
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF480-ELL (FusionGDB2 ID:102184) |
Fusion Gene Summary for ZNF480-ELL |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF480-ELL | Fusion gene ID: 102184 | Hgene | Tgene | Gene symbol | ZNF480 | ELL | Gene ID | 147657 | 8178 |
| Gene name | zinc finger protein 480 | elongation factor for RNA polymerase II | |
| Synonyms | - | C19orf17|ELL1|MEN|PPP1R68 | |
| Cytomap | 19q13.41 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 480 | RNA polymerase II elongation factor ELLELL gene (11-19 lysine-rich leukemia gene)ELL/KMT2A fusionELL/KMT2A fusion proteinKMT2A/ELL fusionKMT2A/ELL fusion proteineleven-nineteen lysine-rich leukemia proteinelongation factor RNA polymerase IIprotein | |
| Modification date | 20200313 | 20200319 | |
| UniProtAcc | . | O00472 | |
| Ensembl transtripts involved in fusion gene | ENST00000335090, ENST00000595962, ENST00000334564, ENST00000490272, | ENST00000262809, ENST00000596124, | |
| Fusion gene scores | * DoF score | 4 X 2 X 4=32 | 11 X 17 X 7=1309 |
| # samples | 4 | 21 | |
| ** MAII score | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(21/1309*10)=-2.64000386427912 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF480 [Title/Abstract] AND ELL [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF480(52819215)-ELL(18572662), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ELL | GO:0010923 | negative regulation of phosphatase activity | 19389623 |
 Fusion gene breakpoints across ZNF480 (5'-gene) Fusion gene breakpoints across ZNF480 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
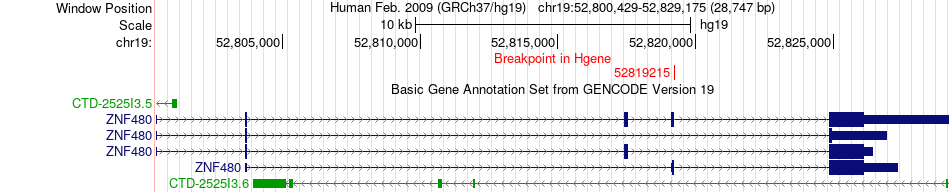 |
 Fusion gene breakpoints across ELL (3'-gene) Fusion gene breakpoints across ELL (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
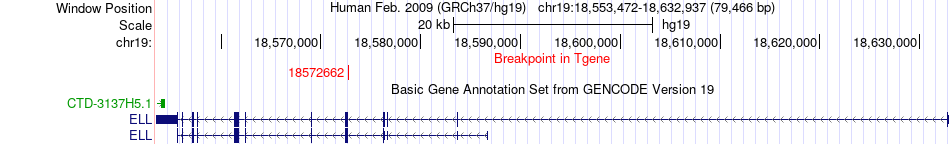 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HF-7134 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
Top |
Fusion Gene ORF analysis for ZNF480-ELL |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000335090 | ENST00000262809 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| In-frame | ENST00000335090 | ENST00000596124 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| In-frame | ENST00000595962 | ENST00000262809 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| In-frame | ENST00000595962 | ENST00000596124 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| intron-3CDS | ENST00000334564 | ENST00000262809 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| intron-3CDS | ENST00000334564 | ENST00000596124 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| intron-3CDS | ENST00000490272 | ENST00000262809 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
| intron-3CDS | ENST00000490272 | ENST00000596124 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000595962 | ZNF480 | chr19 | 52819215 | + | ENST00000262809 | ELL | chr19 | 18572662 | - | 3880 | 394 | 66 | 1790 | 574 |
| ENST00000595962 | ZNF480 | chr19 | 52819215 | + | ENST00000596124 | ELL | chr19 | 18572662 | - | 1792 | 394 | 66 | 1790 | 574 |
| ENST00000335090 | ZNF480 | chr19 | 52819215 | + | ENST00000262809 | ELL | chr19 | 18572662 | - | 3706 | 220 | 108 | 1616 | 502 |
| ENST00000335090 | ZNF480 | chr19 | 52819215 | + | ENST00000596124 | ELL | chr19 | 18572662 | - | 1618 | 220 | 108 | 1616 | 502 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000595962 | ENST00000262809 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - | 0.012542358 | 0.9874577 |
| ENST00000595962 | ENST00000596124 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - | 0.059413243 | 0.94058675 |
| ENST00000335090 | ENST00000262809 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - | 0.015285355 | 0.9847147 |
| ENST00000335090 | ENST00000596124 | ZNF480 | chr19 | 52819215 | + | ELL | chr19 | 18572662 | - | 0.069568254 | 0.9304317 |
Top |
Fusion Genomic Features for ZNF480-ELL |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
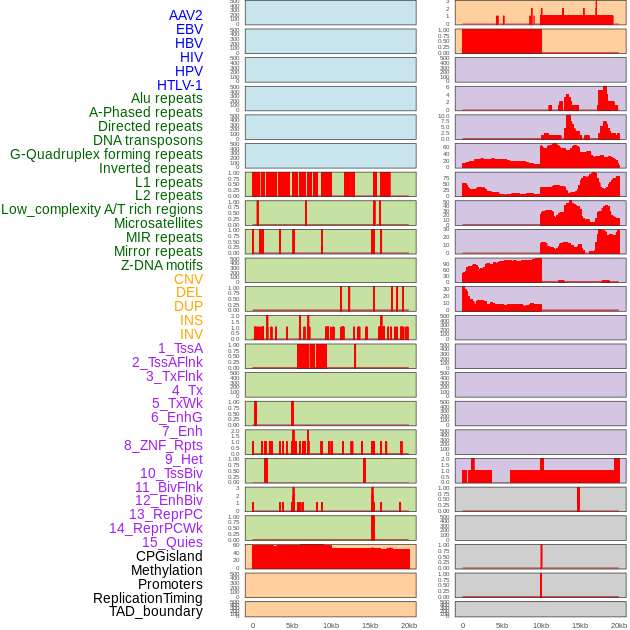 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZNF480-ELL |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:52819215/chr19:18572662) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ELL |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Elongation factor component of the super elongation complex (SEC), a complex required to increase the catalytic rate of RNA polymerase II transcription by suppressing transient pausing by the polymerase at multiple sites along the DNA. Component of the little elongation complex (LEC), a complex required to regulate small nuclear RNA (snRNA) gene transcription by RNA polymerase II and III (PubMed:22195968). Plays a role in immunoglobulin secretion in plasma cells: directs efficient alternative mRNA processing, influencing both proximal poly(A) site choice and exon skipping, as well as immunoglobulin heavy chain (IgH) alternative processing. Probably acts by regulating histone modifications accompanying transition from membrane-specific to secretory IgH mRNA expression. {ECO:0000269|PubMed:20159561, ECO:0000269|PubMed:20471948, ECO:0000269|PubMed:22195968, ECO:0000269|PubMed:23251033}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 27_98 | 109 | 536.0 | Domain | KRAB |
| Tgene | ELL | chr19:52819215 | chr19:18572662 | ENST00000262809 | 3 | 12 | 445_459 | 156 | 622.0 | Motif | Nuclear localization signal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 27_98 | 32 | 459.0 | Domain | KRAB |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 203_225 | 32 | 459.0 | Zinc finger | C2H2-type 1%3B atypical |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 231_253 | 32 | 459.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 259_281 | 32 | 459.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 287_309 | 32 | 459.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 315_337 | 32 | 459.0 | Zinc finger | C2H2-type 5%3B degenerate |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 343_365 | 32 | 459.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 371_393 | 32 | 459.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 399_421 | 32 | 459.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 427_449 | 32 | 459.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 455_477 | 32 | 459.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 483_505 | 32 | 459.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000335090 | + | 2 | 3 | 511_533 | 32 | 459.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 203_225 | 109 | 536.0 | Zinc finger | C2H2-type 1%3B atypical |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 231_253 | 109 | 536.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 259_281 | 109 | 536.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 287_309 | 109 | 536.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 315_337 | 109 | 536.0 | Zinc finger | C2H2-type 5%3B degenerate |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 343_365 | 109 | 536.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 371_393 | 109 | 536.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 399_421 | 109 | 536.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 427_449 | 109 | 536.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 455_477 | 109 | 536.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 483_505 | 109 | 536.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF480 | chr19:52819215 | chr19:18572662 | ENST00000595962 | + | 4 | 5 | 511_533 | 109 | 536.0 | Zinc finger | C2H2-type 12 |
Top |
Fusion Gene Sequence for ZNF480-ELL |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >102184_102184_1_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000335090_ELL_chr19_18572662_ENST00000262809_length(transcript)=3706nt_BP=220nt GATTGACTTCTAAAGAGTCATGCTGTGTGATGAAAAAGCCCAGAAGAGAAGGAAGAGGAAAGCAAAGGAGTCAGGGATGGCTCTTCCTCA GGAATCTCTCTTCCTGACCTGAATATTAACTCCATGTTGGAGCAAAGGAGGGAGCCCTGGTCTGGTGAGAGTGAAGTGAAAATAGCAAAA AATTCAGATGGGAGGGAGTGCATCAAAGGTGTGAACACAGGCAAGAAGGTTCAGTTTCGGAAACCAGCCCCAGGTGCAACAGACGCGGTG CCCTCCCGGAAGCGGGCAACCCCCATCAACTTGGCGAGTGCCATCAGGAAGAGTGGTGCCAGTGCCGTGAGTGGGGGCAGCGGGGTGTCC CAGAGGCCCTTCCGTGACCGAGTGCTGCACCTCCTGGCACTACGGCCCTACCGCAAGGCTGAGCTGCTGCTGCGACTGCAGAAGGACGGC CTGACGCAGGCGGACAAGGACGCGCTGGATGGCCTCCTCCAGCAGGTGGCCAACATGAGTGCTAAGGACGGCACGTGTACACTGCAGGAC TGCATGTACAAGGATGTGCAGAAGGACTGGCCTGGCTACTCGGAGGGGGACCAGCAGCTGCTGAAGCGGGTGCTCGTCCGGAAGCTGTGC CAGCCACAGAGCACTGGCAGCCTCCTTGGAGACCCTGCTGCCTCCAGCCCCCCAGGCGAGCGTGGGCGCTCGGCCTCGCCCCCACAGAAG CGGCTGCAGCCTCCTGATTTCATCGACCCCCTAGCCAACAAGAAACCCCGGATATCGCACTTCACTCAGAGAGCTCAGCCTGCCGTCAAC GGGAAGCTGGGCGTGCCCAATGGCCGTGAGGCCTTGCTGCCCACCCCGGGCCCACCAGCCAGCACGGACACCCTCAGCTCCAGCACTCAC CTGCCCCCGCGGCTGGAGCCCCCGAGGGCCCACGACCCCCTGGCCGATGTCAGCAATGACCTGGGCCACAGCGGCCGAGACTGTGAGCAC GGAGAGGCGGCTGCCCCAGCCCCCACTGTGCGCCTCGGCCTGCCCCTGCTGACGGACTGTGCCCAGCCCAGCAGGCCACACGGCAGCCCC TCGCGCAGCAAGCCCAAGAAGAAGTCCAAGAAGCACAAAGACAAGGAGAGGGCGGCTGAGGACAAGCCCCGGGCCCAGCTTCCAGACTGT GCACCTGCCACCCATGCCACCCCCGGAGCCCCAGCAGACACCCCAGGTTTAAACGGAACCTGCAGCGTTTCCAGTGTTCCCACGTCCACG TCGGAGACGCCTGACTACTTGCTGAAGTACGCAGCCATCTCCTCTTCGGAGCAGCGCCAGAGCTACAAGAACGACTTCAATGCCGAGTAC AGCGAGTACCGCGACCTGCACGCCCGCATTGAGCGCATCACGCGGCGGTTCACCCAGCTCGACGCCCAGCTCCGGCAGCTCTCCCAGGGC TCCGAGGAGTATGAGACTACTCGAGGGCAGATTTTGCAGGAATATCGAAAAATCAAAAAGACCAACACCAACTACAGCCAGGAGAAGCAC CGCTGCGAGTACCTGCACAGCAAGCTGGCCCACATCAAGAGGCTCATCGCCGAGTACGACCAGCGGCAGCTGCAGGCTTGGCCCTAGCCG CCCTCCCCGATGGCGGGGATCTGGGAGGGTCGGGGGAGCAAAAGGCGGTGAGAGAGGATTTATTTAAAAAAATAAACCCGAGGAAGATGC TCATCTGAGCCAGCACCGCCGGCTTTCAGGGCAGCCCCTGCAGACGTCTGGCCCTGGCGGGTGGCTGCAAGCCCACCTCGGCCCTCCCTG CGCTTCCTGAGCAGTCCCTGCTTATGATGGGCTCCCCAGGAAGCCCACTGCCTCCTCCCTGGCTGCAGCCTCCGGGGTTCAGCCTCCTGT CTCGCCAGAAGACTCCTAGGCCCTTGGGGTGGCGCGCCTGCCTTTTCTAGTTTTATACAAAGACAGCCACTTTTAGCTACTGCTAATGAG ACTTGAGTCTATTTTTGTACAAAAGAGAAGCAAAATCTTTTTTCTAAACCTGTGCCTCCCTCTCCTCGGACACCCAGGGTCTAGCCGCTG CCCTGGGTCCTGCCTGCAGTATTACAGAGGTCGTCGAAAGGTGCAGCTGCGTTCTGAGGGCGTGGGGAATGGGCAGGTGGCCTCTGCTGG TCTCTGGCTCTACGTTTAGGCACCCCTTTCCCCAGCCTCTCCTCCTTGGGCAGGGTCTCTGCTCCAGCAAGCAAACAGGGTGGCCCAGGT GGCTATTTTGAGAACTCCAGCTGGTGCCCCCAGACAGCTGCTCAGAGCCAAGGGGGCAGAGGGCTTTCAGCGCCCCCAGGCCTGCCCTGC TATTTCAGGCCCTCAGCTGTCGGGGGCCACTGTGTTTCTGTGCTCCAAGTTGAGACTCGGCCGCAGCGGCGTCAGACTTTTCTTTGCGAT GTCCTCGGTTTCCCATTTGTTGCTGCTGCTGCTCATTCCACACTGTTGAGACCTTGTGGTCTCGATGCTGCTGGCCTCCCTCCGTCCCTC TGTCCACTTGTGGGTCCTGGGGTCGTCTTAGCAGAGCCACCGCCAAGGTTTGCACTCAGGGAGGGGCTGATGGAGGGCTGGGCGCCATGG GGTCAGGGAGGCCCCAGGGACCCATGGGCAGAGCTGGGGAGCCAGGAGGATGGGTCTGCCTGCTCTGGGGGTGCAGGCTCTGCAGACACC CCAATTGCTGCTCTGTTGCTGTGCCTAGTCTAGTGTCCCCCAAAGCATCGGGGGGCCACGCAAGTGTGGGTGTGTGAGTGTGCGTATGTG GGGACCCCCCAGGGCATGGTGGGGTCTGAGGCGTGTGTTCCCATTTAGGGCTGAATGAAGCCATGACTTGGAAAGGTGGTGGGGGGTGGA TGGCGGCTGTGACTCAGGGCCCAGGGATCACATGGGGGTCACTGCTGCCATCAGCAGTCAGGCAGGGAGAACAGAAATGCAAAAATACAG ATGTCTATTTTTCTAAACTGGGGGGTGGAGGGGTGCCTCCTGACAGCTTCCAAAGAGCTGACTGTGGAGGGCCCACAGTGTATCCCACCC CTGCCCGAGAGTCTCTCTCCTTTGGGACCCCCGGGTTCCCAGAGCTCCGAGGAAACTGATCCTTCATGTTTCAATAGCTGTTGTTGTTTT TCTCTTTGGGCCGGGCCGGGCCGGGGAAGGGCCGAGCAGGCTCCAGGAGGCCTCCAGAGTCAGCTGGACCTTGCTGTGTCGCCCGCAGGC AGCCCTGCAGACACCGGTGAGGTGGGATGATTTCAGGAGGTCCCAGCCCGGGACTGCCCGCCCCTCTGCCCTGGGAACCAGTCCAAGGGG CTGTCAGTTGAATTTGCCTTTTTGTTGTTGTTGTTGTTTTTGTTGTTTATTTGTTCTCCTTTTAAGGAATTATGAAGCTAAGAAAAGTTT TGTGTTTTGGGGGTTGGTGGACAAAGTTCTAGCTGATTAAATATGGTCTAACTTATTGATACTATTTTTTTTTCTTTTAATGTTGTACTG TGGAGAAAAACTATTTTGAGCCATGGGGTTGGTGTTTACAAGAACTTTCCACTTTTGCCAAAATGTTACAATTGAAAGGCATCTATCTGC GTCTTCAGTTTCCTGATATTTTCTTAAAAGATCTAGTGTCCTTTTTGTACACTAAACAGGTGAATGCTGACTTTTTTCATTCTCCAATAA >102184_102184_1_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000335090_ELL_chr19_18572662_ENST00000262809_length(amino acids)=502AA_BP=37 MNINSMLEQRREPWSGESEVKIAKNSDGRECIKGVNTGKKVQFRKPAPGATDAVPSRKRATPINLASAIRKSGASAVSGGSGVSQRPFRD RVLHLLALRPYRKAELLLRLQKDGLTQADKDALDGLLQQVANMSAKDGTCTLQDCMYKDVQKDWPGYSEGDQQLLKRVLVRKLCQPQSTG SLLGDPAASSPPGERGRSASPPQKRLQPPDFIDPLANKKPRISHFTQRAQPAVNGKLGVPNGREALLPTPGPPASTDTLSSSTHLPPRLE PPRAHDPLADVSNDLGHSGRDCEHGEAAAPAPTVRLGLPLLTDCAQPSRPHGSPSRSKPKKKSKKHKDKERAAEDKPRAQLPDCAPATHA TPGAPADTPGLNGTCSVSSVPTSTSETPDYLLKYAAISSSEQRQSYKNDFNAEYSEYRDLHARIERITRRFTQLDAQLRQLSQGSEEYET -------------------------------------------------------------- >102184_102184_2_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000335090_ELL_chr19_18572662_ENST00000596124_length(transcript)=1618nt_BP=220nt GATTGACTTCTAAAGAGTCATGCTGTGTGATGAAAAAGCCCAGAAGAGAAGGAAGAGGAAAGCAAAGGAGTCAGGGATGGCTCTTCCTCA GGAATCTCTCTTCCTGACCTGAATATTAACTCCATGTTGGAGCAAAGGAGGGAGCCCTGGTCTGGTGAGAGTGAAGTGAAAATAGCAAAA AATTCAGATGGGAGGGAGTGCATCAAAGGTGTGAACACAGGCAAGAAGGTTCAGTTTCGGAAACCAGCCCCAGGTGCAACAGACGCGGTG CCCTCCCGGAAGCGGGCAACCCCCATCAACTTGGCGAGTGCCATCAGGAAGAGTGGTGCCAGTGCCGTGAGTGGGGGCAGCGGGGTGTCC CAGAGGCCCTTCCGTGACCGAGTGCTGCACCTCCTGGCACTACGGCCCTACCGCAAGGCTGAGCTGCTGCTGCGACTGCAGAAGGACGGC CTGACGCAGGCGGACAAGGACGCGCTGGATGGCCTCCTCCAGCAGGTGGCCAACATGAGTGCTAAGGACGGCACGTGTACACTGCAGGAC TGCATGTACAAGGATGTGCAGAAGGACTGGCCTGGCTACTCGGAGGGGGACCAGCAGCTGCTGAAGCGGGTGCTCGTCCGGAAGCTGTGC CAGCCACAGAGCACTGGCAGCCTCCTTGGAGACCCTGCTGCCTCCAGCCCCCCAGGCGAGCGTGGGCGCTCGGCCTCGCCCCCACAGAAG CGGCTGCAGCCTCCTGATTTCATCGACCCCCTAGCCAACAAGAAACCCCGGATATCGCACTTCACTCAGAGAGCTCAGCCTGCCGTCAAC GGGAAGCTGGGCGTGCCCAATGGCCGTGAGGCCTTGCTGCCCACCCCGGGCCCACCAGCCAGCACGGACACCCTCAGCTCCAGCACTCAC CTGCCCCCGCGGCTGGAGCCCCCGAGGGCCCACGACCCCCTGGCCGATGTCAGCAATGACCTGGGCCACAGCGGCCGAGACTGTGAGCAC GGAGAGGCGGCTGCCCCAGCCCCCACTGTGCGCCTCGGCCTGCCCCTGCTGACGGACTGTGCCCAGCCCAGCAGGCCACACGGCAGCCCC TCGCGCAGCAAGCCCAAGAAGAAGTCCAAGAAGCACAAAGACAAGGAGAGGGCGGCTGAGGACAAGCCCCGGGCCCAGCTTCCAGACTGT GCACCTGCCACCCATGCCACCCCCGGAGCCCCAGCAGACACCCCAGGTTTAAACGGAACCTGCAGCGTTTCCAGTGTTCCCACGTCCACG TCGGAGACGCCTGACTACTTGCTGAAGTACGCAGCCATCTCCTCTTCGGAGCAGCGCCAGAGCTACAAGAACGACTTCAATGCCGAGTAC AGCGAGTACCGCGACCTGCACGCCCGCATTGAGCGCATCACGCGGCGGTTCACCCAGCTCGACGCCCAGCTCCGGCAGCTCTCCCAGGGC TCCGAGGAGTATGAGACTACTCGAGGGCAGATTTTGCAGGAATATCGAAAAATCAAAAAGACCAACACCAACTACAGCCAGGAGAAGCAC >102184_102184_2_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000335090_ELL_chr19_18572662_ENST00000596124_length(amino acids)=502AA_BP=37 MNINSMLEQRREPWSGESEVKIAKNSDGRECIKGVNTGKKVQFRKPAPGATDAVPSRKRATPINLASAIRKSGASAVSGGSGVSQRPFRD RVLHLLALRPYRKAELLLRLQKDGLTQADKDALDGLLQQVANMSAKDGTCTLQDCMYKDVQKDWPGYSEGDQQLLKRVLVRKLCQPQSTG SLLGDPAASSPPGERGRSASPPQKRLQPPDFIDPLANKKPRISHFTQRAQPAVNGKLGVPNGREALLPTPGPPASTDTLSSSTHLPPRLE PPRAHDPLADVSNDLGHSGRDCEHGEAAAPAPTVRLGLPLLTDCAQPSRPHGSPSRSKPKKKSKKHKDKERAAEDKPRAQLPDCAPATHA TPGAPADTPGLNGTCSVSSVPTSTSETPDYLLKYAAISSSEQRQSYKNDFNAEYSEYRDLHARIERITRRFTQLDAQLRQLSQGSEEYET -------------------------------------------------------------- >102184_102184_3_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000595962_ELL_chr19_18572662_ENST00000262809_length(transcript)=3880nt_BP=394nt ACAAACCCGGAAGCGGATCGCGTGGAGTGAAGGTCACGCCGCGGCGCGATTGACTTCTAAAGAGTCATGCTGTGTGATGAAAAAGCCCAG AAGAGAAGGAAGAGGAAAGCAAAGGAGTCAGGGATGGCTCTTCCTCAGGGACACTTAACATTCAGGGACGTGGCCATAGAATTCTCTCAG GCGGAGTGGAAATGCCTGGACCCTGCACAGAGGGCTTTATACAAGGATGTGATGTTGGAGAACTACAGGAACCTGGTCTCCCTGGGAATC TCTCTTCCTGACCTGAATATTAACTCCATGTTGGAGCAAAGGAGGGAGCCCTGGTCTGGTGAGAGTGAAGTGAAAATAGCAAAAAATTCA GATGGGAGGGAGTGCATCAAAGGTGTGAACACAGGCAAGAAGGTTCAGTTTCGGAAACCAGCCCCAGGTGCAACAGACGCGGTGCCCTCC CGGAAGCGGGCAACCCCCATCAACTTGGCGAGTGCCATCAGGAAGAGTGGTGCCAGTGCCGTGAGTGGGGGCAGCGGGGTGTCCCAGAGG CCCTTCCGTGACCGAGTGCTGCACCTCCTGGCACTACGGCCCTACCGCAAGGCTGAGCTGCTGCTGCGACTGCAGAAGGACGGCCTGACG CAGGCGGACAAGGACGCGCTGGATGGCCTCCTCCAGCAGGTGGCCAACATGAGTGCTAAGGACGGCACGTGTACACTGCAGGACTGCATG TACAAGGATGTGCAGAAGGACTGGCCTGGCTACTCGGAGGGGGACCAGCAGCTGCTGAAGCGGGTGCTCGTCCGGAAGCTGTGCCAGCCA CAGAGCACTGGCAGCCTCCTTGGAGACCCTGCTGCCTCCAGCCCCCCAGGCGAGCGTGGGCGCTCGGCCTCGCCCCCACAGAAGCGGCTG CAGCCTCCTGATTTCATCGACCCCCTAGCCAACAAGAAACCCCGGATATCGCACTTCACTCAGAGAGCTCAGCCTGCCGTCAACGGGAAG CTGGGCGTGCCCAATGGCCGTGAGGCCTTGCTGCCCACCCCGGGCCCACCAGCCAGCACGGACACCCTCAGCTCCAGCACTCACCTGCCC CCGCGGCTGGAGCCCCCGAGGGCCCACGACCCCCTGGCCGATGTCAGCAATGACCTGGGCCACAGCGGCCGAGACTGTGAGCACGGAGAG GCGGCTGCCCCAGCCCCCACTGTGCGCCTCGGCCTGCCCCTGCTGACGGACTGTGCCCAGCCCAGCAGGCCACACGGCAGCCCCTCGCGC AGCAAGCCCAAGAAGAAGTCCAAGAAGCACAAAGACAAGGAGAGGGCGGCTGAGGACAAGCCCCGGGCCCAGCTTCCAGACTGTGCACCT GCCACCCATGCCACCCCCGGAGCCCCAGCAGACACCCCAGGTTTAAACGGAACCTGCAGCGTTTCCAGTGTTCCCACGTCCACGTCGGAG ACGCCTGACTACTTGCTGAAGTACGCAGCCATCTCCTCTTCGGAGCAGCGCCAGAGCTACAAGAACGACTTCAATGCCGAGTACAGCGAG TACCGCGACCTGCACGCCCGCATTGAGCGCATCACGCGGCGGTTCACCCAGCTCGACGCCCAGCTCCGGCAGCTCTCCCAGGGCTCCGAG GAGTATGAGACTACTCGAGGGCAGATTTTGCAGGAATATCGAAAAATCAAAAAGACCAACACCAACTACAGCCAGGAGAAGCACCGCTGC GAGTACCTGCACAGCAAGCTGGCCCACATCAAGAGGCTCATCGCCGAGTACGACCAGCGGCAGCTGCAGGCTTGGCCCTAGCCGCCCTCC CCGATGGCGGGGATCTGGGAGGGTCGGGGGAGCAAAAGGCGGTGAGAGAGGATTTATTTAAAAAAATAAACCCGAGGAAGATGCTCATCT GAGCCAGCACCGCCGGCTTTCAGGGCAGCCCCTGCAGACGTCTGGCCCTGGCGGGTGGCTGCAAGCCCACCTCGGCCCTCCCTGCGCTTC CTGAGCAGTCCCTGCTTATGATGGGCTCCCCAGGAAGCCCACTGCCTCCTCCCTGGCTGCAGCCTCCGGGGTTCAGCCTCCTGTCTCGCC AGAAGACTCCTAGGCCCTTGGGGTGGCGCGCCTGCCTTTTCTAGTTTTATACAAAGACAGCCACTTTTAGCTACTGCTAATGAGACTTGA GTCTATTTTTGTACAAAAGAGAAGCAAAATCTTTTTTCTAAACCTGTGCCTCCCTCTCCTCGGACACCCAGGGTCTAGCCGCTGCCCTGG GTCCTGCCTGCAGTATTACAGAGGTCGTCGAAAGGTGCAGCTGCGTTCTGAGGGCGTGGGGAATGGGCAGGTGGCCTCTGCTGGTCTCTG GCTCTACGTTTAGGCACCCCTTTCCCCAGCCTCTCCTCCTTGGGCAGGGTCTCTGCTCCAGCAAGCAAACAGGGTGGCCCAGGTGGCTAT TTTGAGAACTCCAGCTGGTGCCCCCAGACAGCTGCTCAGAGCCAAGGGGGCAGAGGGCTTTCAGCGCCCCCAGGCCTGCCCTGCTATTTC AGGCCCTCAGCTGTCGGGGGCCACTGTGTTTCTGTGCTCCAAGTTGAGACTCGGCCGCAGCGGCGTCAGACTTTTCTTTGCGATGTCCTC GGTTTCCCATTTGTTGCTGCTGCTGCTCATTCCACACTGTTGAGACCTTGTGGTCTCGATGCTGCTGGCCTCCCTCCGTCCCTCTGTCCA CTTGTGGGTCCTGGGGTCGTCTTAGCAGAGCCACCGCCAAGGTTTGCACTCAGGGAGGGGCTGATGGAGGGCTGGGCGCCATGGGGTCAG GGAGGCCCCAGGGACCCATGGGCAGAGCTGGGGAGCCAGGAGGATGGGTCTGCCTGCTCTGGGGGTGCAGGCTCTGCAGACACCCCAATT GCTGCTCTGTTGCTGTGCCTAGTCTAGTGTCCCCCAAAGCATCGGGGGGCCACGCAAGTGTGGGTGTGTGAGTGTGCGTATGTGGGGACC CCCCAGGGCATGGTGGGGTCTGAGGCGTGTGTTCCCATTTAGGGCTGAATGAAGCCATGACTTGGAAAGGTGGTGGGGGGTGGATGGCGG CTGTGACTCAGGGCCCAGGGATCACATGGGGGTCACTGCTGCCATCAGCAGTCAGGCAGGGAGAACAGAAATGCAAAAATACAGATGTCT ATTTTTCTAAACTGGGGGGTGGAGGGGTGCCTCCTGACAGCTTCCAAAGAGCTGACTGTGGAGGGCCCACAGTGTATCCCACCCCTGCCC GAGAGTCTCTCTCCTTTGGGACCCCCGGGTTCCCAGAGCTCCGAGGAAACTGATCCTTCATGTTTCAATAGCTGTTGTTGTTTTTCTCTT TGGGCCGGGCCGGGCCGGGGAAGGGCCGAGCAGGCTCCAGGAGGCCTCCAGAGTCAGCTGGACCTTGCTGTGTCGCCCGCAGGCAGCCCT GCAGACACCGGTGAGGTGGGATGATTTCAGGAGGTCCCAGCCCGGGACTGCCCGCCCCTCTGCCCTGGGAACCAGTCCAAGGGGCTGTCA GTTGAATTTGCCTTTTTGTTGTTGTTGTTGTTTTTGTTGTTTATTTGTTCTCCTTTTAAGGAATTATGAAGCTAAGAAAAGTTTTGTGTT TTGGGGGTTGGTGGACAAAGTTCTAGCTGATTAAATATGGTCTAACTTATTGATACTATTTTTTTTTCTTTTAATGTTGTACTGTGGAGA AAAACTATTTTGAGCCATGGGGTTGGTGTTTACAAGAACTTTCCACTTTTGCCAAAATGTTACAATTGAAAGGCATCTATCTGCGTCTTC AGTTTCCTGATATTTTCTTAAAAGATCTAGTGTCCTTTTTGTACACTAAACAGGTGAATGCTGACTTTTTTCATTCTCCAATAAATTTCA >102184_102184_3_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000595962_ELL_chr19_18572662_ENST00000262809_length(amino acids)=574AA_BP=109 MLCDEKAQKRRKRKAKESGMALPQGHLTFRDVAIEFSQAEWKCLDPAQRALYKDVMLENYRNLVSLGISLPDLNINSMLEQRREPWSGES EVKIAKNSDGRECIKGVNTGKKVQFRKPAPGATDAVPSRKRATPINLASAIRKSGASAVSGGSGVSQRPFRDRVLHLLALRPYRKAELLL RLQKDGLTQADKDALDGLLQQVANMSAKDGTCTLQDCMYKDVQKDWPGYSEGDQQLLKRVLVRKLCQPQSTGSLLGDPAASSPPGERGRS ASPPQKRLQPPDFIDPLANKKPRISHFTQRAQPAVNGKLGVPNGREALLPTPGPPASTDTLSSSTHLPPRLEPPRAHDPLADVSNDLGHS GRDCEHGEAAAPAPTVRLGLPLLTDCAQPSRPHGSPSRSKPKKKSKKHKDKERAAEDKPRAQLPDCAPATHATPGAPADTPGLNGTCSVS SVPTSTSETPDYLLKYAAISSSEQRQSYKNDFNAEYSEYRDLHARIERITRRFTQLDAQLRQLSQGSEEYETTRGQILQEYRKIKKTNTN -------------------------------------------------------------- >102184_102184_4_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000595962_ELL_chr19_18572662_ENST00000596124_length(transcript)=1792nt_BP=394nt ACAAACCCGGAAGCGGATCGCGTGGAGTGAAGGTCACGCCGCGGCGCGATTGACTTCTAAAGAGTCATGCTGTGTGATGAAAAAGCCCAG AAGAGAAGGAAGAGGAAAGCAAAGGAGTCAGGGATGGCTCTTCCTCAGGGACACTTAACATTCAGGGACGTGGCCATAGAATTCTCTCAG GCGGAGTGGAAATGCCTGGACCCTGCACAGAGGGCTTTATACAAGGATGTGATGTTGGAGAACTACAGGAACCTGGTCTCCCTGGGAATC TCTCTTCCTGACCTGAATATTAACTCCATGTTGGAGCAAAGGAGGGAGCCCTGGTCTGGTGAGAGTGAAGTGAAAATAGCAAAAAATTCA GATGGGAGGGAGTGCATCAAAGGTGTGAACACAGGCAAGAAGGTTCAGTTTCGGAAACCAGCCCCAGGTGCAACAGACGCGGTGCCCTCC CGGAAGCGGGCAACCCCCATCAACTTGGCGAGTGCCATCAGGAAGAGTGGTGCCAGTGCCGTGAGTGGGGGCAGCGGGGTGTCCCAGAGG CCCTTCCGTGACCGAGTGCTGCACCTCCTGGCACTACGGCCCTACCGCAAGGCTGAGCTGCTGCTGCGACTGCAGAAGGACGGCCTGACG CAGGCGGACAAGGACGCGCTGGATGGCCTCCTCCAGCAGGTGGCCAACATGAGTGCTAAGGACGGCACGTGTACACTGCAGGACTGCATG TACAAGGATGTGCAGAAGGACTGGCCTGGCTACTCGGAGGGGGACCAGCAGCTGCTGAAGCGGGTGCTCGTCCGGAAGCTGTGCCAGCCA CAGAGCACTGGCAGCCTCCTTGGAGACCCTGCTGCCTCCAGCCCCCCAGGCGAGCGTGGGCGCTCGGCCTCGCCCCCACAGAAGCGGCTG CAGCCTCCTGATTTCATCGACCCCCTAGCCAACAAGAAACCCCGGATATCGCACTTCACTCAGAGAGCTCAGCCTGCCGTCAACGGGAAG CTGGGCGTGCCCAATGGCCGTGAGGCCTTGCTGCCCACCCCGGGCCCACCAGCCAGCACGGACACCCTCAGCTCCAGCACTCACCTGCCC CCGCGGCTGGAGCCCCCGAGGGCCCACGACCCCCTGGCCGATGTCAGCAATGACCTGGGCCACAGCGGCCGAGACTGTGAGCACGGAGAG GCGGCTGCCCCAGCCCCCACTGTGCGCCTCGGCCTGCCCCTGCTGACGGACTGTGCCCAGCCCAGCAGGCCACACGGCAGCCCCTCGCGC AGCAAGCCCAAGAAGAAGTCCAAGAAGCACAAAGACAAGGAGAGGGCGGCTGAGGACAAGCCCCGGGCCCAGCTTCCAGACTGTGCACCT GCCACCCATGCCACCCCCGGAGCCCCAGCAGACACCCCAGGTTTAAACGGAACCTGCAGCGTTTCCAGTGTTCCCACGTCCACGTCGGAG ACGCCTGACTACTTGCTGAAGTACGCAGCCATCTCCTCTTCGGAGCAGCGCCAGAGCTACAAGAACGACTTCAATGCCGAGTACAGCGAG TACCGCGACCTGCACGCCCGCATTGAGCGCATCACGCGGCGGTTCACCCAGCTCGACGCCCAGCTCCGGCAGCTCTCCCAGGGCTCCGAG GAGTATGAGACTACTCGAGGGCAGATTTTGCAGGAATATCGAAAAATCAAAAAGACCAACACCAACTACAGCCAGGAGAAGCACCGCTGC >102184_102184_4_ZNF480-ELL_ZNF480_chr19_52819215_ENST00000595962_ELL_chr19_18572662_ENST00000596124_length(amino acids)=574AA_BP=109 MLCDEKAQKRRKRKAKESGMALPQGHLTFRDVAIEFSQAEWKCLDPAQRALYKDVMLENYRNLVSLGISLPDLNINSMLEQRREPWSGES EVKIAKNSDGRECIKGVNTGKKVQFRKPAPGATDAVPSRKRATPINLASAIRKSGASAVSGGSGVSQRPFRDRVLHLLALRPYRKAELLL RLQKDGLTQADKDALDGLLQQVANMSAKDGTCTLQDCMYKDVQKDWPGYSEGDQQLLKRVLVRKLCQPQSTGSLLGDPAASSPPGERGRS ASPPQKRLQPPDFIDPLANKKPRISHFTQRAQPAVNGKLGVPNGREALLPTPGPPASTDTLSSSTHLPPRLEPPRAHDPLADVSNDLGHS GRDCEHGEAAAPAPTVRLGLPLLTDCAQPSRPHGSPSRSKPKKKSKKHKDKERAAEDKPRAQLPDCAPATHATPGAPADTPGLNGTCSVS SVPTSTSETPDYLLKYAAISSSEQRQSYKNDFNAEYSEYRDLHARIERITRRFTQLDAQLRQLSQGSEEYETTRGQILQEYRKIKKTNTN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF480-ELL |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF480-ELL |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF480-ELL |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
