
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZC3HAV1-BRAF (FusionGDB2 ID:100608) |
Fusion Gene Summary for ZC3HAV1-BRAF |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZC3HAV1-BRAF | Fusion gene ID: 100608 | Hgene | Tgene | Gene symbol | ZC3HAV1 | BRAF | Gene ID | 56829 | 673 |
| Gene name | zinc finger CCCH-type containing, antiviral 1 | B-Raf proto-oncogene, serine/threonine kinase | |
| Synonyms | ARTD13|FLB6421|PARP13|ZAP|ZC3H2|ZC3HDC2 | B-RAF1|B-raf|BRAF1|NS7|RAFB1 | |
| Cytomap | 7q34 | 7q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger CCCH-type antiviral protein 1ADP-ribosyltransferase diphtheria toxin-like 13inactive Poly [ADP-ribose] polymerase 13zinc finger CCCH domain-containing protein 2zinc finger CCCH-type, antiviral 1 | serine/threonine-protein kinase B-raf94 kDa B-raf proteinB-Raf proto-oncogene serine/threonine-protein kinase (p94)B-Raf serine/threonine-proteinmurine sarcoma viral (v-raf) oncogene homolog B1proto-oncogene B-Rafv-raf murine sarcoma viral oncogene | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | P15056 | |
| Ensembl transtripts involved in fusion gene | ENST00000242351, ENST00000464606, ENST00000471652, | ENST00000288602, | |
| Fusion gene scores | * DoF score | 7 X 7 X 6=294 | 48 X 58 X 16=44544 |
| # samples | 8 | 69 | |
| ** MAII score | log2(8/294*10)=-1.877744249949 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(69/44544*10)=-6.0124909441832 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZC3HAV1 [Title/Abstract] AND BRAF [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZC3HAV1(138758602)-BRAF(140481493), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ZC3HAV1 | GO:0006471 | protein ADP-ribosylation | 25043379 |
| Hgene | ZC3HAV1 | GO:0009615 | response to virus | 21876179 |
| Hgene | ZC3HAV1 | GO:0045071 | negative regulation of viral genome replication | 21876179 |
| Hgene | ZC3HAV1 | GO:0061014 | positive regulation of mRNA catabolic process | 21876179 |
| Tgene | BRAF | GO:0000186 | activation of MAPKK activity | 29433126 |
| Tgene | BRAF | GO:0006468 | protein phosphorylation | 17563371 |
| Tgene | BRAF | GO:0010828 | positive regulation of glucose transmembrane transport | 23010278 |
| Tgene | BRAF | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 |
| Tgene | BRAF | GO:0043066 | negative regulation of apoptotic process | 19667065 |
| Tgene | BRAF | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 22065586 |
| Tgene | BRAF | GO:0071277 | cellular response to calcium ion | 18567582 |
| Tgene | BRAF | GO:0090150 | establishment of protein localization to membrane | 23010278 |
 Fusion gene breakpoints across ZC3HAV1 (5'-gene) Fusion gene breakpoints across ZC3HAV1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
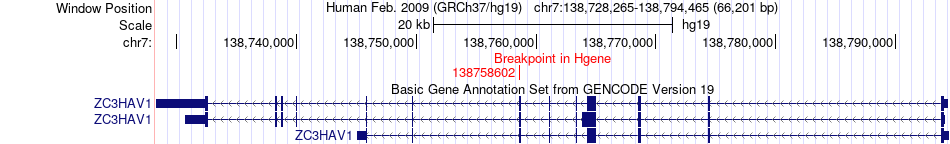 |
 Fusion gene breakpoints across BRAF (3'-gene) Fusion gene breakpoints across BRAF (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
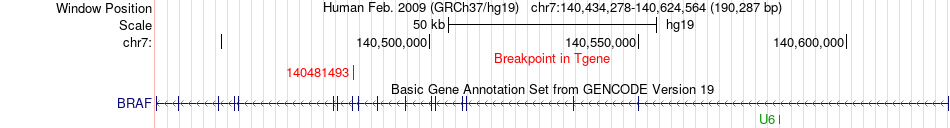 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-KS-A4ID-01A | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - |
Top |
Fusion Gene ORF analysis for ZC3HAV1-BRAF |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000242351 | ENST00000288602 | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - |
| In-frame | ENST00000464606 | ENST00000288602 | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - |
| In-frame | ENST00000471652 | ENST00000288602 | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000242351 | ZC3HAV1 | chr7 | 138758602 | - | ENST00000288602 | BRAF | chr7 | 140481493 | - | 3294 | 2189 | 236 | 3175 | 979 |
| ENST00000464606 | ZC3HAV1 | chr7 | 138758602 | - | ENST00000288602 | BRAF | chr7 | 140481493 | - | 3366 | 2261 | 23 | 3247 | 1074 |
| ENST00000471652 | ZC3HAV1 | chr7 | 138758602 | - | ENST00000288602 | BRAF | chr7 | 140481493 | - | 3365 | 2260 | 307 | 3246 | 979 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000242351 | ENST00000288602 | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - | 0.001133165 | 0.9988669 |
| ENST00000464606 | ENST00000288602 | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - | 0.000791574 | 0.9992085 |
| ENST00000471652 | ENST00000288602 | ZC3HAV1 | chr7 | 138758602 | - | BRAF | chr7 | 140481493 | - | 0.001167497 | 0.9988325 |
Top |
Fusion Genomic Features for ZC3HAV1-BRAF |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
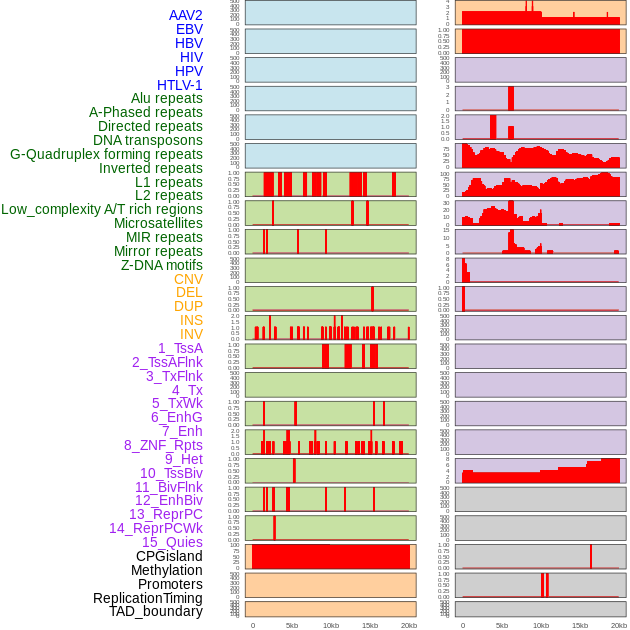 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZC3HAV1-BRAF |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:138758602/chr7:140481493) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | BRAF |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 285_292 | 624 | 903.0 | Motif | Nuclear export signal |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 69_76 | 624 | 903.0 | Motif | Nuclear localization signal |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 285_292 | 624 | 700.0 | Motif | Nuclear export signal |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 69_76 | 624 | 700.0 | Motif | Nuclear localization signal |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 224_254 | 624 | 903.0 | Region | Binding to EXOSC5 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 2_254 | 624 | 903.0 | Region | Note=N-terminal domain |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 224_254 | 624 | 700.0 | Region | Binding to EXOSC5 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 2_254 | 624 | 700.0 | Region | Note=N-terminal domain |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 150_172 | 624 | 903.0 | Zinc finger | C3H1-type 3 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 169_193 | 624 | 903.0 | Zinc finger | C3H1-type 4 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 73_86 | 624 | 903.0 | Zinc finger | C3H1-type 1 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 88_110 | 624 | 903.0 | Zinc finger | C3H1-type 2 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 150_172 | 624 | 700.0 | Zinc finger | C3H1-type 3 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 169_193 | 624 | 700.0 | Zinc finger | C3H1-type 4 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 73_86 | 624 | 700.0 | Zinc finger | C3H1-type 1 |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 88_110 | 624 | 700.0 | Zinc finger | C3H1-type 2 |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 457_717 | 438 | 767.0 | Domain | Protein kinase | |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 463_471 | 438 | 767.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 594_681 | 624 | 903.0 | Domain | WWE |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000242351 | - | 7 | 13 | 716_902 | 624 | 903.0 | Domain | PARP catalytic |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 594_681 | 624 | 700.0 | Domain | WWE |
| Hgene | ZC3HAV1 | chr7:138758602 | chr7:140481493 | ENST00000471652 | - | 7 | 9 | 716_902 | 624 | 700.0 | Domain | PARP catalytic |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 122_129 | 438 | 767.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 428_432 | 438 | 767.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 6_11 | 438 | 767.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 155_227 | 438 | 767.0 | Domain | RBD | |
| Tgene | BRAF | chr7:138758602 | chr7:140481493 | ENST00000288602 | 9 | 18 | 234_280 | 438 | 767.0 | Zinc finger | Phorbol-ester/DAG-type |
Top |
Fusion Gene Sequence for ZC3HAV1-BRAF |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >100608_100608_1_ZC3HAV1-BRAF_ZC3HAV1_chr7_138758602_ENST00000242351_BRAF_chr7_140481493_ENST00000288602_length(transcript)=3294nt_BP=2189nt GAACCGAGAGGGGGCCACCCCAGGCGGTCACCAGCAGATTTGCCCGCGCGTTCTCTTTCTTTCCACCCAGTTGCCCTTGCGGCCGGCTGT AAACCTGCCACTAGGACCCGGTCGGTGAGATCTAGCCTCTTGACCTGAGAGCCGAGAGTGGATCGCTGGGCTGGGCTAACGGCGACGGAG AGCGCGCCCTCGCTGACTCCGGGCGCGCCCAGCAGTAGCACCGCCCGCGCCCGCCCCTGGACACTTGTAAGTTTCGATTTCCGATTTCCG CGGAACCGAGTCCCGCGCCGCGGCAGAGCCAGCACAGCCAGCGCGCCATGGCGGACCCGGAGGTGTGCTGCTTCATCACCAAAATCCTGT GCGCCCACGGGGGCCGCATGGCCCTGGACGCGCTGCTCCAGGAGATCGCGCTGTCTGAGCCGCAGCTCTGTGAGGTGCTGCAGGTGGCCG GGCCCGACCGCTTTGTGGTGTTGGAGACCGGCGGCGAGGCCGGGATCACCCGATCGGTGGTGGCCACCACTCGAGCCCGGGTCTGCCGTC GCAAGTACTGCCAGAGACCCTGCGATAACCTGCATCTCTGCAAACTCAACTTGCTGGGCCGGTGCAACTATTCGCAGTCCGAGCGGAATT TATGCAAATATTCTCATGAGGTTCTCTCAGAAGAGAACTTCAAAGTCCTGAAAAATCACGAACTCTCTGGACTGAACAAAGAGGAATTAG CAGTGCTCCTCCTCCAAAGTGATCCTTTTTTTATGCCCGAGATATGCAAAAGTTATAAGGGAGAGGGTCGGCAGCAGATTTGTAACCAGC AGCCACCGTGTTCAAGACTCCACATCTGTGACCACTTCACCCGAGGGAACTGTCGTTTTCCCAACTGCCTCCGGTCCCATAACCTGATGG ACAGAAAGGTGCTGGCCATCATGAGGGAGCACGGGCTGAACCCCGACGTGGTCCAGAACATCCAGGACATCTGCAACAGCAAGCACATGC AGAAGAATCCCCCAGGGCCCAGAGCTCCTTCTTCACATCGTAGAAACATGGCATATAGGGCTAGAAGCAAGAGTAGAGATCGGTTCTTTC AGGGCAGCCAAGAATTTCTTGCGTCTGCTTCAGCGTCTGCTGAGAGGTCCTGCACACCTAGTCCAGATCAGATCAGCCACAGGGCTTCCC TGGAGGACGCGCCTGTGGACGATCTCACCCGCAAGTTCACGTATCTGGGGAGTCAGGATCGCGCTCGGCCTCCCTCAGGCTCGTCCAAGG CTACTGATCTTGGAGGAACAAGTCAGGCCGGGACAAGCCAGAGGTTTTTAGAGAACGGCAGTCAAGAGGACCTCTTGCATGGAAATCCAG GCAGCACTTACCTTGCTTCCAATTCAACATCAGCCCCCAACTGGAAGAGCCTCACATCCTGGACGAATGACCAAGGCGCCAGGAGAAAGA CTGTGTTTTCTCCCACGCTACCTGCCGCCCGCTCTTCTCTTGGCTCTCTGCAAACACCTGAAGCTGTGACCACCAGAAAGGGCACAGGCT TGCTTTCCTCAGACTACAGGATCATCAATGGCAAAAGTGGAACTCAGGACATCCAGCCTGGCCCTCTTTTTAATAATAATGCTGATGGAG TGGCCACAGATATAACTTCTACCAGATCCTTAAATTACAAAAGCACTAGCAGCGGTCACAGAGAAATATCATCACCTAGGATTCAGGATG CTGGACCTGCTTCCCGAGATGTCCAGGCCACTGGCAGAATCGCAGATGATGCTGACCCAAGAGTAGCACTTGTTAACGATTCTTTATCTG ATGTCACAAGTACCACATCTTCTAGGGTGGATGATCATGACTCAGAGGAAATTTGTCTTGACCATCTGTGTAAGGGTTGTCCGCTTAATG GTAGCTGCAGCAAAGTCCACTTCCATCTGCCTTACCGGTGGCAGATGCTTATTGGTAAAACCTGGACGGACTTTGAGCACATGGAGACGA TCGAGAAAGGCTACTGTAACCCCGGAATCCACCTCTGTTCTGTAGGAAGTTATACAATCAATTTTCGGGTAATGAGTTGTGATTCCTTTC CCATCCGACGCCTCTCCACTCCTTCTTCTGTCACCAAGCCAGCCAATTCTGTCTTCACCACCAAATGGATTTGGTATTGGAAGAATGAAT CTGGCACATGGATTCAGTATGGAGAAGAGAAAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAG TGGGACAAAGAATTGGATCTGGATCATTTGGAACAGTCTACAAGGGAAAGTGGCATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAG CACCTACACCTCAGCAGTTACAAGCCTTCAAAAATGAAGTAGGAGTACTCAGGAAAACACGACATGTGAATATCCTACTCTTCATGGGCT ATTCCACAAAGCCACAACTGGCTATTGTTACCCAGTGGTGTGAGGGCTCCAGCTTGTATCACCATCTCCATATCATTGAGACCAAATTTG AGATGATCAAACTTATAGATATTGCACGACAGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGA GTAATAATATATTTCTTCATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATC AGTTTGAACAGTTGTCTGGATCCATTTTGTGGATGGCACCAGAAGTCATCAGAATGCAAGATAAAAATCCATACAGCTTTCAGTCAGATG TATATGCATTTGGAATTGTTCTGTATGAATTGATGACTGGACAGTTACCTTATTCAAACATCAACAACAGGGACCAGATAATTTTTATGG TGGGACGAGGATACCTGTCTCCAGATCTCAGTAAGGTACGGAGTAACTGTCCAAAAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAA AGAAAAGAGATGAGAGACCACTCTTTCCCCAAATTCTCGCCTCTATTGAGCTGCTGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCAT CAGAACCCTCCTTGAATCGGGCTGGTTTCCAAACAGAGGATTTTAGTCTATATGCTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGG GATATGGTGCGTTTCCTGTCCACTGAAACAAATGAGTGAGAGAGTTCAGGAGAGTAGCAACAAAAGGAAAATAAATGAACATATGTTTGC >100608_100608_1_ZC3HAV1-BRAF_ZC3HAV1_chr7_138758602_ENST00000242351_BRAF_chr7_140481493_ENST00000288602_length(amino acids)=979AA_BP=651 MDTCKFRFPISAEPSPAPRQSQHSQRAMADPEVCCFITKILCAHGGRMALDALLQEIALSEPQLCEVLQVAGPDRFVVLETGGEAGITRS VVATTRARVCRRKYCQRPCDNLHLCKLNLLGRCNYSQSERNLCKYSHEVLSEENFKVLKNHELSGLNKEELAVLLLQSDPFFMPEICKSY KGEGRQQICNQQPPCSRLHICDHFTRGNCRFPNCLRSHNLMDRKVLAIMREHGLNPDVVQNIQDICNSKHMQKNPPGPRAPSSHRRNMAY RARSKSRDRFFQGSQEFLASASASAERSCTPSPDQISHRASLEDAPVDDLTRKFTYLGSQDRARPPSGSSKATDLGGTSQAGTSQRFLEN GSQEDLLHGNPGSTYLASNSTSAPNWKSLTSWTNDQGARRKTVFSPTLPAARSSLGSLQTPEAVTTRKGTGLLSSDYRIINGKSGTQDIQ PGPLFNNNADGVATDITSTRSLNYKSTSSGHREISSPRIQDAGPASRDVQATGRIADDADPRVALVNDSLSDVTSTTSSRVDDHDSEEIC LDHLCKGCPLNGSCSKVHFHLPYRWQMLIGKTWTDFEHMETIEKGYCNPGIHLCSVGSYTINFRVMSCDSFPIRRLSTPSSVTKPANSVF TTKWIWYWKNESGTWIQYGEEKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRK TRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGL ATVKSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPK -------------------------------------------------------------- >100608_100608_2_ZC3HAV1-BRAF_ZC3HAV1_chr7_138758602_ENST00000464606_BRAF_chr7_140481493_ENST00000288602_length(transcript)=3366nt_BP=2261nt AGAGCCAGCACAGCCAGCGCGCCATGGCGGACCCGGAGGTGTGCTGCTTCATCACCAAAATCCTGTGCGCCCACGGGGGCCGCATGGCCC TGGACGCGCTGCTCCAGGAGATCGCGCTGTCTGAGCCGCAGCTCTGTGAGGTGCTGCAGGTGGCCGGGCCCGACCGCTTTGTGGTGTTGG AGACCGGCGGCGAGGCCGGGATCACCCGATCGGTGGTGGCCACCACTCGAGCCCGGGTCTGCCGTCGCAAGTACTGCCAGAGACCCTGCG ATAACCTGCATCTCTGCAAACTCAACTTGCTGGGCCGGTGCAACTATTCGCAGTCCGAGCGGAATTTATGCAAATATTCTCATGAGGTTC TCTCAGAAGAGAACTTCAAAGTCCTGAAAAATCACGAACTCTCTGGACTGAACAAAGAGGAATTAGCAGTGCTCCTCCTCCAAAGTGATC CTTTTTTTATGCCCGAGATATGCAAAAGTTATAAGGGAGAGGGTCGGCAGCAGATTTGTAACCAGCAGCCACCGTGTTCAAGACTCCACA TCTGTGACCACTTCACCCGAGGGAACTGTCGTTTTCCCAACTGCCTCCGGTCCCATAACCTGATGGACAGAAAGGTGCTGGCCATCATGA GGGAGCACGGGCTGAACCCCGACGTGGTCCAGAACATCCAGGACATCTGCAACAGCAAGCACATGCAGAAGAATCCCCCAGGGCCCAGAG CTCCTTCTTCACATCGTAGAAACATGGCATATAGGGCTAGAAGCAAGAGTAGAGATCGGTTCTTTCAGGGCAGCCAAGAATTTCTTGCGT CTGCTTCAGCGTCTGCTGAGAGGTCCTGCACACCTAGTCCAGATCAGATCAGCCACAGGGCTTCCCTGGAGGACGCGCCTGTGGACGATC TCACCCGCAAGTTCACGTATCTGGGGAGTCAGGATCGCGCTCGGCCTCCCTCAGGCTCGTCCAAGGCTACTGATCTTGGAGGAACAAGTC AGGCCGGGACAAGCCAGAGGTTTTTAGAGAACGGCAGTCAAGAGGACCTCTTGCATGGAAATCCAGGCAGCACTTACCTTGCTTCCAATT CAACATCAGCCCCCAACTGGAAGAGCCTCACATCCTGGACGAATGACCAAGGCGCCAGGAGAAAGACTGTGTTTTCTCCCACGCTACCTG CCGCCCGCTCTTCTCTTGGCTCTCTGCAAACACCTGAAGCTGTGACCACCAGAAAGGGCACAGGCTTGCTTTCCTCAGACTACAGGATCA TCAATGGCAAAAGTGGAACTCAGGACATCCAGCCTGGCCCTCTTTTTAATAATAATGCTGATGGAGTGGCCACAGATATAACTTCTACCA GATCCTTAAATTACAAAAGCACTAGCAGCGGTCACAGAGAAATATCATCACCTAGGATTCAGGATGCTGGACCTGCTTCCCGAGATGTCC AGGCCACTGGCAGAATCGCAGATGATGCTGACCCAAGAGTAGCACTTGTTAACGGTAAATACAAAGGGAAGACACTTTGGGCTAGTACAT TTGTTCATGATATACCAAATGGCTCTAGTCAAGTAGTGGATAAAACTACTGATGTAGAAAAAACTGGTGCCACTGGTTTTGGCTTAACAA TGGCAGTCAAGGCAGAAAAAGATATGTTACGTACTGGAAGTCAGAGTCTGAGGAACCTGGTCCCCACCACACCTGGGGAATCCACTGCCC CTGCACAAGTCAGCACTCTGCCTCAGTCACCTGCTGCCCTTTCCTCAAGCAACAGAGCTGCAGTCTGGGGGGCCCAAGGGCAGAACTGCA CCCAGGTTCCTGTTTCCTCTGCCAGCGAGCTCACAAGGAAGACAACAGGCTCTGCTCAGTATTCTTTATCTGATGTCACAAGTACCACAT CTTCTAGGGTGGATGATCATGACTCAGAGGAAATTTGTCTTGACCATCTGTGTAAGGGTTGTCCGCTTAATGGTAGCTGCAGCAAAGTCC ACTTCCATCTGCCTTACCGGTGGCAGATGCTTATTGGTAAAACCTGGACGGACTTTGAGCACATGGAGACGATCGAGAAAGGCTACTGTA ACCCCGGAATCCACCTCTGTTCTGTAGGAAGTTATACAATCAATTTTCGGGTAATGAGTTGTGATTCCTTTCCCATCCGACGCCTCTCCA CTCCTTCTTCTGTCACCAAGCCAGCCAATTCTGTCTTCACCACCAAATGGATTTGGTATTGGAAGAATGAATCTGGCACATGGATTCAGT ATGGAGAAGAGAAAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGACAAAGAATTGGAT CTGGATCATTTGGAACAGTCTACAAGGGAAAGTGGCATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAGCACCTACACCTCAGCAGT TACAAGCCTTCAAAAATGAAGTAGGAGTACTCAGGAAAACACGACATGTGAATATCCTACTCTTCATGGGCTATTCCACAAAGCCACAAC TGGCTATTGTTACCCAGTGGTGTGAGGGCTCCAGCTTGTATCACCATCTCCATATCATTGAGACCAAATTTGAGATGATCAAACTTATAG ATATTGCACGACAGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAATAATATATTTCTTC ATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATCAGTTTGAACAGTTGTCTG GATCCATTTTGTGGATGGCACCAGAAGTCATCAGAATGCAAGATAAAAATCCATACAGCTTTCAGTCAGATGTATATGCATTTGGAATTG TTCTGTATGAATTGATGACTGGACAGTTACCTTATTCAAACATCAACAACAGGGACCAGATAATTTTTATGGTGGGACGAGGATACCTGT CTCCAGATCTCAGTAAGGTACGGAGTAACTGTCCAAAAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAAAGAAAAGAGATGAGAGAC CACTCTTTCCCCAAATTCTCGCCTCTATTGAGCTGCTGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCATCAGAACCCTCCTTGAATC GGGCTGGTTTCCAAACAGAGGATTTTAGTCTATATGCTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGGGATATGGTGCGTTTCCTG TCCACTGAAACAAATGAGTGAGAGAGTTCAGGAGAGTAGCAACAAAAGGAAAATAAATGAACATATGTTTGCTTATATGTTAAATTGAAT >100608_100608_2_ZC3HAV1-BRAF_ZC3HAV1_chr7_138758602_ENST00000464606_BRAF_chr7_140481493_ENST00000288602_length(amino acids)=1074AA_BP=746 MADPEVCCFITKILCAHGGRMALDALLQEIALSEPQLCEVLQVAGPDRFVVLETGGEAGITRSVVATTRARVCRRKYCQRPCDNLHLCKL NLLGRCNYSQSERNLCKYSHEVLSEENFKVLKNHELSGLNKEELAVLLLQSDPFFMPEICKSYKGEGRQQICNQQPPCSRLHICDHFTRG NCRFPNCLRSHNLMDRKVLAIMREHGLNPDVVQNIQDICNSKHMQKNPPGPRAPSSHRRNMAYRARSKSRDRFFQGSQEFLASASASAER SCTPSPDQISHRASLEDAPVDDLTRKFTYLGSQDRARPPSGSSKATDLGGTSQAGTSQRFLENGSQEDLLHGNPGSTYLASNSTSAPNWK SLTSWTNDQGARRKTVFSPTLPAARSSLGSLQTPEAVTTRKGTGLLSSDYRIINGKSGTQDIQPGPLFNNNADGVATDITSTRSLNYKST SSGHREISSPRIQDAGPASRDVQATGRIADDADPRVALVNGKYKGKTLWASTFVHDIPNGSSQVVDKTTDVEKTGATGFGLTMAVKAEKD MLRTGSQSLRNLVPTTPGESTAPAQVSTLPQSPAALSSSNRAAVWGAQGQNCTQVPVSSASELTRKTTGSAQYSLSDVTSTTSSRVDDHD SEEICLDHLCKGCPLNGSCSKVHFHLPYRWQMLIGKTWTDFEHMETIEKGYCNPGIHLCSVGSYTINFRVMSCDSFPIRRLSTPSSVTKP ANSVFTTKWIWYWKNESGTWIQYGEEKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEV GVLRKTRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKI GDFGLATVKSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVR -------------------------------------------------------------- >100608_100608_3_ZC3HAV1-BRAF_ZC3HAV1_chr7_138758602_ENST00000471652_BRAF_chr7_140481493_ENST00000288602_length(transcript)=3365nt_BP=2260nt CTTTTAGTTTCTCTTCTTTCTAAAGAAGGCTCGCGGAGCCCGGCTGGAGAACCTCACCCTCGCCGAGCCTAGAACCGAGAGGGGGCCACC CCAGGCGGTCACCAGCAGATTTGCCCGCGCGTTCTCTTTCTTTCCACCCAGTTGCCCTTGCGGCCGGCTGTAAACCTGCCACTAGGACCC GGTCGGTGAGATCTAGCCTCTTGACCTGAGAGCCGAGAGTGGATCGCTGGGCTGGGCTAACGGCGACGGAGAGCGCGCCCTCGCTGACTC CGGGCGCGCCCAGCAGTAGCACCGCCCGCGCCCGCCCCTGGACACTTGTAAGTTTCGATTTCCGATTTCCGCGGAACCGAGTCCCGCGCC GCGGCAGAGCCAGCACAGCCAGCGCGCCATGGCGGACCCGGAGGTGTGCTGCTTCATCACCAAAATCCTGTGCGCCCACGGGGGCCGCAT GGCCCTGGACGCGCTGCTCCAGGAGATCGCGCTGTCTGAGCCGCAGCTCTGTGAGGTGCTGCAGGTGGCCGGGCCCGACCGCTTTGTGGT GTTGGAGACCGGCGGCGAGGCCGGGATCACCCGATCGGTGGTGGCCACCACTCGAGCCCGGGTCTGCCGTCGCAAGTACTGCCAGAGACC CTGCGATAACCTGCATCTCTGCAAACTCAACTTGCTGGGCCGGTGCAACTATTCGCAGTCCGAGCGGAATTTATGCAAATATTCTCATGA GGTTCTCTCAGAAGAGAACTTCAAAGTCCTGAAAAATCACGAACTCTCTGGACTGAACAAAGAGGAATTAGCAGTGCTCCTCCTCCAAAG TGATCCTTTTTTTATGCCCGAGATATGCAAAAGTTATAAGGGAGAGGGTCGGCAGCAGATTTGTAACCAGCAGCCACCGTGTTCAAGACT CCACATCTGTGACCACTTCACCCGAGGGAACTGTCGTTTTCCCAACTGCCTCCGGTCCCATAACCTGATGGACAGAAAGGTGCTGGCCAT CATGAGGGAGCACGGGCTGAACCCCGACGTGGTCCAGAACATCCAGGACATCTGCAACAGCAAGCACATGCAGAAGAATCCCCCAGGGCC CAGAGCTCCTTCTTCACATCGTAGAAACATGGCATATAGGGCTAGAAGCAAGAGTAGAGATCGGTTCTTTCAGGGCAGCCAAGAATTTCT TGCGTCTGCTTCAGCGTCTGCTGAGAGGTCCTGCACACCTAGTCCAGATCAGATCAGCCACAGGGCTTCCCTGGAGGACGCGCCTGTGGA CGATCTCACCCGCAAGTTCACGTATCTGGGGAGTCAGGATCGCGCTCGGCCTCCCTCAGGCTCGTCCAAGGCTACTGATCTTGGAGGAAC AAGTCAGGCCGGGACAAGCCAGAGGTTTTTAGAGAACGGCAGTCAAGAGGACCTCTTGCATGGAAATCCAGGCAGCACTTACCTTGCTTC CAATTCAACATCAGCCCCCAACTGGAAGAGCCTCACATCCTGGACGAATGACCAAGGCGCCAGGAGAAAGACTGTGTTTTCTCCCACGCT ACCTGCCGCCCGCTCTTCTCTTGGCTCTCTGCAAACACCTGAAGCTGTGACCACCAGAAAGGGCACAGGCTTGCTTTCCTCAGACTACAG GATCATCAATGGCAAAAGTGGAACTCAGGACATCCAGCCTGGCCCTCTTTTTAATAATAATGCTGATGGAGTGGCCACAGATATAACTTC TACCAGATCCTTAAATTACAAAAGCACTAGCAGCGGTCACAGAGAAATATCATCACCTAGGATTCAGGATGCTGGACCTGCTTCCCGAGA TGTCCAGGCCACTGGCAGAATCGCAGATGATGCTGACCCAAGAGTAGCACTTGTTAACGATTCTTTATCTGATGTCACAAGTACCACATC TTCTAGGGTGGATGATCATGACTCAGAGGAAATTTGTCTTGACCATCTGTGTAAGGGTTGTCCGCTTAATGGTAGCTGCAGCAAAGTCCA CTTCCATCTGCCTTACCGGTGGCAGATGCTTATTGGTAAAACCTGGACGGACTTTGAGCACATGGAGACGATCGAGAAAGGCTACTGTAA CCCCGGAATCCACCTCTGTTCTGTAGGAAGTTATACAATCAATTTTCGGGTAATGAGTTGTGATTCCTTTCCCATCCGACGCCTCTCCAC TCCTTCTTCTGTCACCAAGCCAGCCAATTCTGTCTTCACCACCAAATGGATTTGGTATTGGAAGAATGAATCTGGCACATGGATTCAGTA TGGAGAAGAGAAAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGACAAAGAATTGGATC TGGATCATTTGGAACAGTCTACAAGGGAAAGTGGCATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAGCACCTACACCTCAGCAGTT ACAAGCCTTCAAAAATGAAGTAGGAGTACTCAGGAAAACACGACATGTGAATATCCTACTCTTCATGGGCTATTCCACAAAGCCACAACT GGCTATTGTTACCCAGTGGTGTGAGGGCTCCAGCTTGTATCACCATCTCCATATCATTGAGACCAAATTTGAGATGATCAAACTTATAGA TATTGCACGACAGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAATAATATATTTCTTCA TGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATCAGTTTGAACAGTTGTCTGG ATCCATTTTGTGGATGGCACCAGAAGTCATCAGAATGCAAGATAAAAATCCATACAGCTTTCAGTCAGATGTATATGCATTTGGAATTGT TCTGTATGAATTGATGACTGGACAGTTACCTTATTCAAACATCAACAACAGGGACCAGATAATTTTTATGGTGGGACGAGGATACCTGTC TCCAGATCTCAGTAAGGTACGGAGTAACTGTCCAAAAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAAAGAAAAGAGATGAGAGACC ACTCTTTCCCCAAATTCTCGCCTCTATTGAGCTGCTGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCATCAGAACCCTCCTTGAATCG GGCTGGTTTCCAAACAGAGGATTTTAGTCTATATGCTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGGGATATGGTGCGTTTCCTGT CCACTGAAACAAATGAGTGAGAGAGTTCAGGAGAGTAGCAACAAAAGGAAAATAAATGAACATATGTTTGCTTATATGTTAAATTGAATA >100608_100608_3_ZC3HAV1-BRAF_ZC3HAV1_chr7_138758602_ENST00000471652_BRAF_chr7_140481493_ENST00000288602_length(amino acids)=979AA_BP=651 MDTCKFRFPISAEPSPAPRQSQHSQRAMADPEVCCFITKILCAHGGRMALDALLQEIALSEPQLCEVLQVAGPDRFVVLETGGEAGITRS VVATTRARVCRRKYCQRPCDNLHLCKLNLLGRCNYSQSERNLCKYSHEVLSEENFKVLKNHELSGLNKEELAVLLLQSDPFFMPEICKSY KGEGRQQICNQQPPCSRLHICDHFTRGNCRFPNCLRSHNLMDRKVLAIMREHGLNPDVVQNIQDICNSKHMQKNPPGPRAPSSHRRNMAY RARSKSRDRFFQGSQEFLASASASAERSCTPSPDQISHRASLEDAPVDDLTRKFTYLGSQDRARPPSGSSKATDLGGTSQAGTSQRFLEN GSQEDLLHGNPGSTYLASNSTSAPNWKSLTSWTNDQGARRKTVFSPTLPAARSSLGSLQTPEAVTTRKGTGLLSSDYRIINGKSGTQDIQ PGPLFNNNADGVATDITSTRSLNYKSTSSGHREISSPRIQDAGPASRDVQATGRIADDADPRVALVNDSLSDVTSTTSSRVDDHDSEEIC LDHLCKGCPLNGSCSKVHFHLPYRWQMLIGKTWTDFEHMETIEKGYCNPGIHLCSVGSYTINFRVMSCDSFPIRRLSTPSSVTKPANSVF TTKWIWYWKNESGTWIQYGEEKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRK TRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGL ATVKSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZC3HAV1-BRAF |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZC3HAV1-BRAF |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZC3HAV1-BRAF |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
