
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF302-ANKRD27 (FusionGDB2 ID:101838) |
Fusion Gene Summary for ZNF302-ANKRD27 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF302-ANKRD27 | Fusion gene ID: 101838 | Hgene | Tgene | Gene symbol | ZNF302 | ANKRD27 | Gene ID | 55900 | 84079 |
| Gene name | zinc finger protein 302 | ankyrin repeat domain 27 | |
| Synonyms | HSD16|MST154|MSTP154|ZNF135L|ZNF140L|ZNF327 | PP12899|VARP | |
| Cytomap | 19q13.11 | 19q13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 302zinc finger protein 327 | ankyrin repeat domain-containing protein 27VPS9 domain-containing proteinVPS9-ankyrin-repeat proteinVps9 domain and ankyrin-repeat-containing proteinankyrin repeat domain 27 (VPS9 domain) | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q96NW4 | |
| Ensembl transtripts involved in fusion gene | ENST00000423823, ENST00000446502, ENST00000457781, ENST00000505242, ENST00000505365, ENST00000507959, ENST00000509528, | ENST00000587352, ENST00000306065, | |
| Fusion gene scores | * DoF score | 4 X 3 X 4=48 | 5 X 7 X 3=105 |
| # samples | 4 | 7 | |
| ** MAII score | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/105*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF302 [Title/Abstract] AND ANKRD27 [Title/Abstract] AND fusion [Title/Abstract] | ||
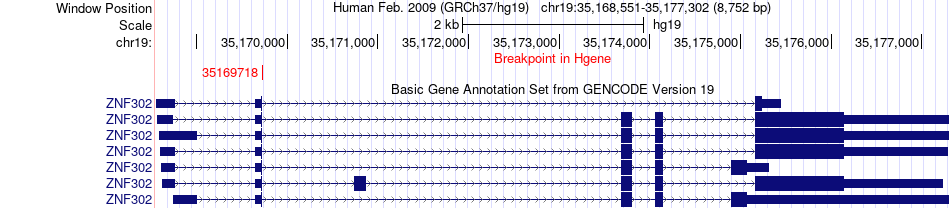
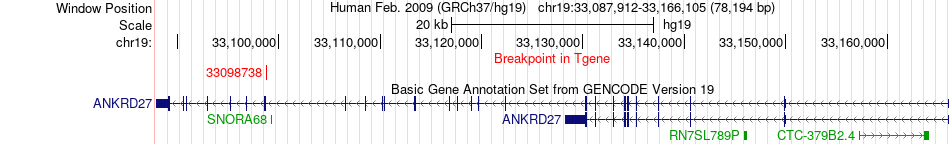
| Most frequent breakpoint | ZNF302(35169718)-ANKRD27(33098738), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ANKRD27 | GO:0035544 | negative regulation of SNARE complex assembly | 24856514 |
| Tgene | ANKRD27 | GO:0045022 | early endosome to late endosome transport | 16525121 |
 Fusion gene breakpoints across ZNF302 (5'-gene) Fusion gene breakpoints across ZNF302 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across ANKRD27 (3'-gene) Fusion gene breakpoints across ANKRD27 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-D1-A3JP-01A | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
Top |
Fusion Gene ORF analysis for ZNF302-ANKRD27 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000423823 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| 5CDS-intron | ENST00000446502 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| 5CDS-intron | ENST00000457781 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| 5CDS-intron | ENST00000505242 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| 5CDS-intron | ENST00000505365 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| 5CDS-intron | ENST00000507959 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| 5CDS-intron | ENST00000509528 | ENST00000587352 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000423823 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000446502 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000457781 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000505242 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000505365 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000507959 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
| In-frame | ENST00000509528 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000509528 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2403 | 287 | 251 | 1264 | 337 |
| ENST00000457781 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2365 | 249 | 213 | 1226 | 337 |
| ENST00000505242 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2619 | 503 | 467 | 1480 | 337 |
| ENST00000423823 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2361 | 245 | 209 | 1222 | 337 |
| ENST00000507959 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2348 | 232 | 196 | 1209 | 337 |
| ENST00000446502 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2333 | 217 | 181 | 1194 | 337 |
| ENST00000505365 | ZNF302 | chr19 | 35169718 | - | ENST00000306065 | ANKRD27 | chr19 | 33098738 | - | 2461 | 345 | 309 | 1322 | 337 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000509528 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.005677007 | 0.994323 |
| ENST00000457781 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.00592466 | 0.9940753 |
| ENST00000505242 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.005846426 | 0.99415356 |
| ENST00000423823 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.006171498 | 0.9938285 |
| ENST00000507959 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.005827624 | 0.9941724 |
| ENST00000446502 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.005706662 | 0.99429333 |
| ENST00000505365 | ENST00000306065 | ZNF302 | chr19 | 35169718 | - | ANKRD27 | chr19 | 33098738 | - | 0.005887054 | 0.9941129 |
Top |
Fusion Genomic Features for ZNF302-ANKRD27 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
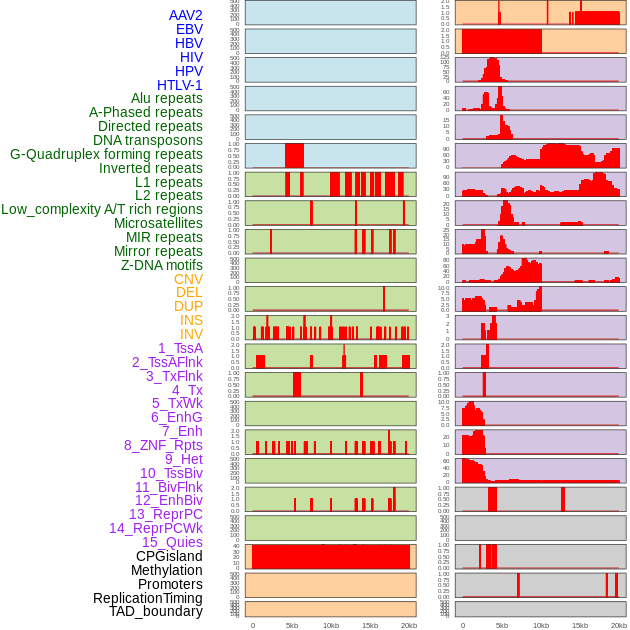
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZNF302-ANKRD27 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:35169718/chr19:33098738) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ANKRD27 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: May be a guanine exchange factor (GEF) for Rab21, Rab32 and Rab38 and regulate endosome dynamics (PubMed:16525121, PubMed:18477474). May regulate the participation of VAMP7 in membrane fusion events; in vitro inhibits VAMP7-mediated SNARE complex formation by trapping VAMP7 in a closed, fusogenically inactive conformation (PubMed:23104059). Involved in peripheral melanosomal distribution of TYRP1 in melanocytes; the function, which probably is implicating vesicle-trafficking, includes cooperation with Rab32, Rab38 and VAMP7 (By similarity). Involved in the regulation of neurite growth; the function seems to require its GEF activity, probably towards Rab21, and VAMP7 but not Rab32/38 (By similarity). Proposed to be involved in Golgi sorting of VAMP7 and transport of VAMP7 vesicles to the cell surface; the function seems to implicate kinesin heavy chain isoform 5 proteins, GOLGA4, RAB21 and MACF1 (PubMed:22705394). Required for the colocalization of VAMP7 and Rab21, probably on TGN sites (PubMed:19745841). Involved in GLUT1 endosome-to-plasma membrane trafficking; the function is dependent of association with VPS29 (PubMed:24856514). Regulates the proper trafficking of melanogenic enzymes TYR, TYRP1 and DCT/TYRP2 to melanosomes in melanocytes (By similarity). {ECO:0000250|UniProtKB:Q3UMR0, ECO:0000269|PubMed:23104059, ECO:0000269|PubMed:24856514, ECO:0000305|PubMed:16525121, ECO:0000305|PubMed:18477474, ECO:0000305|PubMed:22705394}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 743_772 | 725 | 1051.0 | Repeat | Note=ANK 8 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 776_805 | 725 | 1051.0 | Repeat | Note=ANK 9 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 809_838 | 725 | 1051.0 | Repeat | Note=ANK 10 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 842_871 | 725 | 1051.0 | Repeat | Note=ANK 11 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 4_75 | 3 | 400.0 | Domain | KRAB |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 4_75 | 3 | 400.0 | Domain | KRAB |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 4_75 | 3 | 400.0 | Domain | KRAB |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 280_302 | 3 | 400.0 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 308_330 | 3 | 400.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 336_358 | 3 | 400.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 364_386 | 3 | 400.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 392_414 | 3 | 400.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 420_442 | 3 | 400.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000423823 | - | 2 | 5 | 448_470 | 3 | 400.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 280_302 | 3 | 400.0 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 308_330 | 3 | 400.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 336_358 | 3 | 400.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 364_386 | 3 | 400.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 392_414 | 3 | 400.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 420_442 | 3 | 400.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000457781 | - | 2 | 5 | 448_470 | 3 | 400.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 280_302 | 3 | 400.0 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 308_330 | 3 | 400.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 336_358 | 3 | 400.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 364_386 | 3 | 400.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 392_414 | 3 | 400.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 420_442 | 3 | 400.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF302 | chr19:35169718 | chr19:33098738 | ENST00000505242 | - | 2 | 5 | 448_470 | 3 | 400.0 | Zinc finger | C2H2-type 7 |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 233_371 | 725 | 1051.0 | Domain | VPS9 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 1_372 | 725 | 1051.0 | Region | Sufficient for GEF activity towards RAB21 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 396_426 | 725 | 1051.0 | Repeat | Note=ANK 1 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 462_491 | 725 | 1051.0 | Repeat | Note=ANK 2 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 495_524 | 725 | 1051.0 | Repeat | Note=ANK 3 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 528_560 | 725 | 1051.0 | Repeat | Note=ANK 4 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 564_593 | 725 | 1051.0 | Repeat | Note=ANK 5 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 597_627 | 725 | 1051.0 | Repeat | Note=ANK 6 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 668_698 | 725 | 1051.0 | Repeat | Note=ANK 7 |
Top |
Fusion Gene Sequence for ZNF302-ANKRD27 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >101838_101838_1_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000423823_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2361nt_BP=245nt GATAGCTTCAGGCCTGGCGGCGGCTTCCAAGCTAAGGAACGGTTTGGGGCAGTGTCGTTCCCGGAGGTCGGCCGCCGTTACCCGCTCACC AGCTACGCGGCGCGTCAGGTCCGCGGAGGCGCGGGCTCGGGGCGCCTGCGGGACGGTGAGGCCCTGCTGAGGACTCCGGACACTGCCCAT CTCTAAGATAAGAACCTGGAAAGGGGACTCTGTTGGCCATTGGAAATTGCAGAATAATGTCTCAGAGGCTGGCGAAGGTTCCTGCCAGTG GGCTTGGTGTGAACGTGACCAGCCAGGACGGCTCCTCCCCGCTGCATGTCGCCGCCCTGCACGGCCGGGCGGACCTCATCCCCCTCCTGC TGAAGCACGGGGCCAACGCAGGTGCCAGGAACGCAGACCAAGCCGTCCCGCTCCACCTGGCCTGCCAGCAGGGCCACTTTCAGGTGGTGA AGTGTCTGTTAGATTCGAATGCAAAACCCAATAAGAAGGACCTCAGTGGAAACACGCCCCTCATTTACGCCTGCTCCGGTGGCCATCACG AGCTTGTGGCACTGCTGCTACAGCACGGGGCCTCCATTAACGCTTCTAACAATAAGGGCAACACAGCGCTGCACGAGGCTGTGATTGAAA AGCACGTCTTCGTGGTAGAGCTGCTTCTGCTCCACGGAGCGTCAGTTCAGGTGCTGAACAAGCGGCAGCGCACGGCTGTAGACTGTGCTG AACAGAATTCAAAAATAATGGAATTGCTTCAGGTGGTACCAAGCTGTGTTGCTTCATTAGATGATGTGGCTGAAACTGACCGCAAGGAGT ATGTCACTGTTAAGATCAGGAAAAAATGGAACTCAAAACTGTATGATCTACCAGATGAGCCTTTTACAAGACAGTTTTACTTTGTCCACT CAGCTGGTCAGTTTAAGGGAAAGACTTCAAGGGAGATTATGGCAAGAGATAGAAGTGTCCCTAATTTAACCGAAGGTTCTTTGCATGAGC CAGGGAGGCAAAGTGTCACACTGAGACAGAATAACCTGCCAGCTCAGAGTGGATCTCATGCTGCTGAGAAAGGCAACAGCGACTGGCCAG AGAGGCCTGGACTGACACAGACTGGCCCTGGACACAGACGGATGCTGCGGAGACACACGGTAGAGGATGCGGTCGTGTCCCAGGGCCCGG AGGCTGCTGGCCCCCTCTCCACTCCCCAAGAGGTTAGTGCTTCCCGGTCCTAACAGGAATGAGGAGTTGTTGAACCCACTGCTAGGAAGC AAGGATGCAACAAGATGATGCTGAGCGTGAACACATCTGAGAACTAAATGTGCTTCCATGAGACTGGCTTGAGAAGTCTTCAGCACCAAG TTCCTGAAAGCTTTTCTGTGGCAGGAAAGAATGCAACAAAAAAGTTAACCACCACCATCTCTCTCCTCTTCAAAGCTAATGAATACAATT GAAACAGACAAAAATTCCAGTAGCATCCAGATCCTTAAGCCAGAGGTGCATGCTTCTTTTTAAGTATGAGGGTTTGTTGGTCACAGTGGG AGAGGTTTCACCACCGCATTCTGACCTCCTCCTCCCAAAAGGTGCTAAACCTCTCTGACCTGTGTACATTCACAAACCACAGCTAGAATT CCTCCACCTAGGATTAAGCTGGAGAGAAGTAAGTAATTTAGGTTTCATGGTACTGTAGAGGCCAGGCTGAAATGTCATATCTGAAGGAAG AAAGCAGCAGCTGGACAATGTTTCTTTGCAAAGCAACACTCGAACCAAAAGATGCCTCAATCCCATTTTGATATTCATTTTAGTGAAAGG ATGCATCAGACCTGTTCCACATCATGCACATGGGAAAGGGTGGTTATCATTTTCCTTCTAACAAGTAGGTACAGATATTCGGTTACTACA CGTGCACCTGTAGCAGTATTTCTAGAAACATCCCTTTTTGTTGAGAACCTCCCTTGAATGTCTGTCACACTCACACCTGACGGGATGGTT ACTGGATTAGAGAGTAGATTTGGCACATCTTTTCTTAGTCTTTTGATTCAAATTCAAAACTTAACAGCACAAACCAGGTCAGAGTTACTT TCGGTTAGAATTTATTGCCATTTATTCCTTTTTATAAATTTCTATAGATTATACTGTTATTTTTATGTTATTGGCCTAGAGCTACACGTA TATGGGTTTGTCCTGAGTCCGTTTTCAAATGACCTTGTGATAGGGAAATGGTTTTGTCCATGTTCTTGGAAATACTTGTGTATGTACAGA AGGAAGGGAGGGATTATTTTTCTACAAAGTAATTTATGATTTCTAATTTTCTAATGTGCCTTGGATATGTGCCAAATGATGGAAAAGAAA >101838_101838_1_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000423823_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- >101838_101838_2_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000446502_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2333nt_BP=217nt AAGCTAAGGAACGGTTTGGGGCAGTGTCGTTCCCGGAGGTCGGCCGCCGTTACCCGCTCACCAGCTACGCGGCGCGTCAGGTCCGCGGAG GCGCGGGCTCGGGGCGCCTGCGGGACGGTGAGGCCCTGCTGAGGACTCCGGACACTGCCCATCTCTAAGATAAGAACCTGGAAAGGGGAC TCTGTTGGCCATTGGAAATTGCAGAATAATGTCTCAGAGGCTGGCGAAGGTTCCTGCCAGTGGGCTTGGTGTGAACGTGACCAGCCAGGA CGGCTCCTCCCCGCTGCATGTCGCCGCCCTGCACGGCCGGGCGGACCTCATCCCCCTCCTGCTGAAGCACGGGGCCAACGCAGGTGCCAG GAACGCAGACCAAGCCGTCCCGCTCCACCTGGCCTGCCAGCAGGGCCACTTTCAGGTGGTGAAGTGTCTGTTAGATTCGAATGCAAAACC CAATAAGAAGGACCTCAGTGGAAACACGCCCCTCATTTACGCCTGCTCCGGTGGCCATCACGAGCTTGTGGCACTGCTGCTACAGCACGG GGCCTCCATTAACGCTTCTAACAATAAGGGCAACACAGCGCTGCACGAGGCTGTGATTGAAAAGCACGTCTTCGTGGTAGAGCTGCTTCT GCTCCACGGAGCGTCAGTTCAGGTGCTGAACAAGCGGCAGCGCACGGCTGTAGACTGTGCTGAACAGAATTCAAAAATAATGGAATTGCT TCAGGTGGTACCAAGCTGTGTTGCTTCATTAGATGATGTGGCTGAAACTGACCGCAAGGAGTATGTCACTGTTAAGATCAGGAAAAAATG GAACTCAAAACTGTATGATCTACCAGATGAGCCTTTTACAAGACAGTTTTACTTTGTCCACTCAGCTGGTCAGTTTAAGGGAAAGACTTC AAGGGAGATTATGGCAAGAGATAGAAGTGTCCCTAATTTAACCGAAGGTTCTTTGCATGAGCCAGGGAGGCAAAGTGTCACACTGAGACA GAATAACCTGCCAGCTCAGAGTGGATCTCATGCTGCTGAGAAAGGCAACAGCGACTGGCCAGAGAGGCCTGGACTGACACAGACTGGCCC TGGACACAGACGGATGCTGCGGAGACACACGGTAGAGGATGCGGTCGTGTCCCAGGGCCCGGAGGCTGCTGGCCCCCTCTCCACTCCCCA AGAGGTTAGTGCTTCCCGGTCCTAACAGGAATGAGGAGTTGTTGAACCCACTGCTAGGAAGCAAGGATGCAACAAGATGATGCTGAGCGT GAACACATCTGAGAACTAAATGTGCTTCCATGAGACTGGCTTGAGAAGTCTTCAGCACCAAGTTCCTGAAAGCTTTTCTGTGGCAGGAAA GAATGCAACAAAAAAGTTAACCACCACCATCTCTCTCCTCTTCAAAGCTAATGAATACAATTGAAACAGACAAAAATTCCAGTAGCATCC AGATCCTTAAGCCAGAGGTGCATGCTTCTTTTTAAGTATGAGGGTTTGTTGGTCACAGTGGGAGAGGTTTCACCACCGCATTCTGACCTC CTCCTCCCAAAAGGTGCTAAACCTCTCTGACCTGTGTACATTCACAAACCACAGCTAGAATTCCTCCACCTAGGATTAAGCTGGAGAGAA GTAAGTAATTTAGGTTTCATGGTACTGTAGAGGCCAGGCTGAAATGTCATATCTGAAGGAAGAAAGCAGCAGCTGGACAATGTTTCTTTG CAAAGCAACACTCGAACCAAAAGATGCCTCAATCCCATTTTGATATTCATTTTAGTGAAAGGATGCATCAGACCTGTTCCACATCATGCA CATGGGAAAGGGTGGTTATCATTTTCCTTCTAACAAGTAGGTACAGATATTCGGTTACTACACGTGCACCTGTAGCAGTATTTCTAGAAA CATCCCTTTTTGTTGAGAACCTCCCTTGAATGTCTGTCACACTCACACCTGACGGGATGGTTACTGGATTAGAGAGTAGATTTGGCACAT CTTTTCTTAGTCTTTTGATTCAAATTCAAAACTTAACAGCACAAACCAGGTCAGAGTTACTTTCGGTTAGAATTTATTGCCATTTATTCC TTTTTATAAATTTCTATAGATTATACTGTTATTTTTATGTTATTGGCCTAGAGCTACACGTATATGGGTTTGTCCTGAGTCCGTTTTCAA ATGACCTTGTGATAGGGAAATGGTTTTGTCCATGTTCTTGGAAATACTTGTGTATGTACAGAAGGAAGGGAGGGATTATTTTTCTACAAA >101838_101838_2_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000446502_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- >101838_101838_3_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000457781_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2365nt_BP=249nt GTGGGCGGTGCACTGGGTCAGTCCCGCGATAGCTTCAGGCCTGGCGGCGGCTTCCAAGCTAAGGAACGGTTTGGGGCAGTGTCGTTCCCG GAGGTCGGCCGCCGTTACCCGCTCACCAGCTACGCGGCGCGTCAGGTCCGCGGAGGCGCGGGCTCGGGGCGCCTGCGGGACGGACACTGC CCATCTCTAAGATAAGAACCTGGAAAGGGGACTCTGTTGGCCATTGGAAATTGCAGAATAATGTCTCAGAGGCTGGCGAAGGTTCCTGCC AGTGGGCTTGGTGTGAACGTGACCAGCCAGGACGGCTCCTCCCCGCTGCATGTCGCCGCCCTGCACGGCCGGGCGGACCTCATCCCCCTC CTGCTGAAGCACGGGGCCAACGCAGGTGCCAGGAACGCAGACCAAGCCGTCCCGCTCCACCTGGCCTGCCAGCAGGGCCACTTTCAGGTG GTGAAGTGTCTGTTAGATTCGAATGCAAAACCCAATAAGAAGGACCTCAGTGGAAACACGCCCCTCATTTACGCCTGCTCCGGTGGCCAT CACGAGCTTGTGGCACTGCTGCTACAGCACGGGGCCTCCATTAACGCTTCTAACAATAAGGGCAACACAGCGCTGCACGAGGCTGTGATT GAAAAGCACGTCTTCGTGGTAGAGCTGCTTCTGCTCCACGGAGCGTCAGTTCAGGTGCTGAACAAGCGGCAGCGCACGGCTGTAGACTGT GCTGAACAGAATTCAAAAATAATGGAATTGCTTCAGGTGGTACCAAGCTGTGTTGCTTCATTAGATGATGTGGCTGAAACTGACCGCAAG GAGTATGTCACTGTTAAGATCAGGAAAAAATGGAACTCAAAACTGTATGATCTACCAGATGAGCCTTTTACAAGACAGTTTTACTTTGTC CACTCAGCTGGTCAGTTTAAGGGAAAGACTTCAAGGGAGATTATGGCAAGAGATAGAAGTGTCCCTAATTTAACCGAAGGTTCTTTGCAT GAGCCAGGGAGGCAAAGTGTCACACTGAGACAGAATAACCTGCCAGCTCAGAGTGGATCTCATGCTGCTGAGAAAGGCAACAGCGACTGG CCAGAGAGGCCTGGACTGACACAGACTGGCCCTGGACACAGACGGATGCTGCGGAGACACACGGTAGAGGATGCGGTCGTGTCCCAGGGC CCGGAGGCTGCTGGCCCCCTCTCCACTCCCCAAGAGGTTAGTGCTTCCCGGTCCTAACAGGAATGAGGAGTTGTTGAACCCACTGCTAGG AAGCAAGGATGCAACAAGATGATGCTGAGCGTGAACACATCTGAGAACTAAATGTGCTTCCATGAGACTGGCTTGAGAAGTCTTCAGCAC CAAGTTCCTGAAAGCTTTTCTGTGGCAGGAAAGAATGCAACAAAAAAGTTAACCACCACCATCTCTCTCCTCTTCAAAGCTAATGAATAC AATTGAAACAGACAAAAATTCCAGTAGCATCCAGATCCTTAAGCCAGAGGTGCATGCTTCTTTTTAAGTATGAGGGTTTGTTGGTCACAG TGGGAGAGGTTTCACCACCGCATTCTGACCTCCTCCTCCCAAAAGGTGCTAAACCTCTCTGACCTGTGTACATTCACAAACCACAGCTAG AATTCCTCCACCTAGGATTAAGCTGGAGAGAAGTAAGTAATTTAGGTTTCATGGTACTGTAGAGGCCAGGCTGAAATGTCATATCTGAAG GAAGAAAGCAGCAGCTGGACAATGTTTCTTTGCAAAGCAACACTCGAACCAAAAGATGCCTCAATCCCATTTTGATATTCATTTTAGTGA AAGGATGCATCAGACCTGTTCCACATCATGCACATGGGAAAGGGTGGTTATCATTTTCCTTCTAACAAGTAGGTACAGATATTCGGTTAC TACACGTGCACCTGTAGCAGTATTTCTAGAAACATCCCTTTTTGTTGAGAACCTCCCTTGAATGTCTGTCACACTCACACCTGACGGGAT GGTTACTGGATTAGAGAGTAGATTTGGCACATCTTTTCTTAGTCTTTTGATTCAAATTCAAAACTTAACAGCACAAACCAGGTCAGAGTT ACTTTCGGTTAGAATTTATTGCCATTTATTCCTTTTTATAAATTTCTATAGATTATACTGTTATTTTTATGTTATTGGCCTAGAGCTACA CGTATATGGGTTTGTCCTGAGTCCGTTTTCAAATGACCTTGTGATAGGGAAATGGTTTTGTCCATGTTCTTGGAAATACTTGTGTATGTA CAGAAGGAAGGGAGGGATTATTTTTCTACAAAGTAATTTATGATTTCTAATTTTCTAATGTGCCTTGGATATGTGCCAAATGATGGAAAA >101838_101838_3_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000457781_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- >101838_101838_4_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000505242_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2619nt_BP=503nt GTCAGTCCCGCGATAGCTTCAGGCCTGGCGGCGGCTTCCAAGCTAAGGAACGGTTTGGGGCAGTGTCGTTCCCGGAGGTCGGCCGCCGTT ACCCGCTCACCAGCTACGCGGCGCGTCAGGTCCGCGGAGGCGCGGGCTCGGGGCGCCTGCGGGACGGTGAGGCCCTGCTGAGGACTCCGG CAAGTGTGGGTCGCGGCGACGGCGGGGCTAAGGCCCTGGGTCCGCGCGCGGTTTGACCACGGCCGGGGCCTTGGGCATTTCCTGGCCTTC CTGTTGAGCCGTGTAAACGCGGGGTGATGACGGCGCCGACCTCTTGGCACTGTTGTGAGAGCGAAGTGGGCGCGAGAGCAGACGCCAGCT ACAGTTTTTTTTGGGTTATGTCGTCATGAAGCCGGCGCTTTCAGTTGTGCAACCTTGAACAAATGGGACACTGCCCATCTCTAAGATAAG AACCTGGAAAGGGGACTCTGTTGGCCATTGGAAATTGCAGAATAATGTCTCAGAGGCTGGCGAAGGTTCCTGCCAGTGGGCTTGGTGTGA ACGTGACCAGCCAGGACGGCTCCTCCCCGCTGCATGTCGCCGCCCTGCACGGCCGGGCGGACCTCATCCCCCTCCTGCTGAAGCACGGGG CCAACGCAGGTGCCAGGAACGCAGACCAAGCCGTCCCGCTCCACCTGGCCTGCCAGCAGGGCCACTTTCAGGTGGTGAAGTGTCTGTTAG ATTCGAATGCAAAACCCAATAAGAAGGACCTCAGTGGAAACACGCCCCTCATTTACGCCTGCTCCGGTGGCCATCACGAGCTTGTGGCAC TGCTGCTACAGCACGGGGCCTCCATTAACGCTTCTAACAATAAGGGCAACACAGCGCTGCACGAGGCTGTGATTGAAAAGCACGTCTTCG TGGTAGAGCTGCTTCTGCTCCACGGAGCGTCAGTTCAGGTGCTGAACAAGCGGCAGCGCACGGCTGTAGACTGTGCTGAACAGAATTCAA AAATAATGGAATTGCTTCAGGTGGTACCAAGCTGTGTTGCTTCATTAGATGATGTGGCTGAAACTGACCGCAAGGAGTATGTCACTGTTA AGATCAGGAAAAAATGGAACTCAAAACTGTATGATCTACCAGATGAGCCTTTTACAAGACAGTTTTACTTTGTCCACTCAGCTGGTCAGT TTAAGGGAAAGACTTCAAGGGAGATTATGGCAAGAGATAGAAGTGTCCCTAATTTAACCGAAGGTTCTTTGCATGAGCCAGGGAGGCAAA GTGTCACACTGAGACAGAATAACCTGCCAGCTCAGAGTGGATCTCATGCTGCTGAGAAAGGCAACAGCGACTGGCCAGAGAGGCCTGGAC TGACACAGACTGGCCCTGGACACAGACGGATGCTGCGGAGACACACGGTAGAGGATGCGGTCGTGTCCCAGGGCCCGGAGGCTGCTGGCC CCCTCTCCACTCCCCAAGAGGTTAGTGCTTCCCGGTCCTAACAGGAATGAGGAGTTGTTGAACCCACTGCTAGGAAGCAAGGATGCAACA AGATGATGCTGAGCGTGAACACATCTGAGAACTAAATGTGCTTCCATGAGACTGGCTTGAGAAGTCTTCAGCACCAAGTTCCTGAAAGCT TTTCTGTGGCAGGAAAGAATGCAACAAAAAAGTTAACCACCACCATCTCTCTCCTCTTCAAAGCTAATGAATACAATTGAAACAGACAAA AATTCCAGTAGCATCCAGATCCTTAAGCCAGAGGTGCATGCTTCTTTTTAAGTATGAGGGTTTGTTGGTCACAGTGGGAGAGGTTTCACC ACCGCATTCTGACCTCCTCCTCCCAAAAGGTGCTAAACCTCTCTGACCTGTGTACATTCACAAACCACAGCTAGAATTCCTCCACCTAGG ATTAAGCTGGAGAGAAGTAAGTAATTTAGGTTTCATGGTACTGTAGAGGCCAGGCTGAAATGTCATATCTGAAGGAAGAAAGCAGCAGCT GGACAATGTTTCTTTGCAAAGCAACACTCGAACCAAAAGATGCCTCAATCCCATTTTGATATTCATTTTAGTGAAAGGATGCATCAGACC TGTTCCACATCATGCACATGGGAAAGGGTGGTTATCATTTTCCTTCTAACAAGTAGGTACAGATATTCGGTTACTACACGTGCACCTGTA GCAGTATTTCTAGAAACATCCCTTTTTGTTGAGAACCTCCCTTGAATGTCTGTCACACTCACACCTGACGGGATGGTTACTGGATTAGAG AGTAGATTTGGCACATCTTTTCTTAGTCTTTTGATTCAAATTCAAAACTTAACAGCACAAACCAGGTCAGAGTTACTTTCGGTTAGAATT TATTGCCATTTATTCCTTTTTATAAATTTCTATAGATTATACTGTTATTTTTATGTTATTGGCCTAGAGCTACACGTATATGGGTTTGTC CTGAGTCCGTTTTCAAATGACCTTGTGATAGGGAAATGGTTTTGTCCATGTTCTTGGAAATACTTGTGTATGTACAGAAGGAAGGGAGGG ATTATTTTTCTACAAAGTAATTTATGATTTCTAATTTTCTAATGTGCCTTGGATATGTGCCAAATGATGGAAAAGAAACAGTAAACTTTA >101838_101838_4_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000505242_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- >101838_101838_5_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000505365_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2461nt_BP=345nt GAGGCCCTGCTGAGGACTCCGGCAAGTGTGGGTCGCGGCGACGGCGGGGCTAAGGCCCTGGGTCCGCGCGCGGTTTGACCACGGCCGGGG CCTTGGGCATTTCCTGGCCTTCCTGTTGAGCCGTGTAAACGCGGGGTGATGACGGCGCCGACCTCTTGGCACTGTTGTGAGAGCGAAGTG GGCGCGAGAGCAGACGCCAGCTACAGTTTTTTTTGGGTTATGTCGTCATGAAGCCGGCGCTTTCAGTTGTGCAACCTTGAACAAATGGGA CACTGCCCATCTCTAAGATAAGAACCTGGAAAGGGGACTCTGTTGGCCATTGGAAATTGCAGAATAATGTCTCAGAGGCTGGCGAAGGTT CCTGCCAGTGGGCTTGGTGTGAACGTGACCAGCCAGGACGGCTCCTCCCCGCTGCATGTCGCCGCCCTGCACGGCCGGGCGGACCTCATC CCCCTCCTGCTGAAGCACGGGGCCAACGCAGGTGCCAGGAACGCAGACCAAGCCGTCCCGCTCCACCTGGCCTGCCAGCAGGGCCACTTT CAGGTGGTGAAGTGTCTGTTAGATTCGAATGCAAAACCCAATAAGAAGGACCTCAGTGGAAACACGCCCCTCATTTACGCCTGCTCCGGT GGCCATCACGAGCTTGTGGCACTGCTGCTACAGCACGGGGCCTCCATTAACGCTTCTAACAATAAGGGCAACACAGCGCTGCACGAGGCT GTGATTGAAAAGCACGTCTTCGTGGTAGAGCTGCTTCTGCTCCACGGAGCGTCAGTTCAGGTGCTGAACAAGCGGCAGCGCACGGCTGTA GACTGTGCTGAACAGAATTCAAAAATAATGGAATTGCTTCAGGTGGTACCAAGCTGTGTTGCTTCATTAGATGATGTGGCTGAAACTGAC CGCAAGGAGTATGTCACTGTTAAGATCAGGAAAAAATGGAACTCAAAACTGTATGATCTACCAGATGAGCCTTTTACAAGACAGTTTTAC TTTGTCCACTCAGCTGGTCAGTTTAAGGGAAAGACTTCAAGGGAGATTATGGCAAGAGATAGAAGTGTCCCTAATTTAACCGAAGGTTCT TTGCATGAGCCAGGGAGGCAAAGTGTCACACTGAGACAGAATAACCTGCCAGCTCAGAGTGGATCTCATGCTGCTGAGAAAGGCAACAGC GACTGGCCAGAGAGGCCTGGACTGACACAGACTGGCCCTGGACACAGACGGATGCTGCGGAGACACACGGTAGAGGATGCGGTCGTGTCC CAGGGCCCGGAGGCTGCTGGCCCCCTCTCCACTCCCCAAGAGGTTAGTGCTTCCCGGTCCTAACAGGAATGAGGAGTTGTTGAACCCACT GCTAGGAAGCAAGGATGCAACAAGATGATGCTGAGCGTGAACACATCTGAGAACTAAATGTGCTTCCATGAGACTGGCTTGAGAAGTCTT CAGCACCAAGTTCCTGAAAGCTTTTCTGTGGCAGGAAAGAATGCAACAAAAAAGTTAACCACCACCATCTCTCTCCTCTTCAAAGCTAAT GAATACAATTGAAACAGACAAAAATTCCAGTAGCATCCAGATCCTTAAGCCAGAGGTGCATGCTTCTTTTTAAGTATGAGGGTTTGTTGG TCACAGTGGGAGAGGTTTCACCACCGCATTCTGACCTCCTCCTCCCAAAAGGTGCTAAACCTCTCTGACCTGTGTACATTCACAAACCAC AGCTAGAATTCCTCCACCTAGGATTAAGCTGGAGAGAAGTAAGTAATTTAGGTTTCATGGTACTGTAGAGGCCAGGCTGAAATGTCATAT CTGAAGGAAGAAAGCAGCAGCTGGACAATGTTTCTTTGCAAAGCAACACTCGAACCAAAAGATGCCTCAATCCCATTTTGATATTCATTT TAGTGAAAGGATGCATCAGACCTGTTCCACATCATGCACATGGGAAAGGGTGGTTATCATTTTCCTTCTAACAAGTAGGTACAGATATTC GGTTACTACACGTGCACCTGTAGCAGTATTTCTAGAAACATCCCTTTTTGTTGAGAACCTCCCTTGAATGTCTGTCACACTCACACCTGA CGGGATGGTTACTGGATTAGAGAGTAGATTTGGCACATCTTTTCTTAGTCTTTTGATTCAAATTCAAAACTTAACAGCACAAACCAGGTC AGAGTTACTTTCGGTTAGAATTTATTGCCATTTATTCCTTTTTATAAATTTCTATAGATTATACTGTTATTTTTATGTTATTGGCCTAGA GCTACACGTATATGGGTTTGTCCTGAGTCCGTTTTCAAATGACCTTGTGATAGGGAAATGGTTTTGTCCATGTTCTTGGAAATACTTGTG TATGTACAGAAGGAAGGGAGGGATTATTTTTCTACAAAGTAATTTATGATTTCTAATTTTCTAATGTGCCTTGGATATGTGCCAAATGAT >101838_101838_5_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000505365_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- >101838_101838_6_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000507959_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2348nt_BP=232nt CTGGCGGCGGCTTCCAAGCTAAGGAACGGTTTGGGGCAGTGTCGTTCCCGGAGGTCGGCCGCCGTTACCCGCTCACCAGCTACGCGGCGC GTCAGGTCCGCGGAGGCGCGGGCTCGGGGCGCCTGCGGGACGGTGAGGCCCTGCTGAGGACTCCGGACACTGCCCATCTCTAAGATAAGA ACCTGGAAAGGGGACTCTGTTGGCCATTGGAAATTGCAGAATAATGTCTCAGAGGCTGGCGAAGGTTCCTGCCAGTGGGCTTGGTGTGAA CGTGACCAGCCAGGACGGCTCCTCCCCGCTGCATGTCGCCGCCCTGCACGGCCGGGCGGACCTCATCCCCCTCCTGCTGAAGCACGGGGC CAACGCAGGTGCCAGGAACGCAGACCAAGCCGTCCCGCTCCACCTGGCCTGCCAGCAGGGCCACTTTCAGGTGGTGAAGTGTCTGTTAGA TTCGAATGCAAAACCCAATAAGAAGGACCTCAGTGGAAACACGCCCCTCATTTACGCCTGCTCCGGTGGCCATCACGAGCTTGTGGCACT GCTGCTACAGCACGGGGCCTCCATTAACGCTTCTAACAATAAGGGCAACACAGCGCTGCACGAGGCTGTGATTGAAAAGCACGTCTTCGT GGTAGAGCTGCTTCTGCTCCACGGAGCGTCAGTTCAGGTGCTGAACAAGCGGCAGCGCACGGCTGTAGACTGTGCTGAACAGAATTCAAA AATAATGGAATTGCTTCAGGTGGTACCAAGCTGTGTTGCTTCATTAGATGATGTGGCTGAAACTGACCGCAAGGAGTATGTCACTGTTAA GATCAGGAAAAAATGGAACTCAAAACTGTATGATCTACCAGATGAGCCTTTTACAAGACAGTTTTACTTTGTCCACTCAGCTGGTCAGTT TAAGGGAAAGACTTCAAGGGAGATTATGGCAAGAGATAGAAGTGTCCCTAATTTAACCGAAGGTTCTTTGCATGAGCCAGGGAGGCAAAG TGTCACACTGAGACAGAATAACCTGCCAGCTCAGAGTGGATCTCATGCTGCTGAGAAAGGCAACAGCGACTGGCCAGAGAGGCCTGGACT GACACAGACTGGCCCTGGACACAGACGGATGCTGCGGAGACACACGGTAGAGGATGCGGTCGTGTCCCAGGGCCCGGAGGCTGCTGGCCC CCTCTCCACTCCCCAAGAGGTTAGTGCTTCCCGGTCCTAACAGGAATGAGGAGTTGTTGAACCCACTGCTAGGAAGCAAGGATGCAACAA GATGATGCTGAGCGTGAACACATCTGAGAACTAAATGTGCTTCCATGAGACTGGCTTGAGAAGTCTTCAGCACCAAGTTCCTGAAAGCTT TTCTGTGGCAGGAAAGAATGCAACAAAAAAGTTAACCACCACCATCTCTCTCCTCTTCAAAGCTAATGAATACAATTGAAACAGACAAAA ATTCCAGTAGCATCCAGATCCTTAAGCCAGAGGTGCATGCTTCTTTTTAAGTATGAGGGTTTGTTGGTCACAGTGGGAGAGGTTTCACCA CCGCATTCTGACCTCCTCCTCCCAAAAGGTGCTAAACCTCTCTGACCTGTGTACATTCACAAACCACAGCTAGAATTCCTCCACCTAGGA TTAAGCTGGAGAGAAGTAAGTAATTTAGGTTTCATGGTACTGTAGAGGCCAGGCTGAAATGTCATATCTGAAGGAAGAAAGCAGCAGCTG GACAATGTTTCTTTGCAAAGCAACACTCGAACCAAAAGATGCCTCAATCCCATTTTGATATTCATTTTAGTGAAAGGATGCATCAGACCT GTTCCACATCATGCACATGGGAAAGGGTGGTTATCATTTTCCTTCTAACAAGTAGGTACAGATATTCGGTTACTACACGTGCACCTGTAG CAGTATTTCTAGAAACATCCCTTTTTGTTGAGAACCTCCCTTGAATGTCTGTCACACTCACACCTGACGGGATGGTTACTGGATTAGAGA GTAGATTTGGCACATCTTTTCTTAGTCTTTTGATTCAAATTCAAAACTTAACAGCACAAACCAGGTCAGAGTTACTTTCGGTTAGAATTT ATTGCCATTTATTCCTTTTTATAAATTTCTATAGATTATACTGTTATTTTTATGTTATTGGCCTAGAGCTACACGTATATGGGTTTGTCC TGAGTCCGTTTTCAAATGACCTTGTGATAGGGAAATGGTTTTGTCCATGTTCTTGGAAATACTTGTGTATGTACAGAAGGAAGGGAGGGA TTATTTTTCTACAAAGTAATTTATGATTTCTAATTTTCTAATGTGCCTTGGATATGTGCCAAATGATGGAAAAGAAACAGTAAACTTTAT >101838_101838_6_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000507959_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- >101838_101838_7_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000509528_ANKRD27_chr19_33098738_ENST00000306065_length(transcript)=2403nt_BP=287nt GAAATGTGGTTCGCCGTGGGCGGTGCACTGGGTCAGTCCCGCGATAGCTTCAGGCCTGGCGGCGGCTTCCAAGCTAAGGAACGGTTTGGG GCAGTGTCGTTCCCGGAGGTCGGCCGCCGTTACCCGCTCACCAGCTACGCGGCGCGTCAGGTCCGCGGAGGCGCGGGCTCGGGGCGCCTG CGGGACGGTGAGGCCCTGCTGAGGACTCCGGACACTGCCCATCTCTAAGATAAGAACCTGGAAAGGGGACTCTGTTGGCCATTGGAAATT GCAGAATAATGTCTCAGAGGCTGGCGAAGGTTCCTGCCAGTGGGCTTGGTGTGAACGTGACCAGCCAGGACGGCTCCTCCCCGCTGCATG TCGCCGCCCTGCACGGCCGGGCGGACCTCATCCCCCTCCTGCTGAAGCACGGGGCCAACGCAGGTGCCAGGAACGCAGACCAAGCCGTCC CGCTCCACCTGGCCTGCCAGCAGGGCCACTTTCAGGTGGTGAAGTGTCTGTTAGATTCGAATGCAAAACCCAATAAGAAGGACCTCAGTG GAAACACGCCCCTCATTTACGCCTGCTCCGGTGGCCATCACGAGCTTGTGGCACTGCTGCTACAGCACGGGGCCTCCATTAACGCTTCTA ACAATAAGGGCAACACAGCGCTGCACGAGGCTGTGATTGAAAAGCACGTCTTCGTGGTAGAGCTGCTTCTGCTCCACGGAGCGTCAGTTC AGGTGCTGAACAAGCGGCAGCGCACGGCTGTAGACTGTGCTGAACAGAATTCAAAAATAATGGAATTGCTTCAGGTGGTACCAAGCTGTG TTGCTTCATTAGATGATGTGGCTGAAACTGACCGCAAGGAGTATGTCACTGTTAAGATCAGGAAAAAATGGAACTCAAAACTGTATGATC TACCAGATGAGCCTTTTACAAGACAGTTTTACTTTGTCCACTCAGCTGGTCAGTTTAAGGGAAAGACTTCAAGGGAGATTATGGCAAGAG ATAGAAGTGTCCCTAATTTAACCGAAGGTTCTTTGCATGAGCCAGGGAGGCAAAGTGTCACACTGAGACAGAATAACCTGCCAGCTCAGA GTGGATCTCATGCTGCTGAGAAAGGCAACAGCGACTGGCCAGAGAGGCCTGGACTGACACAGACTGGCCCTGGACACAGACGGATGCTGC GGAGACACACGGTAGAGGATGCGGTCGTGTCCCAGGGCCCGGAGGCTGCTGGCCCCCTCTCCACTCCCCAAGAGGTTAGTGCTTCCCGGT CCTAACAGGAATGAGGAGTTGTTGAACCCACTGCTAGGAAGCAAGGATGCAACAAGATGATGCTGAGCGTGAACACATCTGAGAACTAAA TGTGCTTCCATGAGACTGGCTTGAGAAGTCTTCAGCACCAAGTTCCTGAAAGCTTTTCTGTGGCAGGAAAGAATGCAACAAAAAAGTTAA CCACCACCATCTCTCTCCTCTTCAAAGCTAATGAATACAATTGAAACAGACAAAAATTCCAGTAGCATCCAGATCCTTAAGCCAGAGGTG CATGCTTCTTTTTAAGTATGAGGGTTTGTTGGTCACAGTGGGAGAGGTTTCACCACCGCATTCTGACCTCCTCCTCCCAAAAGGTGCTAA ACCTCTCTGACCTGTGTACATTCACAAACCACAGCTAGAATTCCTCCACCTAGGATTAAGCTGGAGAGAAGTAAGTAATTTAGGTTTCAT GGTACTGTAGAGGCCAGGCTGAAATGTCATATCTGAAGGAAGAAAGCAGCAGCTGGACAATGTTTCTTTGCAAAGCAACACTCGAACCAA AAGATGCCTCAATCCCATTTTGATATTCATTTTAGTGAAAGGATGCATCAGACCTGTTCCACATCATGCACATGGGAAAGGGTGGTTATC ATTTTCCTTCTAACAAGTAGGTACAGATATTCGGTTACTACACGTGCACCTGTAGCAGTATTTCTAGAAACATCCCTTTTTGTTGAGAAC CTCCCTTGAATGTCTGTCACACTCACACCTGACGGGATGGTTACTGGATTAGAGAGTAGATTTGGCACATCTTTTCTTAGTCTTTTGATT CAAATTCAAAACTTAACAGCACAAACCAGGTCAGAGTTACTTTCGGTTAGAATTTATTGCCATTTATTCCTTTTTATAAATTTCTATAGA TTATACTGTTATTTTTATGTTATTGGCCTAGAGCTACACGTATATGGGTTTGTCCTGAGTCCGTTTTCAAATGACCTTGTGATAGGGAAA TGGTTTTGTCCATGTTCTTGGAAATACTTGTGTATGTACAGAAGGAAGGGAGGGATTATTTTTCTACAAAGTAATTTATGATTTCTAATT >101838_101838_7_ZNF302-ANKRD27_ZNF302_chr19_35169718_ENST00000509528_ANKRD27_chr19_33098738_ENST00000306065_length(amino acids)=337AA_BP=10 MLAIGNCRIMSQRLAKVPASGLGVNVTSQDGSSPLHVAALHGRADLIPLLLKHGANAGARNADQAVPLHLACQQGHFQVVKCLLDSNAKP NKKDLSGNTPLIYACSGGHHELVALLLQHGASINASNNKGNTALHEAVIEKHVFVVELLLLHGASVQVLNKRQRTAVDCAEQNSKIMELL QVVPSCVASLDDVAETDRKEYVTVKIRKKWNSKLYDLPDEPFTRQFYFVHSAGQFKGKTSREIMARDRSVPNLTEGSLHEPGRQSVTLRQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF302-ANKRD27 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 451_730 | 725.0 | 1051.0 | RAB32 | |
| Tgene | ANKRD27 | chr19:35169718 | chr19:33098738 | ENST00000306065 | 21 | 29 | 451_600 | 725.0 | 1051.0 | RAB38 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF302-ANKRD27 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF302-ANKRD27 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
