAGAAAAGGAAAAGAATCCCGCCCACTAAGACGCGACCCCTCAGACAGGGGTCCAAGAAGCCCCTGCTGG AGGACGACCCTCAGGGTGCCAGGAAGATCTCGTCGGAGCCTGTTCCTGGCGAGATCATTGCTGTGCGTGTGAAGGAAGAGCATCTAGACG TGGCCTCGCCCGACAAGGCTTCAAGTCCAGAGCTGCCTGTCTCCGTCGAGAACATCAAGCAGGAAACAGACGACTGAGCCTTCCTGCCTC CAGCCTGGCTTCTAGCTGGAAGCCAGCCCAGCGTTTCTCTACCACCACCACCATGCCTCCACCTGACTTTGGCTTGGAGACTGATCCTCT CTGTGTAAATTCTGCCCGGTGCTGTGAAGGCTGGACGGTGGAGGACCTGCTGGGGTCTCCTGGGACCCGCCTGTTGCTTCTGCCCTCCCC TGTGGAAAGGTCTATATGACGGGCCGCCTGAGGCCCCAGAACTCGTCTGTGAACCACCTTTTCCAGCCAGAGTTCCCAAAGCTGGAACGC TAGCTGCCTGCTCTTCCTTAAGATGGCCTCCCCCCGACCCGCCACGGCCCTCAGTTGCCAGGGATGGGGCCACCACTGTCACACTGTGGA ATACAAGACAGTGAACTCTGTCTGCCTGAACGAGCCATGTAAATTAAGTTCTAGAGCAGCTCTCTGAGCAGGATAAGGTCCCCTGACAGT GAGTTGTGTGGTGGGGGCAGCCTCTGCCTCAAAAATTCACCAAGCAGAATGCCTCTCAGCCTCATGTGTTGGTCCTCTGCTCCTCCTAGC TCCCCAGGGATGTTGGGGACCCAGCTTGTCTCGGCAGCTAAGAAGCAGTGACCAGGATGTGGATTTTGGCGACCTGTGTGGTGGCCTTGA GCTGCTTTCTGTGTTTGTGAGGACTGACTCCCATTTCCTAAAGGAAATGCCCCCGGGGAGGACATTGGGAGGAAGATGGCCTGAGTGTGC ACTTTGGCTCTGCTACCTGCTCCTGAAGCCCCGCTAAAAATAATTCATCCAAGATTCCTTTGTAGTTAAAGGGTCCAGTTCTGACTGGAG CCTCTAGAGAGCTGGGCTTGTATGTTCTTTTGGCCTTTTGTTCCTACCTAAATGAAGAAACCATGCCTGGAGGGGCCGTGAACACAGAAC CCTCAAGACAAGGATGACAGAGCTGGAGGACACATCTAGCTGCCATTGCAACCTCACTGGGCTCCCCAGACTCTGTGTGTGAGAAATTAA >100591_100591_4_ZC3H7B-L3MBTL2_ZC3H7B_chr22_41723368_ENST00000352645_L3MBTL2_chr22_41620024_ENST00000216237_length(amino acids)=540AA_BP=149 MMERQKRKADIEKGLQFIQSTLPLKQEEYEAFLLKLVQNLFAEGNDLFREKDYKQALVQYMEGLNVADYAASDQVALPRELLCKLHVNRA ACYFTMGLYEKALEDSEKALGLDSESIRALFRKARALNELGRHKEAYECSSRCSLALPHMVESMKYPFRQGMRLEVVDKSQVSRTRMAVV DTVIGGRLRLLYEDGDSDDDFWCHMWSPLIHPVGWSRRVGHGIKMSERRSDMAHHPTFRKIYCDAVPYLFKKVRAVYTEGGWFEEGMKLE AIDPLNLGNICVATVCKVLLDGYLMICVDGGPSTDGLDWFCYHASSHAIFPATFCQKNDIELTPPKGYEAQTFNWENYLEKTKSKAAPSR LFNMDCPNHGFKVGMKLEAVDLMEPRLICVATVKRVVHRLLSIHFDGWDSEYDQWVDCESPDIYPVGWCELTGYQLQPPVAAEPATPLKA KEATKKKKKQFGKKRKRIPPTKTRPLRQGSKKPLLEDDPQGARKISSEPVPGEIIAVRVKEEHLDVASPDKASSPELPVSVENIKQETDD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZC3H7B-L3MBTL2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZC3H7B-L3MBTL2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZC3H7B-L3MBTL2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
