
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BRAF-TRIM24 (FusionGDB2 ID:10136) |
Fusion Gene Summary for BRAF-TRIM24 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BRAF-TRIM24 | Fusion gene ID: 10136 | Hgene | Tgene | Gene symbol | BRAF | TRIM24 | Gene ID | 673 | 8805 |
| Gene name | B-Raf proto-oncogene, serine/threonine kinase | tripartite motif containing 24 | |
| Synonyms | B-RAF1|B-raf|BRAF1|NS7|RAFB1 | PTC6|RNF82|TF1A|TIF1|TIF1A|TIF1ALPHA|hTIF1 | |
| Cytomap | 7q34 | 7q33-q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase B-raf94 kDa B-raf proteinB-Raf proto-oncogene serine/threonine-protein kinase (p94)B-Raf serine/threonine-proteinmurine sarcoma viral (v-raf) oncogene homolog B1proto-oncogene B-Rafv-raf murine sarcoma viral oncogene | transcription intermediary factor 1-alphaE3 ubiquitin-protein ligase TRIM24RING finger protein 82RING-type E3 ubiquitin transferase TIF1-alphaTIF1-alphatranscriptional intermediary factor 1 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P15056 | O15164 | |
| Ensembl transtripts involved in fusion gene | ENST00000288602, | ENST00000497516, ENST00000343526, ENST00000415680, | |
| Fusion gene scores | * DoF score | 28 X 21 X 11=6468 | 10 X 9 X 7=630 |
| # samples | 32 | 11 | |
| ** MAII score | log2(32/6468*10)=-4.3371758685863 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/630*10)=-2.51784830486262 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: BRAF [Title/Abstract] AND TRIM24 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BRAF(140624366)-TRIM24(138258252), # samples:1 TRIM24(138255748)-BRAF(140550012), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BRAF | GO:0000186 | activation of MAPKK activity | 29433126 |
| Hgene | BRAF | GO:0006468 | protein phosphorylation | 17563371 |
| Hgene | BRAF | GO:0010828 | positive regulation of glucose transmembrane transport | 23010278 |
| Hgene | BRAF | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 |
| Hgene | BRAF | GO:0043066 | negative regulation of apoptotic process | 19667065 |
| Hgene | BRAF | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 22065586 |
| Hgene | BRAF | GO:0071277 | cellular response to calcium ion | 18567582 |
| Hgene | BRAF | GO:0090150 | establishment of protein localization to membrane | 23010278 |
| Tgene | TRIM24 | GO:0016567 | protein ubiquitination | 19556538 |
| Tgene | TRIM24 | GO:0071391 | cellular response to estrogen stimulus | 21164480 |
 Fusion gene breakpoints across BRAF (5'-gene) Fusion gene breakpoints across BRAF (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
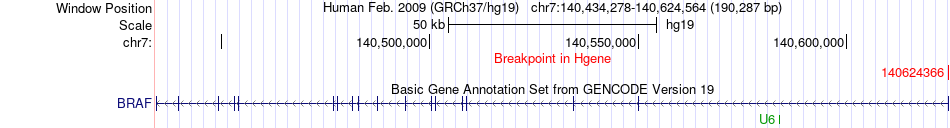 |
 Fusion gene breakpoints across TRIM24 (3'-gene) Fusion gene breakpoints across TRIM24 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
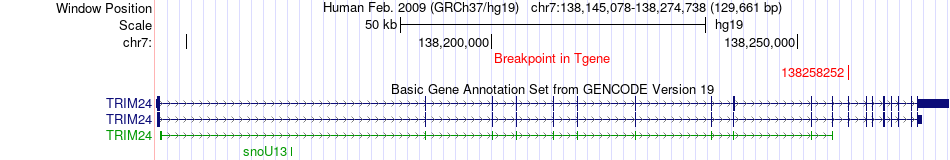 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | READ | TCGA-F5-6464-01A | BRAF | chr7 | 140624366 | - | TRIM24 | chr7 | 138258252 | + |
Top |
Fusion Gene ORF analysis for BRAF-TRIM24 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000288602 | ENST00000497516 | BRAF | chr7 | 140624366 | - | TRIM24 | chr7 | 138258252 | + |
| In-frame | ENST00000288602 | ENST00000343526 | BRAF | chr7 | 140624366 | - | TRIM24 | chr7 | 138258252 | + |
| In-frame | ENST00000288602 | ENST00000415680 | BRAF | chr7 | 140624366 | - | TRIM24 | chr7 | 138258252 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000288602 | BRAF | chr7 | 140624366 | - | ENST00000343526 | TRIM24 | chr7 | 138258252 | + | 6516 | 199 | 61 | 1473 | 470 |
| ENST00000288602 | BRAF | chr7 | 140624366 | - | ENST00000415680 | TRIM24 | chr7 | 138258252 | + | 2108 | 199 | 61 | 1473 | 470 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000288602 | ENST00000343526 | BRAF | chr7 | 140624366 | - | TRIM24 | chr7 | 138258252 | + | 0.00017269 | 0.9998273 |
| ENST00000288602 | ENST00000415680 | BRAF | chr7 | 140624366 | - | TRIM24 | chr7 | 138258252 | + | 0.000462664 | 0.99953735 |
Top |
Fusion Genomic Features for BRAF-TRIM24 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| BRAF | chr7 | 140624365 | - | TRIM24 | chr7 | 138258251 | + | 1.12E-09 | 1 |
| BRAF | chr7 | 140624365 | - | TRIM24 | chr7 | 138258251 | + | 1.12E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
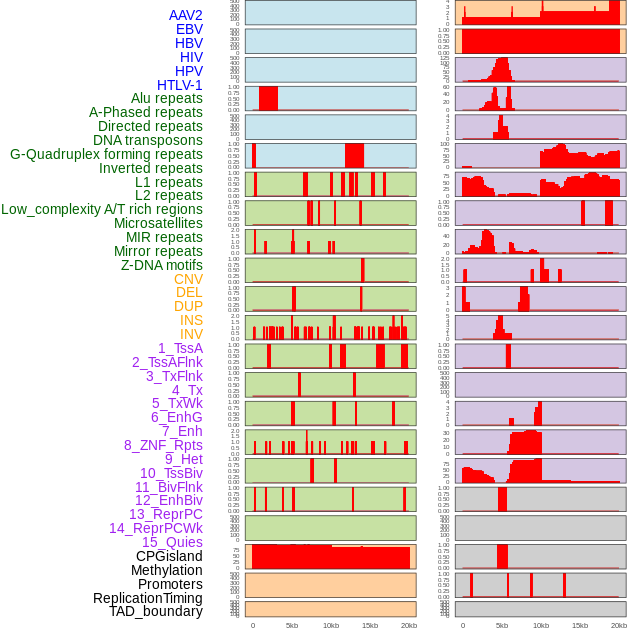 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
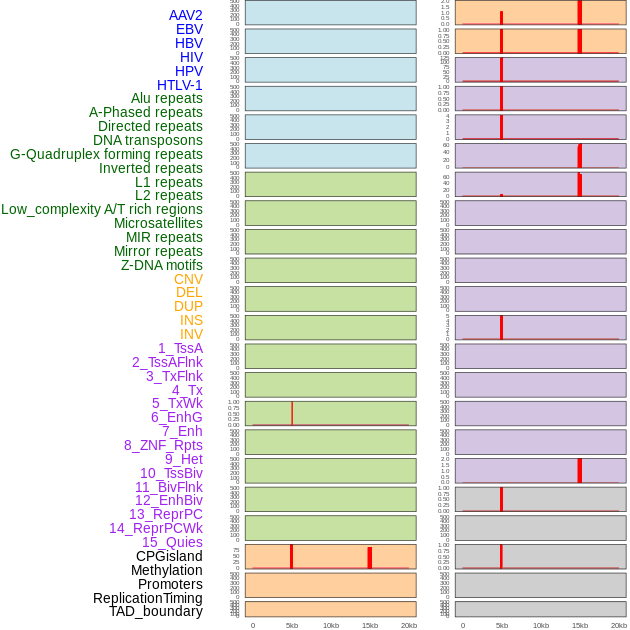 |
Top |
Fusion Protein Features for BRAF-TRIM24 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:140624366/chr7:138258252) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BRAF | TRIM24 |
| FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000305}. | FUNCTION: Transcriptional coactivator that interacts with numerous nuclear receptors and coactivators and modulates the transcription of target genes. Interacts with chromatin depending on histone H3 modifications, having the highest affinity for histone H3 that is both unmodified at 'Lys-4' (H3K4me0) and acetylated at 'Lys-23' (H3K23ac). Has E3 protein-ubiquitin ligase activity. Promotes ubiquitination and proteasomal degradation of p53/TP53. Plays a role in the regulation of cell proliferation and apoptosis, at least in part via its effects on p53/TP53 levels. Up-regulates ligand-dependent transcription activation by AR, GCR/NR3C1, thyroid hormone receptor (TR) and ESR1. Modulates transcription activation by retinoic acid (RA) receptors, including RARA. Plays a role in regulating retinoic acid-dependent proliferation of hepatocytes (By similarity). {ECO:0000250, ECO:0000269|PubMed:16322096, ECO:0000269|PubMed:19556538, ECO:0000269|PubMed:21164480}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 6_11 | 46 | 767.0 | Compositional bias | Note=Poly-Gly |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 932_987 | 626 | 1051.0 | Domain | Bromo | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 932_987 | 592 | 1017.0 | Domain | Bromo | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 891_907 | 626 | 1051.0 | Motif | Nuclear localization signal | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 891_907 | 592 | 1017.0 | Motif | Nuclear localization signal | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 754_779 | 626 | 1051.0 | Region | Note=Nuclear receptor binding site (NRBS) | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 754_779 | 592 | 1017.0 | Region | Note=Nuclear receptor binding site (NRBS) | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 826_873 | 626 | 1051.0 | Zinc finger | PHD-type | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 826_873 | 592 | 1017.0 | Zinc finger | PHD-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 122_129 | 46 | 767.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 428_432 | 46 | 767.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 155_227 | 46 | 767.0 | Domain | RBD |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 457_717 | 46 | 767.0 | Domain | Protein kinase |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 463_471 | 46 | 767.0 | Nucleotide binding | ATP |
| Hgene | BRAF | chr7:140624366 | chr7:138258252 | ENST00000288602 | - | 1 | 18 | 234_280 | 46 | 767.0 | Zinc finger | Phorbol-ester/DAG-type |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 289_359 | 626 | 1051.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 289_359 | 592 | 1017.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 344_347 | 626 | 1051.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 9_15 | 626 | 1051.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 344_347 | 592 | 1017.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 9_15 | 592 | 1017.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 158_211 | 626 | 1051.0 | Zinc finger | B box-type 1 | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 218_259 | 626 | 1051.0 | Zinc finger | B box-type 2 | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 56_82 | 626 | 1051.0 | Zinc finger | RING-type | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 158_211 | 592 | 1017.0 | Zinc finger | B box-type 1 | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 218_259 | 592 | 1017.0 | Zinc finger | B box-type 2 | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 56_82 | 592 | 1017.0 | Zinc finger | RING-type |
Top |
Fusion Gene Sequence for BRAF-TRIM24 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >10136_10136_1_BRAF-TRIM24_BRAF_chr7_140624366_ENST00000288602_TRIM24_chr7_138258252_ENST00000343526_length(transcript)=6516nt_BP=199nt CGCCTCCCTTCCCCCTCCCCGCCCGACAGCGGCCGCTCGGGCCCCGGCTCTCGGTTATAAGATGGCGGCGCTGAGCGGTGGCGGTGGTGG CGGCGCGGAGCCGGGCCAGGCTCTGTTCAACGGGGACATGGAGCCCGAGGCCGGCGCCGGCGCCGGCGCCGCGGCCTCTTCGGCTGCGGA CCCTGCCATTCCGGAGGAGATTGACTGTTCAAGTACTATTATGCTGGACAATATTGTGAGGAAAGATACTAATATAGATCATGGCCAGCC AAGACCACCCTCAAACAGAACGGTCCAGTCACCAAATTCATCAGTGCCATCTCCAGGCCTTGCAGGACCTGTTACTATGACTAGTGTACA CCCCCCAATACGTTCACCTAGTGCCTCCAGCGTTGGAAGCCGAGGAAGCTCTGGCTCTTCCAGCAAACCAGCAGGAGCTGACTCTACACA CAAAGTCCCAGTGGTCATGCTGGAGCCAATTCGAATAAAACAAGAAAACAGTGGACCACCGGAAAATTATGATTTCCCTGTTGTTATAGT GAAGCAAGAATCAGATGAAGAATCTAGGCCTCAAAATGCCAATTATCCAAGAAGCATACTCACCTCCCTGCTCTTAAATAGCAGTCAGAG CTCTACTTCTGAGGAGACTGTGCTAAGATCAGATGCCCCTGATAGTACAGGAGATCAACCTGGACTTCACCAGGACAATTCCTCAAATGG AAAGTCTGAATGGTTGGATCCTTCCCAGAAGTCACCTCTTCATGTTGGAGAGACAAGGAAAGAGGATGACCCCAATGAGGACTGGTGTGC AGTTTGTCAAAACGGAGGGGAACTCCTCTGCTGTGAAAAGTGCCCCAAAGTATTCCATCTTTCTTGTCATGTGCCCACATTGACAAATTT TCCAAGTGGAGAGTGGATTTGCACTTTCTGCCGAGACTTATCTAAACCAGAAGTTGAATATGATTGTGATGCTCCCAGTCACAACTCAGA AAAAAAGAAAACTGAAGGCCTTGTTAAGTTAACACCTATAGATAAAAGGAAGTGTGAGCGCCTACTTTTATTTCTTTACTGCCATGAAAT GAGCCTGGCTTTTCAAGACCCTGTTCCTCTAACTGTGCCTGATTATTACAAAATAATTAAAAATCCAATGGATTTGTCAACCATCAAGAA AAGACTACAAGAAGATTATTCCATGTACTCAAAACCTGAAGATTTTGTAGCTGATTTTAGATTGATCTTTCAAAACTGTGCTGAATTCAA TGAGCCTGATTCAGAAGTAGCCAATGCTGGTATAAAACTTGAAAATTATTTTGAAGAACTTCTAAAGAACCTCTATCCAGAAAAAAGGTT TCCCAAACCAGAATTCAGGAATGAATCAGAAGATAATAAATTTAGTGATGATTCAGATGATGACTTTGTACAGCCCCGGAAGAAACGCCT CAAAAGCATTGAAGAACGCCAGTTGCTTAAATAATATGCAGCACCACTAGCTTGTGCTGGTTTTTAGATTTTTTTGTTTTCAAAAAAACA TTTGTCAGTAATTTAACATCACTACAAAAAGAAGAGTTTGTGACTATTCTCATCTCTGTTTTGGACGTTTACTAGACTTTGATTTCCTTA ATAGCCCATTTCTGTTAACCTCTTATCACTAAGAAAGAAAGGAAAGAAGGAGATGAATAGAAGAAAGAAAATGGAAAGAAGGAAAAAAGG AGGATAGAAAAAGGATGGAAGAAAGAAGCATTGAAAACAAAGACATTCTTCCCACTTCTTGGATTTTTAAACCACAGTCTGGAGTGATAG CTACTGTAGAAAGGAAATAGACTTTGTATGAACTCTTTAAGTTGAAAAGTAAAAAATATATGTGGTTTGGATGTGTGCTTTAATTCAGCT TTAGAAATTAATACCACTACCCGTGAATTATATGGCCTGACAATATGAATTAGGTGTACTGTACTGAAGAACAGTACTCCACAAACATGG GTGGTAACAAGAGTTCCATCCCAGGAGGCCAAACGGTGCAACAGAAGGGTAGGTTAGATGCTATTAAGAAGGCACTTAATAGTACATCAT GTAAGATGGCAACTGTATTAAAGAAAAATCCGGAAAACAAATGTTTGATTTTTGTTTTTGTTTTTATCTTGTCTGTAGAGGTATTTTGGT ATAGCAGGTTTTCAAGGCCATTTTTTATACATTTCTAGATCTAGATTTTCAACTTCTTCCACTGAGGGAAGTATATACATTTGGGTTTGC TGTGTGTCTATGTGAGGTTTAATTGTACAGGTGATCCTTTTACAACAAGCCTCATTGTTTGCAGTATAGCTTTTAGTGGAACTACCCAAA ATATAAATACAGCAGCGTGCACTGTATTTGATGTGAGGGTTCTTCATCATATACCCTACTGGGCATTAAATATAAGTTCCTCTGAAAGGG ACTCGTTTTTGTGGTTTTCATCTGTCTATAATTTGGAATGAAAATGTGTTGTAGGATTTTGGGAGCAGGCAGCTGGGGGGAATTAATAGT GATTTTTTTTTTTTCCTGAAGCATCTATCTCATGTTTTTCTTTTGAGAGTCAGAACATCAAACTTAATCTTTGATCTGACTTCTGATTTT ATTCTTCTGATTGATTGATAGAGGTACAAAAGACTTATCTTCTGAGGACAAGCATATTCTTAATGTGCCAGACCTACCCGGTTCAGCTGA TATAGATAGATAGATAGATAGAAGAAAATTGCTGTGCCATACATTAATCCAGCATTTGACACAATATCTAAATGGTTTGCCGAAGTTAAT CTGTATTTATAAAACATTAACTGGAGTAAATTTTTCTCCTTAGGATGATAGAATAAAAAGAGCTCACTTGAAAGAAGGCTATTATTTGCA TTATATCACCTGCCATAAATTTAACATAGAAGACTTGGTTTTATCTACATGGCTTTACTTCTTAAAGTGGAAAATATCCTGTGGCTAATT CATTGGGTTTTCAGGTACTGCTAAAGATAAAATACAGGGAGAAAATAACTTGTTAGCAATAGATCCCCATTGTTTATATATATAGGTCTT GTTCATAATATGTCAATTATGTATTGTTAAAAAGTCCTACTCACTTTTCAAATATGTGTTACATGGTAATGTTTGTCATTGTTGTTTTAA AGTTGCATTTGACATTTGTTCTCCAAAGAGTGTTTGAACAGATTTTGATAACAGTGCATACACCTTTTGTCTTTTTTTGCTCCAGCATTT TATTTTTAGCACAGCAGCTGTGGAGCTTTTGCTGATAATTTTATTGTTGGTATCTTAGAATACTGTTCGTCTGACAGTTTATTCTTCCAT ACTTGTCTCCCAAGTTAAAACAGAGCAAAACAGATAAATTATTTTGAGAATCATCACAAAAGGGAGTTAGAACTTGTGAGACTAAAACTA TAATTTCCTAAATGTGACTAATGAACCCCAAACTGTCACTATTACATTCAGTGCCATATTTATCTTTCAAAACAGTATTTGTTTCACCCT AACCATCTTTGGCTGAATGAAAGGCAGTTGAGAACCATCTTTTGGTCATACATAATATAATTATATTAAAAGTAAAGATCGGGGCTGGGC GCAGTGGCTCACACCTGCAATCCCACCAGCACTTTGGGAGGCCAAGGTGGGCGGATCACAAAGTCAAGAGTTCGAGATCAGCCTGGCCAA CATTGTGAAACCCCGCCTCTACAAAAAATACAAAAATTAGCCGGACATGGTGGCGTGCAACTGTAATCCCAACTACTCAGGAGGCTGAGA CAGGAGAATCACTTGAACGCAGGAGGCAGAAGTTGCAGTGAGCCAAGATCTCACCATTGCACTCCAGCCTGAGTGACAGAGACTCAGTCT CAAAAACAAAAAGTAAAGGTCTTGGGTTTCTCTCTGTAAATAGCCTTTGTGTAATGGTCTTTAGATTAATATCCTTTGACTTTATTAATA AGTACCTGTGTGCCCAGGTGGATCTCAGCTAAGTCATCCTTAAATTCTGTTCAAATAGAAATAAACCATAAAATGAGCCTAACTATAACC CACACTCACTTATCAAGCATTCCCCCAAAAGTTCAGTGCGATTCAGGTAACCTTGACCTAAACTTGTACACTCCGGTGTACATCTTTGAC CATAAACTCTGGGAGATTTCCCTCTGGATTTGGGATCAGGAGCCCAGTAGGGCCCCATTGCTGAATGTTTATCACACTTCTGGCTAGGGC TTATGGATGAGCAGACCACGTAGGAGCTAGGATTTACTCAGTTCCACCTCACTGACAGCTTTTCTAGGTGCCAAGCAACTGACTTTTCCT GTGTAAGGAAGCCCAAGTACTTGACCAGTGATATATATGGCCATGGCAAAAGTATAAATGGATGGATGTAGACCATATTCAACGCAGAAT ACATTTTAGATCGGTATCTGGCATTGTAATCATTTTGTCTTTTGAATTAACATGCCCTGCTTGTCTTTTCTGAATGGAAGGGTAGGAAAG TGGAAGGTACTAAAAAGGAAAATGTAGTTTTAGTTTCTTTTCTAAGCTAAAGAACTGGCTTTACTACTGCTATATTTTACTACCACATAG TAGTGTAGATGTAGTATAACTTGCTTAGGAATGCTTTCAAAAAGGTTACCTCCATAGTCAAAAAAAGGGGGAGTATACAGCTGATTGTTG CATTTATGAAACTGCATTTCAATATGCTATGCTGTTACTGTGCTTAGACACTTCCTTACGTTTGAGTTCCAAGGAGAGAGAGTGAGAAGA GGAACTCCTTTTGCTGAGCTCTGGTCCAAATTAATGTGAATCCCCAAAGTAAATTGCTGCATATGAACAACAGGTTTTAGGTTTAAATTG AAGATACATTCATTTTATATGTATTTGTTGTGAATGAGAGAAATTGAGTGATCTGATTGGCCATTAAAATTATGGTGAATGGGTCGGGCG CAGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGGTGGGTAGATACCTGAGGTCAGGAGTTTGAGACCAGCCTGACCAACAT GGTGAAACCCCATCTCTACTAAAAATACCAAAATCACCAGGGTGTGGTGGCCCATGCCTGTAATCCTAGCTACTCGGGAGGCTGAGGCAG GAGAATCGGCTTGAACCCAGGAGGCAGAGGTTGCAGTGAGCTGAGATCGCACCACTGTAGTCCAGCCTGGGTGTCAGAGCAAGACTCTGT CACAAAAAGAAAAATTATGGGGAATGGCCGGGCGCGGTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCCGAGGTGGGTGGATCACC TGAAGTCAGGAGCTCAAGACCAGCCTGGCCAACATGGTGAAACCCCACCTCAACAGAATATACAAAAAAATTAGCTGGGTGTGGTGGCGG GTGCCTGTAATCTCAGCTACTTTGGGATGCTGAGGCAGGAGAATAGCTTGAACCCAGGAGGTGGAGGTTGCAGTGAGTCGAGATTGCACC ATTGCACTCCAGCCTGGGCAACAAGAGCAAAACTCTGTCTCAAAAAAAAAAAAAAATGGTGACCATGCGTTCAACCATATAATTTCTTAC TGAAATTGAGCTCCTCCTTTTGAATAACCATGTACATATTTTTAAAAGTATTTTATAGCCCTATGTAAATTAATAGTTGATGTCTTTTTA TAAATGTTTATTGCATATTCATCTTGAATGTGAATTTACTGTTGTAATTCACATACTGGTGTCATGTTTCTGAGTGAGAATAATGCTAGT CAGTCAGATCACAGATTTTTCTCCATGAAGGTTTTACTTATTGAGAGGGACATCTGAGGCAGTAGTCTTCGTCTTTAATTTTTGGGGGCA GATTGGTGCTTTGGTTTTAATAAAAACTAACTTTGATGGATTTTTACTGTCAATATTTTACCACAAGGATTTTTTAATAGAAAAATTACT GAATGTTACTTCAGCAGAGTTCTATCTACTTTGAGTCTCCATTACTACTACTGTTGTTCCCCTAGCTGTAACATTGATCCATATAGGGGC AAGCTACTAGTCCATAAATAGCTGTAATAGGAAAACAGTTTTTTAAAATTCTAACACGTGTATTTCACCAGCCTTAAAAGATTTAAAAAA AAATCTGACAGGAAGGAAAGCGGGTGGTAAGTATCTTGGATTATTCTGCCTCATACCAGCCACTCCCAAAGTAGGAGGACCAAAGATGCC ATGAAGACTTCTAGTTGTCTGGAAGTTATTTGGGGAAGGGGGGAAAGACACAGAGTGGGGACCTTTGGAAGTCATTTTAAAAATTTAGCT ATGTTAATTCCACTTGTAAAGTTTGAAAGGTTTTGATTCTTGTAATTTCTGAGAATTACTATGTTTTAAATTTACTATTGGATATTTAAC AGATCTTTCATGGGGCCAACCTATTTGCTTCCTTAATTTTCAAATTAAGTCCTAATTTGAGTCCTAAAAAAATCTTTCCTTGTTAGTTGC >10136_10136_1_BRAF-TRIM24_BRAF_chr7_140624366_ENST00000288602_TRIM24_chr7_138258252_ENST00000343526_length(amino acids)=470AA_BP=46 MAALSGGGGGGAEPGQALFNGDMEPEAGAGAGAAASSAADPAIPEEIDCSSTIMLDNIVRKDTNIDHGQPRPPSNRTVQSPNSSVPSPGL AGPVTMTSVHPPIRSPSASSVGSRGSSGSSSKPAGADSTHKVPVVMLEPIRIKQENSGPPENYDFPVVIVKQESDEESRPQNANYPRSIL TSLLLNSSQSSTSEETVLRSDAPDSTGDQPGLHQDNSSNGKSEWLDPSQKSPLHVGETRKEDDPNEDWCAVCQNGGELLCCEKCPKVFHL SCHVPTLTNFPSGEWICTFCRDLSKPEVEYDCDAPSHNSEKKKTEGLVKLTPIDKRKCERLLLFLYCHEMSLAFQDPVPLTVPDYYKIIK NPMDLSTIKKRLQEDYSMYSKPEDFVADFRLIFQNCAEFNEPDSEVANAGIKLENYFEELLKNLYPEKRFPKPEFRNESEDNKFSDDSDD -------------------------------------------------------------- >10136_10136_2_BRAF-TRIM24_BRAF_chr7_140624366_ENST00000288602_TRIM24_chr7_138258252_ENST00000415680_length(transcript)=2108nt_BP=199nt CGCCTCCCTTCCCCCTCCCCGCCCGACAGCGGCCGCTCGGGCCCCGGCTCTCGGTTATAAGATGGCGGCGCTGAGCGGTGGCGGTGGTGG CGGCGCGGAGCCGGGCCAGGCTCTGTTCAACGGGGACATGGAGCCCGAGGCCGGCGCCGGCGCCGGCGCCGCGGCCTCTTCGGCTGCGGA CCCTGCCATTCCGGAGGAGATTGACTGTTCAAGTACTATTATGCTGGACAATATTGTGAGGAAAGATACTAATATAGATCATGGCCAGCC AAGACCACCCTCAAACAGAACGGTCCAGTCACCAAATTCATCAGTGCCATCTCCAGGCCTTGCAGGACCTGTTACTATGACTAGTGTACA CCCCCCAATACGTTCACCTAGTGCCTCCAGCGTTGGAAGCCGAGGAAGCTCTGGCTCTTCCAGCAAACCAGCAGGAGCTGACTCTACACA CAAAGTCCCAGTGGTCATGCTGGAGCCAATTCGAATAAAACAAGAAAACAGTGGACCACCGGAAAATTATGATTTCCCTGTTGTTATAGT GAAGCAAGAATCAGATGAAGAATCTAGGCCTCAAAATGCCAATTATCCAAGAAGCATACTCACCTCCCTGCTCTTAAATAGCAGTCAGAG CTCTACTTCTGAGGAGACTGTGCTAAGATCAGATGCCCCTGATAGTACAGGAGATCAACCTGGACTTCACCAGGACAATTCCTCAAATGG AAAGTCTGAATGGTTGGATCCTTCCCAGAAGTCACCTCTTCATGTTGGAGAGACAAGGAAAGAGGATGACCCCAATGAGGACTGGTGTGC AGTTTGTCAAAACGGAGGGGAACTCCTCTGCTGTGAAAAGTGCCCCAAAGTATTCCATCTTTCTTGTCATGTGCCCACATTGACAAATTT TCCAAGTGGAGAGTGGATTTGCACTTTCTGCCGAGACTTATCTAAACCAGAAGTTGAATATGATTGTGATGCTCCCAGTCACAACTCAGA AAAAAAGAAAACTGAAGGCCTTGTTAAGTTAACACCTATAGATAAAAGGAAGTGTGAGCGCCTACTTTTATTTCTTTACTGCCATGAAAT GAGCCTGGCTTTTCAAGACCCTGTTCCTCTAACTGTGCCTGATTATTACAAAATAATTAAAAATCCAATGGATTTGTCAACCATCAAGAA AAGACTACAAGAAGATTATTCCATGTACTCAAAACCTGAAGATTTTGTAGCTGATTTTAGATTGATCTTTCAAAACTGTGCTGAATTCAA TGAGCCTGATTCAGAAGTAGCCAATGCTGGTATAAAACTTGAAAATTATTTTGAAGAACTTCTAAAGAACCTCTATCCAGAAAAAAGGTT TCCCAAACCAGAATTCAGGAATGAATCAGAAGATAATAAATTTAGTGATGATTCAGATGATGACTTTGTACAGCCCCGGAAGAAACGCCT CAAAAGCATTGAAGAACGCCAGTTGCTTAAATAATATGCAGCACCACTAGCTTGTGCTGGTTTTTAGATTTTTTTGTTTTCAAAAAAACA TTTGTCAGTAATTTAACATCACTACAAAAAGAAGAGTTTGTGACTATTCTCATCTCTGTTTTGGACGTTTACTAGACTTTGATTTCCTTA ATAGCCCATTTCTGTTAACCTCTTATCACTAAGAAAGAAAGGAAAGAAGGAGATGAATAGAAGAAAGAAAATGGAAAGAAGGAAAAAAGG AGGATAGAAAAAGGATGGAAGAAAGAAGCATTGAAAACAAAGACATTCTTCCCACTTCTTGGATTTTTAAACCACAGTCTGGAGTGATAG CTACTGTAGAAAGGAAATAGACTTTGTATGAACTCTTTAAGTTGAAAAGTAAAAAATATATGTGGTTTGGATGTGTGCTTTAATTCAGCT TTAGAAATTAATACCACTACCCGTGAATTATATGGCCTGACAATATGAATTAGGTGTACTGTACTGAAGAACAGTACTCCACAAACATGG GTGGTAACAAGAGTTCCATCCCAGGAGGCCAAACGGTGCAACAGAAGGGTAGGTTAGATGCTATTAAGAAGGCACTTAATAGTACATCAT >10136_10136_2_BRAF-TRIM24_BRAF_chr7_140624366_ENST00000288602_TRIM24_chr7_138258252_ENST00000415680_length(amino acids)=470AA_BP=46 MAALSGGGGGGAEPGQALFNGDMEPEAGAGAGAAASSAADPAIPEEIDCSSTIMLDNIVRKDTNIDHGQPRPPSNRTVQSPNSSVPSPGL AGPVTMTSVHPPIRSPSASSVGSRGSSGSSSKPAGADSTHKVPVVMLEPIRIKQENSGPPENYDFPVVIVKQESDEESRPQNANYPRSIL TSLLLNSSQSSTSEETVLRSDAPDSTGDQPGLHQDNSSNGKSEWLDPSQKSPLHVGETRKEDDPNEDWCAVCQNGGELLCCEKCPKVFHL SCHVPTLTNFPSGEWICTFCRDLSKPEVEYDCDAPSHNSEKKKTEGLVKLTPIDKRKCERLLLFLYCHEMSLAFQDPVPLTVPDYYKIIK NPMDLSTIKKRLQEDYSMYSKPEDFVADFRLIFQNCAEFNEPDSEVANAGIKLENYFEELLKNLYPEKRFPKPEFRNESEDNKFSDDSDD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BRAF-TRIM24 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000343526 | 10 | 19 | 834_840 | 626.0 | 1051.0 | histone H3 that is not methylated at 'Lys-4' (H3K4me0) | |
| Tgene | TRIM24 | chr7:140624366 | chr7:138258252 | ENST00000415680 | 10 | 19 | 834_840 | 592.0 | 1017.0 | histone H3 that is not methylated at 'Lys-4' (H3K4me0) |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BRAF-TRIM24 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BRAF-TRIM24 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
