
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF136-NFIC (FusionGDB2 ID:101484) |
Fusion Gene Summary for ZNF136-NFIC |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF136-NFIC | Fusion gene ID: 101484 | Hgene | Tgene | Gene symbol | ZNF136 | NFIC | Gene ID | 7695 | 4782 |
| Gene name | zinc finger protein 136 | nuclear factor I C | |
| Synonyms | pHZ-20 | CTF|CTF5|NF-I|NFI | |
| Cytomap | 19p13.2 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 136 | nuclear factor 1 C-typeCCAAT-box-binding transcription factorNF-I/CNF1-CTGGCA-binding proteinnuclear factor I/C (CCAAT-binding transcription factor) | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | P08651 | |
| Ensembl transtripts involved in fusion gene | ENST00000343979, ENST00000398616, | ENST00000588839, ENST00000341919, ENST00000346156, ENST00000395111, ENST00000443272, ENST00000586919, ENST00000589123, ENST00000590282, | |
| Fusion gene scores | * DoF score | 8 X 2 X 6=96 | 24 X 21 X 9=4536 |
| # samples | 10 | 27 | |
| ** MAII score | log2(10/96*10)=0.0588936890535686 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(27/4536*10)=-4.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF136 [Title/Abstract] AND NFIC [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF136(12274021)-NFIC(3381709), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NFIC | GO:0000122 | negative regulation of transcription by RNA polymerase II | 19706729 |
| Tgene | NFIC | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1524678|19706729 |
 Fusion gene breakpoints across ZNF136 (5'-gene) Fusion gene breakpoints across ZNF136 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
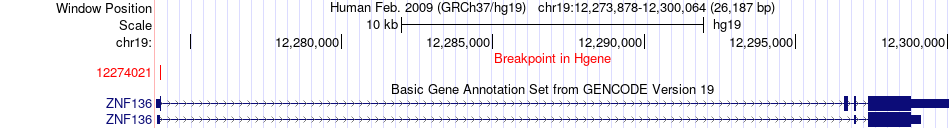 |
 Fusion gene breakpoints across NFIC (3'-gene) Fusion gene breakpoints across NFIC (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
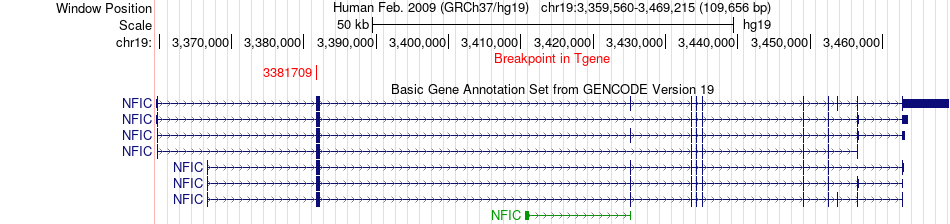 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-HT-7604 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
Top |
Fusion Gene ORF analysis for ZNF136-NFIC |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000343979 | ENST00000588839 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000341919 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000346156 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000395111 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000443272 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000586919 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000589123 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-3CDS | ENST00000398616 | ENST00000590282 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| 5UTR-intron | ENST00000398616 | ENST00000588839 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000341919 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000346156 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000395111 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000443272 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000586919 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000589123 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
| In-frame | ENST00000343979 | ENST00000590282 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000589123 | NFIC | chr19 | 3381709 | + | 8088 | 143 | 140 | 1639 | 499 |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000346156 | NFIC | chr19 | 3381709 | + | 2250 | 143 | 140 | 1360 | 406 |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000395111 | NFIC | chr19 | 3381709 | + | 1859 | 143 | 140 | 1432 | 430 |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000586919 | NFIC | chr19 | 3381709 | + | 1361 | 143 | 140 | 1360 | 407 |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000341919 | NFIC | chr19 | 3381709 | + | 1598 | 143 | 140 | 1399 | 419 |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000590282 | NFIC | chr19 | 3381709 | + | 1617 | 143 | 140 | 1432 | 430 |
| ENST00000343979 | ZNF136 | chr19 | 12274021 | + | ENST00000443272 | NFIC | chr19 | 3381709 | + | 1778 | 143 | 140 | 1639 | 499 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000343979 | ENST00000589123 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.005044813 | 0.9949551 |
| ENST00000343979 | ENST00000346156 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.025534028 | 0.9744659 |
| ENST00000343979 | ENST00000395111 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.027277797 | 0.9727222 |
| ENST00000343979 | ENST00000586919 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.033361603 | 0.9666384 |
| ENST00000343979 | ENST00000341919 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.027274553 | 0.97272545 |
| ENST00000343979 | ENST00000590282 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.028399237 | 0.97160083 |
| ENST00000343979 | ENST00000443272 | ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 0.032238852 | 0.96776116 |
Top |
Fusion Genomic Features for ZNF136-NFIC |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 1.65E-16 | 1 |
| ZNF136 | chr19 | 12274021 | + | NFIC | chr19 | 3381709 | + | 1.65E-16 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
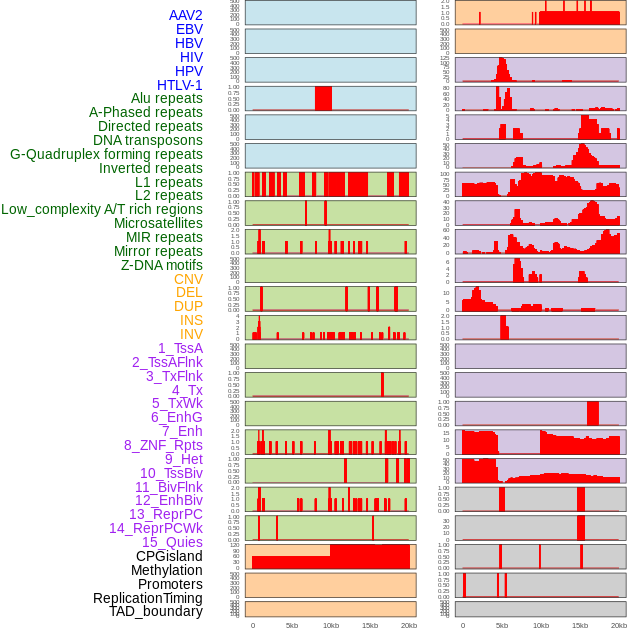 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
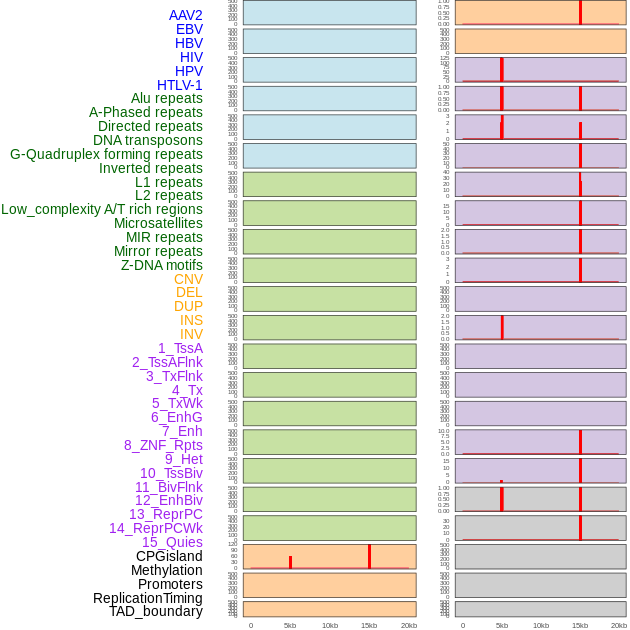 |
Top |
Fusion Protein Features for ZNF136-NFIC |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:12274021/chr19:3381709) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NFIC |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Recognizes and binds the palindromic sequence 5'-TTGGCNNNNNGCCAA-3' present in viral and cellular promoters and in the origin of replication of adenovirus type 2. These proteins are individually capable of activating transcription and replication. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000346156 | 0 | 9 | 1_195 | 1 | 669.0 | DNA binding | CTF/NF-I | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000395111 | 0 | 10 | 1_195 | 1 | 548.3333333333334 | DNA binding | CTF/NF-I | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000586919 | 0 | 8 | 1_195 | 1 | 407.0 | DNA binding | CTF/NF-I | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000589123 | 0 | 11 | 1_195 | 1 | 500.0 | DNA binding | CTF/NF-I | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000341919 | 0 | 9 | 404_412 | 10 | 429.0 | Motif | 9aaTAD | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000346156 | 0 | 9 | 404_412 | 1 | 669.0 | Motif | 9aaTAD | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000395111 | 0 | 10 | 404_412 | 1 | 548.3333333333334 | Motif | 9aaTAD | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000443272 | 0 | 11 | 404_412 | 10 | 509.0 | Motif | 9aaTAD | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000586919 | 0 | 8 | 404_412 | 1 | 407.0 | Motif | 9aaTAD | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000589123 | 0 | 11 | 404_412 | 1 | 500.0 | Motif | 9aaTAD | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000590282 | 0 | 10 | 404_412 | 10 | 461.6666666666667 | Motif | 9aaTAD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 4_90 | 1 | 541.0 | Domain | KRAB |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 140_162 | 1 | 541.0 | Zinc finger | C2H2-type 1%3B degenerate |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 168_190 | 1 | 541.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 196_218 | 1 | 541.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 224_246 | 1 | 541.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 252_274 | 1 | 541.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 280_302 | 1 | 541.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 308_330 | 1 | 541.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 336_358 | 1 | 541.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 364_386 | 1 | 541.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 392_414 | 1 | 541.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 420_442 | 1 | 541.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 448_470 | 1 | 541.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 476_498 | 1 | 541.0 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF136 | chr19:12274021 | chr19:3381709 | ENST00000343979 | + | 1 | 4 | 504_526 | 1 | 541.0 | Zinc finger | C2H2-type 14 |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000341919 | 0 | 9 | 1_195 | 10 | 429.0 | DNA binding | CTF/NF-I | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000443272 | 0 | 11 | 1_195 | 10 | 509.0 | DNA binding | CTF/NF-I | |
| Tgene | NFIC | chr19:12274021 | chr19:3381709 | ENST00000590282 | 0 | 10 | 1_195 | 10 | 461.6666666666667 | DNA binding | CTF/NF-I |
Top |
Fusion Gene Sequence for ZNF136-NFIC |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >101484_101484_1_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000341919_length(transcript)=1598nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACC AGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCC AAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAG CCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACA CGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGGCATCTCGT CCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCTCCAGCTTCACCCAGCACC ACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTACGACGTCCATCCTACCCC AGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCAAAGATCTTGTCTCGCTGG CCTGCGACCCAGCCAGCCAGCAACCTGGACCGTCCTGGTATCTGGGATAGCAAAGGTCTTCTTCCCTCGCCCCTTCTCCATCGTCCCAGG AATCCCAGGGGGCAGCACAGCCGGCCCCCGGCCCACGTTTTCGGTGGAAAATTAGAGTGAACAAGAACACCCCTGCCGACTCCCAGCCCG >101484_101484_1_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000341919_length(amino acids)=419AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDA EQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGS KRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHP -------------------------------------------------------------- >101484_101484_2_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000346156_length(transcript)=2250nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCA GCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGG CATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGA TGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACA TGGAAGGAGGCATCTCGTCCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCT CCAGCTTCACCCAGCACCACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTA CGACGTCCATCCTACCCCAGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCA AAGATCTTGTCTCGCTGGCCTGCGACCCAGCCAGCCAGCAACCTGGACCGCCTACTCTCCGCCCGACACGTCCCCTGCAAACCGTTCCTT TGTGGGATTAGGACCAAGGGATCCTGCGGGCATTTATCAGGCACAGTCCTGGTATCTGGGATAGCAAAGGTCTTCTTCCCTCGCCCCTTC TCCATCGTCCCAGGAATCCCAGGGGGCAGCACAGCCGGCCCCCGGCCCACGTTTTCGGTGGAAAATTAGAGTGAACAAGAACACCCCTGC CGACTCCCAGCCCGGCCAAAAAGACAAAACACATAGACGCACACACTCAGGAGGAAAAGAAAAAACAAAGGCAGAAGAAGAAGAAGAAGA AATAAAAACCCACCCAAGCAAGAAGACAAAAGGTAAAGACGCAACGTTTCCAACTCTCGGGACGCCAAGGCCGCAGGACTGGAGGGCCAG GCCCCGCCACCCCCACGGGAGACCCGGGACAGGGCGTCTTCCTAAGTTATTCATCTCCTCTCCGCCTGCTGCTCGGGAAGGACAGACGCC GGCCGCCCGCCCGCGCCCCGGAGGCCCTGGCTCTGTCCGGAGACCAGGTGAGCACAGCCTGGAGCCTGTGCCCAGGGCCGACAGGCGCGA CACCCAGCAAGGCCACCTCTCCCCGGGCCCCCGCGCCTCTGCCGGACACGGACCGGCCCCTCAGCCCCCACCGAGGACGCAGCCACTGGG GGGAAAGGGAGACACAGCGGACCCCGGCCGGGCAGCGGAGACCGCAGAGGCGGGCAGGGTGGGGCAGGCGAGTGGTGTCGCGGGGGTGCG TGGCGCTTGCGAGCCCTGGCCAGGGGAGGAAGTGAGGCCCAGGCACCTGCTGCCCCTCGAGGGGGCCCTGCCTGCCGCGGGGCCTCCCCA CAAGCCCCTCCCAAAGCGCCGGCCGACTCGCTGTCTCGCTGGGGACTCTTTCAGCCCTCGCGCCCGCCCGTTTGGGAGGAGAAGTCTCTA >101484_101484_2_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000346156_length(amino acids)=406AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDT TDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSP SSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHPSSALHFPTTSILPQTASTYFPHTA -------------------------------------------------------------- >101484_101484_3_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000395111_length(transcript)=1859nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACC AGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCC AAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAG CCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACA CGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGGCATCTCGT CCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCTCCAGCTTCACCCAGCACC ACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTACGACGTCCATCCTACCCC AGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCAAAGATCTTGTCTCGCTGG CCTGCGACCCAGCCAGCCAGCAACCTGGACCGCCTACTCTCCGCCCGACACGTCCCCTGCAAACCGTTCCTTTGTGGGATTAGGACCAAG GGATCCTGCGGGCATTTATCAGGCACAGTCCTGGTATCTGGGATAGCAAAGGTCTTCTTCCCTCGCCCCTTCTCCATCGTCCCAGGAATC CCAGGGGGCAGCACAGCCGGCCCCCGGCCCACGTTTTCGGTGGAAAATTAGAGTGAACAAGAACACCCCTGCCGACTCCCAGCCCGGCCA AAAAGACAAAACACATAGACGCACACACTCAGGAGGAAAAGAAAAAACAAAGGCAGAAGAAGAAGAAGAAGAAATAAAAACCCACCCAAG CAAGAAGACAAAAGGTAAAGACGCAACGTTTCCAACTCTCGGGACGCCAAGGCCGCAGGACTGGAGGGCCAGGCCCCGCCACCCCCACGG >101484_101484_3_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000395111_length(amino acids)=430AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDA EQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGS KRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHP -------------------------------------------------------------- >101484_101484_4_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000443272_length(transcript)=1778nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACC AGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCC AAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAG CCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACA CGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGGCATCTCGT CCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCTCCAGCTTCACCCAGCACC ACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTACGACGTCCATCCTACCCC AGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCAAAGATCTTGTCTCGCTGG CCTGCGACCCAGCCAGCCAGCAACCTGGACCGTTAAATGGAAGTGGTCAGCTCAAAATGCCCAGCCACTGCCTTTCTGCTCAGATGCTGG CACCTCCGCCCCCGGGGCTGCCACGGCTGGCGCTCCCCCCTGCCACCAAACCCGCCACCACCTCCGAGGGAGGAGCCACGTCGCCGACCT CGCCTTCCTACTCTCCGCCCGACACGTCCCCTGCAAACCGTTCCTTTGTGGGATTAGGACCAAGGGATCCTGCGGGCATTTATCAGGCAC AGTCCTGGTATCTGGGATAGCAAAGGTCTTCTTCCCTCGCCCCTTCTCCATCGTCCCAGGAATCCCAGGGGGCAGCACAGCCGGCCCCCG >101484_101484_4_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000443272_length(amino acids)=499AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDA EQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGS KRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHP SSALHFPTTSILPQTASTYFPHTAIRYPPHLNPQDPLKDLVSLACDPASQQPGPLNGSGQLKMPSHCLSAQMLAPPPPGLPRLALPPATK -------------------------------------------------------------- >101484_101484_5_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000586919_length(transcript)=1361nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCA GCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGG CATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGA TGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACA TGGAAGGAGGCATCTCGTCCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCT CCAGCTTCACCCAGCACCACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTA CGACGTCCATCCTACCCCAGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCA AAGATCTTGTCTCGCTGGCCTGCGACCCAGCCAGCCAGCAACCTGGACCGCCTACTCTCCGCCCGACACGTCCCCTGCAAACCGTTCCTT >101484_101484_5_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000586919_length(amino acids)=407AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDT TDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSP SSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHPSSALHFPTTSILPQTASTYFPHTA -------------------------------------------------------------- >101484_101484_6_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000589123_length(transcript)=8088nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACC AGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCC AAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAG CCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACA CGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGGCATCTCGT CCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCTCCAGCTTCACCCAGCACC ACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTACGACGTCCATCCTACCCC AGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCAAAGATCTTGTCTCGCTGG CCTGCGACCCAGCCAGCCAGCAACCTGGACCGTTAAATGGAAGTGGTCAGCTCAAAATGCCCAGCCACTGCCTTTCTGCTCAGATGCTGG CACCTCCGCCCCCGGGGCTGCCACGGCTGGCGCTCCCCCCTGCCACCAAACCCGCCACCACCTCCGAGGGAGGAGCCACGTCGCCGACCT CGCCTTCCTACTCTCCGCCCGACACGTCCCCTGCAAACCGTTCCTTTGTGGGATTAGGACCAAGGGATCCTGCGGGCATTTATCAGGCAC AGTCCTGGTATCTGGGATAGCAAAGGTCTTCTTCCCTCGCCCCTTCTCCATCGTCCCAGGAATCCCAGGGGGCAGCACAGCCGGCCCCCG GCCCACGTTTTCGGTGGAAAATTAGAGTGAACAAGAACACCCCTGCCGACTCCCAGCCCGGCCAAAAAGACAAAACACATAGACGCACAC ACTCAGGAGGAAAAGAAAAAACAAAGGCAGAAGAAGAAGAAGAAGAAATAAAAACCCACCCAAGCAAGAAGACAAAAGGTAAAGACGCAA CGTTTCCAACTCTCGGGACGCCAAGGCCGCAGGACTGGAGGGCCAGGCCCCGCCACCCCCACGGGAGACCCGGGACAGGGCGTCTTCCTA AGTTATTCATCTCCTCTCCGCCTGCTGCTCGGGAAGGACAGACGCCGGCCGCCCGCCCGCGCCCCGGAGGCCCTGGCTCTGTCCGGAGAC CAGGTGAGCACAGCCTGGAGCCTGTGCCCAGGGCCGACAGGCGCGACACCCAGCAAGGCCACCTCTCCCCGGGCCCCCGCGCCTCTGCCG GACACGGACCGGCCCCTCAGCCCCCACCGAGGACGCAGCCACTGGGGGGAAAGGGAGACACAGCGGACCCCGGCCGGGCAGCGGAGACCG CAGAGGCGGGCAGGGTGGGGCAGGCGAGTGGTGTCGCGGGGGTGCGTGGCGCTTGCGAGCCCTGGCCAGGGGAGGAAGTGAGGCCCAGGC ACCTGCTGCCCCTCGAGGGGGCCCTGCCTGCCGCGGGGCCTCCCCACAAGCCCCTCCCAAAGCGCCGGCCGACTCGCTGTCTCGCTGGGG ACTCTTTCAGCCCTCGCGCCCGCCCGTTTGGGAGGAGAAGTCTCTATGCAATTGGCCCCGGCCCCTCCACCCCCCACCCCCGGCATAGGA GGCCCCCCCACCTCGCCCGGCTCACACCCCCAAAGGGAGGGACCCACATTGCACACACTGTAAGAAATGCACTTTCCGAGGAAGGGGATG GGGGAGCCCGGACACCCAGAGCTCCCCGAGTTGGGGGTGCCCGTCTGGAGCGCCCCCGTCAGCCCCTGGCGGTGGGAGGTGAGAGCGAGT GGTTTAAGTGCCTGATTACCACCACCCGCCCCCCCCTTTGTCCAGCTGGGACACGGAATGGCCGCGGGCCTCCTCCCCCTCCCCTCCAGC CTCTCCACCAGCCCCTCCAGTCAACCCTCATCGCCGTGCCCCCCCAGAGCTAGAGAGATGGGGCCCCTGCGTGGCCCGAGGGGCAGAGCT GGGCGTCACTTCGCAAGCGTCCTGCCCTGCCGGGGCGCGGGGGTGGGCTCTGGGGAAGCCGGTGCGCCCCCCACGCCTCCGCTGCCAGTG CCTTACATTCTGGAGCGACCCCCCTCCCTGGTGCCTCCCAGCGAAGGGGGACCGCCGTTTGCACTTTCATCGCCTACCCCGACGCGGGGC CCAGCTGCGGGACGTGCATCACGGCTGGGCCCCCAGAGGAGAGAGGAGGCCGACGCCAGCGGTCCCCGCTCGGAACGGGGAGGGTTTTCG GGGGGTTCGGCGTCGCACCTTGGGGCCCCCCGCAGCCGTGTAGGGGGCCTCCCATCTGCTAAGCGTTTTTCCGTTGAGCCGCTCCAAAAA CACTAAGCTGGGGACGCCAGGTGCCCCCCCACCCCGGCTCCCTGGCCCTATCCACACCTCCACCCCCACCCCAGGATCGCCATCTTTAGG GGAGGCCTGGGAGGGGGTGTTAGGTGTTTTAGGGCCACCGAGCTCAAACACAAGGACCCCTCCCCGGCCCACCCAGCCCAGCCCCAACTG ACCTCCATGCCTAGGGAAAAACTCCCCCCACCACTGCCCCCTCCCCCGACCCAGGCCAAAGCCAGGGCAGGTCTCCGGGTCTCACCTGCT CCTAGCCTCACCCCCCTGCCCCCGAAAACCAGACTCTCCTCCCAAACTAGCCTCAGGAGCTTGGCGAACCCGCTCGCTCCTAAAGAGAAA GACCCAGGACCCTCCCCCATCACCCCCAAGAGAGGTTCGCCATCCTCTGGCCTCGAGCCCTTGGTCCCTCCGTCCGTCTGTCCTCGGGGC CCGCTCCCCCGGTGGCCCTTGGGGATCAAAGCGTGGGCCGCTCTCCGGGAGGGCGGGCGGGGGAGGGGGTGGTCGGGTTGTGCCATTGGG GTGTCCGGAAGCTTCTCAGCCAGGGTGGGGGTCGTGGAGTGGGGGAGGGAGGCCAGCCGGGCTCCAGAGGGGTCAGGGCGCGACGAGAAC CAACTCTTTACCTAACTTTGCATGGTGCTTAGTCAAGGACTCCTGCGACCTGGCTCCCGAGGTCAGCTGGCGGCGCTGACACACATGCAT GGCAGACTATCCCTGGCTCTATCTCCCTGTTCCTCGCCCCCTCCACCCCCCACTTCCTCTTTAAAAAAAAAAAAAAAAAAAAAAAGATAC AAGAAAAACCTTTAAAAAAATTCCATGTTTCCTAATTTGCACGAAATTTTCTACCACAAGATGTGCCTTGCCTTCCGAGAATAAGTATTA CCTTTAAACAATATCAGCGCACACACATAGCTGCATGTTCTGCTCGTGTAGTTTAAAAAAAAAAAGACAAAACAGTGACATGAAATAAAA AATAAAAATTGAAAAGGGATGTATTTCTATTTGTAAAAAAAATAAAATAAAAAATAAGAAAGTGAGAATCTAAAAAAAAAAAAAAAAAAA AAAAAGGAAGAAAAACCACGCTAAAAATCAAGCCACTGAAAACAATTGCCCCCAGGTCTACCCAGCCCCTGGCTGTCCTTGGTCCTGTCT CCCCTCCTGCTGTATTCAGGGGTGCCCCCTGGTGCTCAGCCTCTACCACCCCCAACCCTGCTCTTGGGTACCCAGAGGGGTCATTTCTGA ATCCCTTGCCCAGAGGACAGACCTCCGGGGCCCATCTTGGCCCTGGGAAAGGGCTCTCCTCTCTGATTGGTCCCTAGGCCACGGGCCGGC CCCCAGACACCATTCACCGACCCACTGCAGGCTGTCCTCCAACCATGGGGTGGCCACTCCACCCGCAGCCAGACTCCCCGCTCCCCACTT TTCATGCAGGCTGGCATACCCCTGGCTCAGGGTCAAATGCTGTTCCACACCCACCTCAGAGGCACCCCCTCTCCCCTGCCCCGTGCATCC CCACCCTTCTTGCCAAAGGACCTCTTTTCCCCTATCCAGAGACCACCCCAGGTGGCATTCTCTCCCACCTTCTCCTTTGTCCCCCATCCC CTGTCTCTGTCTTCCAGCTGTGAATATGAAGGGTATCCTGTATGAAACAAAAACAAAACCTGATATATGCAATATCTGTCTGTCTGTCTG TACCCATGGGCCTGGCTCAGCCATTGGAGGCCCAGCCGAGGGTCCGGCAGGGCACAGGGACAGCCAGGTGGCACCGAGTCACAGGCTGTG GTCCGGTGGCTGAGCATGCTGTTGTCTTGTCCTTGATTTTATTTTCTTTTGTTCTTTTTTTTTTTCTTTTCTTTTTGTTTTTAACTCCAG CTTCCTTTGCTTTTTACTTGACCAAAGCTAAGACAATAGCCAGATGGTTAGTGGGGCAGCCAGGCAGGGAGGACCCAGGGCTGGGATTCT CCAACCTTAGGCCATTCCTGCAGCCCTCACCACCTCCAGCCCCTCCAAGCATCTCGTGTAGGGACCCACGCAGATGGTCCCATTCATTCA CTATTGCCCCCAACCCCGGGATTTTGGGTGGTCTCCACAGCCACCATCATACACTCATCCCGTGTTTTCTTCCAAAAAGTCACCTCAGCA GCCTCCCCAGGCGATACAGAGGGAGAGCCCAGACCACCACAGCTGGCCACGACATTGCCCTTAAGTAATATGCATTGGCCAGAGAGCCCG GGCTGGCTGTGCACAGCATTCATGTAGCTGATTTCTAGCTTTTTTTTTTTTTCTGCCCCACTCCTGAGCAAATCTGTCTTGCCAAGGAAC TAGGAGCAACCGGAGGCAAAGGGAGTGGGTGGCCCCATCACTATTGGGACCATCGCGTCCCTGCACAGCCCACACCCGGGGGCCCAGAGT CCTGGGCTGGACGCCACCCTTCTCACCCCGAGCTTGCCTCCTTGGCTCACTTGGCACCTTGGCTGAGTACAGCAGGCAAAAGCCCATACC AGGCAGCATGTTGTGGATGGTTTAGTTCTCCCCGCCTCCCTGTTTCTTGGAAAAGCTACAGGGTCCCTGTAGGGCAAAATTCCCAGGCGC CTTGCTGCAGACAGAGTAAGACAAAAACACCAGGAAGCAGGATTCCGTGCCCATCTCTGCAGTTTGGGTTCACAAAAGGGGGTGCCGTCA TCCCTGGGTGGAGGAGGGAGTGTTGGTTTTTTGTTTTTGTTTTTTTAACATGTATGAAACTGACATCTTCTCAAATCTTGTTCCACCCCC CTCTGGAAGCCCCCATCACCCACCCCTGCTATGGACACCACACCTATGCCAGGCCCCCCCCCCCACCCCAGTCTCATTCTGGGGTCTGCC CATGCTGTGGGAAAGAATAGGGAGGCCTCCCAAATATATGCAAATTGTCCCCATTCCGTGGGGGCACCTGACAATGACCCGGGTGGAGAT GGGGCATGGAGGAGTAGGAAGACCCAGCCCTATTTGACTGGGGAGAGGAGGATCTGGAGTCCTTCATGCCCAGGTCTGGAACCCAGGTTC TGACCCCAGGGCCCCACCCTGGGCTGGACAATCAGATCCCAAAGGAATGCCAAAGGGGACTCGGTTGGGAGAGCCGCTTAGGGGCCAGAC CTGGGTCCCCCTGCAGGTCCCCAGGCAGCAGACAATTCCACCTTCCCTGCCCCAGGACCTTGAGAGACAGCAGCATTCCAGGCACAGACA GACTTGGCTGCACCCCACTGTCCCTTGCAAGACAGGTTCTGGAGCCAGGAGCAACTGTCCAGCCCTCCAGAAGAGACAGCAAGCAGCCCC CCTACCCACTCTGGCCTCCCCAATGGTACTTTGACCTCCAGTGTAGGGCTATACTATACATATATATATATATATATATATATATATATA ATTTTGGAATTTGTTTCTCATAATACAGAATATATAGTGGCTACCTTGTATCTTGGTCTGGATTCTCTCTCTGAGACCCCGGATTTTACT TTCTCTTTGGAGGGCGCTGGGACATACATCTCTCAATCCAGCTTCCTCCGCATCCTCCCATCTTGCCCCATTTCTGCCACGTCAGACACT TCCTGAGAGTCTCACCTTCAAAATGACACCGCTGCCCATCCATTGCTCAATGGTACAGAGTGTGGGGTCAGTCCACCACCCTTGACCTCC CGGCAGGGCAAGGTGAGGAGGCGGACCCAAAGCAGTACCAGCAGGACTTGTTGCCAGTGATACCAAAACAGACTTTTCCCAAGCAGTGCC TCACATGTCTGCTGGTGTGGCTTTGGGATTCTCCTGCCCCACCCCCCCGTCCATGGCAGCCCCCTCCCCAAGGCTTTGCTCACACCTGAG ACAGGAAGGAGGAAGGGGATCCAATAGGAATATGGGCCCCGGAGGGGAAGTCATGCACCCCCAAGCCACCACCCCCCAGCCTTCCACGCA CATCTCCTGGCTGGAAGAGAGCCCTCCAAAAAGGGGACACAGGCTGCCCCGGCCCCTCAACTGCATCCACACCCCATCCTCTCATCTTGG GTCCCAGCCAGGCCCCCCCAAAACCAAAGCCCCCTCAAGTCCTGGGGTCCCAGCCTGTGCCCCCAGCTTCCTGCCCACCCAGCCCTGAGC ATTCTCACACAGAGAAAGAACAAGCAAGGGCTCCAGGGGGACAGGATGGGGCAGGGCATACAGTGGGGGGTGGGGGGGCAGCTGGGAGGA GGGAGGGACAAAACAAAACATTTTCCTTTGGGTTTTTTTTTTCTTTCTTTTTTCTCCCCTTTACTCTTTGGGTGGTGTTGCTTTTCCTTT CCTTTTCCCTTTGAGATTTTTTTGTTGTTGTTTCCTTTTTGTATTTTACTGATATCACCAGGATAGTTTACTCTCCTTCTAGCTTTCTGC TTACCGCACACTGGATAACACACACATACACACCCACAAAAATGCTCATGAACCCAATCCGGAGAAGGTTCCAGCAGGTCCCCCACCCTC CCCTCCTCCTCCTACTTCTCCTCTTGACAGCGAGGACAGGAGGGGGACAAGGGGACACCTGGGCAGACCCGCCGGCTCTCCCCCCACCCC ACCCCGCCCCTCACATCATACTCCAATCATAACCTTGTATATTATGCAGTCATTTTGGTTTTCGCGGACGCGCCTACCTAAGTACCATTT ACAGAAAGTGACTCTGGCTGTCATTATTTTGTTTATTTGTTCCCTATGCAAAAAAAAAATGAAAATGAAAAAAGGGGGATTCCATAAAAG >101484_101484_6_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000589123_length(amino acids)=499AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDA EQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGS KRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHP SSALHFPTTSILPQTASTYFPHTAIRYPPHLNPQDPLKDLVSLACDPASQQPGPLNGSGQLKMPSHCLSAQMLAPPPPGLPRLALPPATK -------------------------------------------------------------- >101484_101484_7_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000590282_length(transcript)=1617nt_BP=143nt TTTCTCGCGTCTGCCGTACTTGGTTTCGCTTCGCTAGTCCCAGAGGCCCAGAGTGGCTCGCCTGGAGTCTCTGTGGCGCGGTTTCCTGTA CCTGCCTTGGGATCCGGAGGGAGGAAGCTGGGACACCCGGGAGTCAGGAAATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTC ACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACG AGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGG ACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGG GCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGC TGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCG TCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACC AGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCC AAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAG CCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACA CGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGGCATCTCGT CCCCGGTGAAGAAGACAGAGATGGACAAGTCACCATTCAACAGCCCGTCCCCCCAGGACTCTCCCCGCCTCTCCAGCTTCACCCAGCACC ACCGGCCCGTCATCGCCGTGCACAGCGGGATCGCCCGGAGCCCACACCCGTCCTCCGCTCTGCATTTCCCTACGACGTCCATCCTACCCC AGACGGCCTCCACCTACTTCCCCCACACGGCCATCCGCTACCCACCTCATCTCAACCCCCAGGACCCGCTCAAAGATCTTGTCTCGCTGG CCTGCGACCCAGCCAGCCAGCAACCTGGACCGCCTACTCTCCGCCCGACACGTCCCCTGCAAACCGTTCCTTTGTGGGATTAGGACCAAG GGATCCTGCGGGCATTTATCAGGCACAGTCCTGGTATCTGGGATAGCAAAGGTCTTCTTCCCTCGCCCCTTCTCCATCGTCCCAGGAATC >101484_101484_7_ZNF136-NFIC_ZNF136_chr19_12274021_ENST00000343979_NFIC_chr19_3381709_ENST00000590282_length(amino acids)=430AA_BP=1 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDA EQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGS KRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGGISSPVKKTEMDKSPFNSPSPQDSPRLSSFTQHHRPVIAVHSGIARSPHP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF136-NFIC |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF136-NFIC |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF136-NFIC |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
