
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF511-PPAPDC1A (FusionGDB2 ID:102230) |
Fusion Gene Summary for ZNF511-PPAPDC1A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF511-PPAPDC1A | Fusion gene ID: 102230 | Hgene | Tgene | Gene symbol | ZNF511 | PPAPDC1A | Gene ID | 118472 | 196051 |
| Gene name | zinc finger protein 511 | phospholipid phosphatase 4 | |
| Synonyms | Zfp511 | DPPL2|PPAPDC1|PPAPDC1A | |
| Cytomap | 10q26.3 | 10q26.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 511 | phospholipid phosphatase 4diacylglycerol pyrophosphate like 2diacylglycerol pyrophosphate phosphatase-like 2phosphatidate phosphatase PPAPDC1Aphosphatidic acid phosphatase type 2 domain containing 1Aphosphatidic acid phosphatase type 2 domain-contain | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000463816, ENST00000359035, ENST00000361518, ENST00000368554, | ENST00000369073, ENST00000398248, ENST00000398250, ENST00000439221, ENST00000541332, ENST00000496437, | |
| Fusion gene scores | * DoF score | 2 X 1 X 2=4 | 8 X 6 X 6=288 |
| # samples | 2 | 13 | |
| ** MAII score | log2(2/4*10)=2.32192809488736 | log2(13/288*10)=-1.14755718841386 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF511 [Title/Abstract] AND PPAPDC1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF511(135125345)-PPAPDC1A(122263330), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ZNF511-PPAPDC1A seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ZNF511-PPAPDC1A seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | PPAPDC1A | GO:0046839 | phospholipid dephosphorylation | 17590538 |
 Fusion gene breakpoints across ZNF511 (5'-gene) Fusion gene breakpoints across ZNF511 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
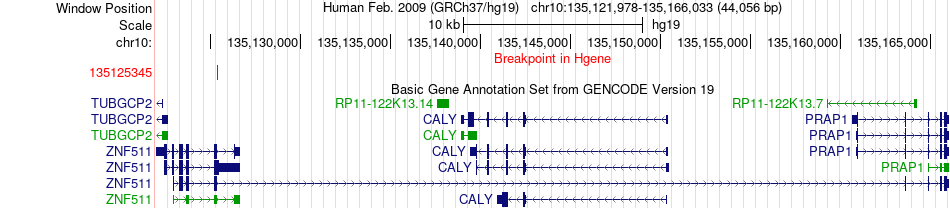 |
 Fusion gene breakpoints across PPAPDC1A (3'-gene) Fusion gene breakpoints across PPAPDC1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
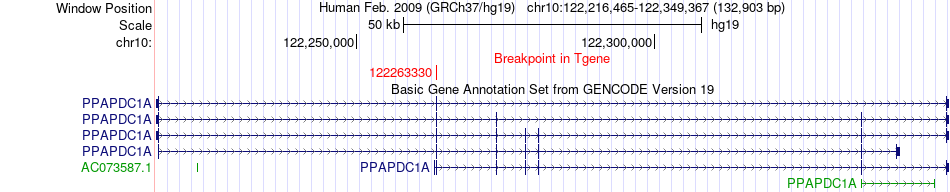 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A8BM-01A | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
Top |
Fusion Gene ORF analysis for ZNF511-PPAPDC1A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000463816 | ENST00000369073 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 3UTR-3CDS | ENST00000463816 | ENST00000398248 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 3UTR-3CDS | ENST00000463816 | ENST00000398250 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 3UTR-3CDS | ENST00000463816 | ENST00000439221 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 3UTR-3CDS | ENST00000463816 | ENST00000541332 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 3UTR-intron | ENST00000463816 | ENST00000496437 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 5CDS-intron | ENST00000359035 | ENST00000496437 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 5CDS-intron | ENST00000361518 | ENST00000496437 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| 5CDS-intron | ENST00000368554 | ENST00000496437 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| Frame-shift | ENST00000359035 | ENST00000369073 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| Frame-shift | ENST00000359035 | ENST00000398248 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| Frame-shift | ENST00000359035 | ENST00000398250 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| Frame-shift | ENST00000359035 | ENST00000439221 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| Frame-shift | ENST00000359035 | ENST00000541332 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000361518 | ENST00000369073 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000361518 | ENST00000398248 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000361518 | ENST00000398250 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000361518 | ENST00000439221 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000361518 | ENST00000541332 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000368554 | ENST00000369073 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000368554 | ENST00000398248 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000368554 | ENST00000398250 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000368554 | ENST00000439221 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
| In-frame | ENST00000368554 | ENST00000541332 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000368554 | ZNF511 | chr10 | 135125345 | - | ENST00000398250 | PPAPDC1A | chr10 | 122263330 | + | 1620 | 507 | 22 | 1266 | 414 |
| ENST00000368554 | ZNF511 | chr10 | 135125345 | - | ENST00000439221 | PPAPDC1A | chr10 | 122263330 | + | 1431 | 507 | 22 | 1077 | 351 |
| ENST00000368554 | ZNF511 | chr10 | 135125345 | - | ENST00000398248 | PPAPDC1A | chr10 | 122263330 | + | 1169 | 507 | 22 | 741 | 239 |
| ENST00000368554 | ZNF511 | chr10 | 135125345 | - | ENST00000541332 | PPAPDC1A | chr10 | 122263330 | + | 1816 | 507 | 22 | 1152 | 376 |
| ENST00000368554 | ZNF511 | chr10 | 135125345 | - | ENST00000369073 | PPAPDC1A | chr10 | 122263330 | + | 1616 | 507 | 22 | 1266 | 414 |
| ENST00000361518 | ZNF511 | chr10 | 135125345 | - | ENST00000398250 | PPAPDC1A | chr10 | 122263330 | + | 2262 | 1149 | 445 | 1908 | 487 |
| ENST00000361518 | ZNF511 | chr10 | 135125345 | - | ENST00000439221 | PPAPDC1A | chr10 | 122263330 | + | 2073 | 1149 | 445 | 1719 | 424 |
| ENST00000361518 | ZNF511 | chr10 | 135125345 | - | ENST00000398248 | PPAPDC1A | chr10 | 122263330 | + | 1811 | 1149 | 445 | 1383 | 312 |
| ENST00000361518 | ZNF511 | chr10 | 135125345 | - | ENST00000541332 | PPAPDC1A | chr10 | 122263330 | + | 2458 | 1149 | 445 | 1794 | 449 |
| ENST00000361518 | ZNF511 | chr10 | 135125345 | - | ENST00000369073 | PPAPDC1A | chr10 | 122263330 | + | 2258 | 1149 | 445 | 1908 | 487 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000368554 | ENST00000398250 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.004437678 | 0.9955623 |
| ENST00000368554 | ENST00000439221 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.004211773 | 0.9957883 |
| ENST00000368554 | ENST00000398248 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.016174484 | 0.9838255 |
| ENST00000368554 | ENST00000541332 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.023075469 | 0.97692454 |
| ENST00000368554 | ENST00000369073 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.004597897 | 0.9954021 |
| ENST00000361518 | ENST00000398250 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.005767835 | 0.9942321 |
| ENST00000361518 | ENST00000439221 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.008003203 | 0.9919968 |
| ENST00000361518 | ENST00000398248 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.023834774 | 0.9761653 |
| ENST00000361518 | ENST00000541332 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.024196023 | 0.975804 |
| ENST00000361518 | ENST00000369073 | ZNF511 | chr10 | 135125345 | - | PPAPDC1A | chr10 | 122263330 | + | 0.00591819 | 0.9940818 |
Top |
Fusion Genomic Features for ZNF511-PPAPDC1A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZNF511-PPAPDC1A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:135125345/chr10:122263330) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF511 | chr10:135125345 | chr10:122263330 | ENST00000361518 | - | 5 | 6 | 107_130 | 226 | 253.0 | Zinc finger | Note=C2H2-type 2 |
| Hgene | ZNF511 | chr10:135125345 | chr10:122263330 | ENST00000361518 | - | 5 | 6 | 144_169 | 226 | 253.0 | Zinc finger | Note=C2H2-type 3 |
| Hgene | ZNF511 | chr10:135125345 | chr10:122263330 | ENST00000361518 | - | 5 | 6 | 80_105 | 226 | 253.0 | Zinc finger | Note=C2H2-type 1 |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 102_110 | 18 | 97.0 | Region | Phosphatase sequence motif I | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 143_146 | 18 | 97.0 | Region | Phosphatase sequence motif II | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 195_205 | 18 | 97.0 | Region | Phosphatase sequence motif III | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 102_110 | 18 | 272.0 | Region | Phosphatase sequence motif I | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 143_146 | 18 | 272.0 | Region | Phosphatase sequence motif II | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 195_205 | 18 | 272.0 | Region | Phosphatase sequence motif III | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 102_110 | 18 | 209.0 | Region | Phosphatase sequence motif I | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 143_146 | 18 | 209.0 | Region | Phosphatase sequence motif II | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 195_205 | 18 | 209.0 | Region | Phosphatase sequence motif III | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 142_162 | 18 | 97.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 179_199 | 18 | 97.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 202_222 | 18 | 97.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 49_69 | 18 | 97.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 84_104 | 18 | 97.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 142_162 | 18 | 272.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 179_199 | 18 | 272.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 202_222 | 18 | 272.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 49_69 | 18 | 272.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 84_104 | 18 | 272.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 142_162 | 18 | 209.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 179_199 | 18 | 209.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 202_222 | 18 | 209.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 49_69 | 18 | 209.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 84_104 | 18 | 209.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF511 | chr10:135125345 | chr10:122263330 | ENST00000359035 | - | 1 | 5 | 107_130 | 0 | 263.0 | Zinc finger | Note=C2H2-type 2 |
| Hgene | ZNF511 | chr10:135125345 | chr10:122263330 | ENST00000359035 | - | 1 | 5 | 144_169 | 0 | 263.0 | Zinc finger | Note=C2H2-type 3 |
| Hgene | ZNF511 | chr10:135125345 | chr10:122263330 | ENST00000359035 | - | 1 | 5 | 80_105 | 0 | 263.0 | Zinc finger | Note=C2H2-type 1 |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398248 | 0 | 3 | 4_24 | 18 | 97.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000398250 | 0 | 7 | 4_24 | 18 | 272.0 | Transmembrane | Helical | |
| Tgene | PPAPDC1A | chr10:135125345 | chr10:122263330 | ENST00000439221 | 0 | 5 | 4_24 | 18 | 209.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ZNF511-PPAPDC1A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >102230_102230_1_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000369073_length(transcript)=2258nt_BP=1149nt CCAGGGACTGCAGCGGGAGACGACAGGCGGGGCAAGGCGGACGGGAGAGGGCGTGCGAGAGGGGGACCTGCGACCCGGCTTCCCAGGGCT GGGAGGGGACACCCGGGACGACCCGGAGGCCCGCTCCGCCCCGCCCCGGCTGAGTGTCGGGCCGCGGGGCAAGACGCTGGCCTGGCCGGT GCAGGCCCGAAGGGCTTCGGCCCCAGAGTTCGGGGACCGCTGCACAGCCAGAGGGAGCGCGCGGGAAGAGCCGCGTCCCGCCAGCCCCGT CGCCTCGTGGGCCCCCCCGGGCCTGGCAGTCCCCCAACCCCCTCATCACGCCCGCGCCCGCGTTCGGCCAGGACTCACCGCAGTCCCGGA GCCACAGCCCCCGCGCAGCCCCCGACGGCGGCGGAAGTGAGCGTGACGTCACGTCCGGCTCGGCTCGCGGACGGTGGCCGACAGGCTGCG CCCGCCCGCGCCCGGGGTGATGCAGTTGCCCCCCGCGCTGTGCGCCCGCCTCGCTGCGGGGCCCGGGGCGGCGGAGCCGCTGCCTGTAGA GCGGGATCCCGCGGCTGGGGCCGCGCCCTTTCGCTTCGTTGCGCGCCCCGTGCGCTTCCCGCGGGAGCACCAGTTCTTCGAGGATGGGGA CGTGCAGCGCCACCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGT GGCCGGCTGCTGCCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCG GGCCTTCCCTTCCGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTA TCAGTGCTTGGTAGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGA CTTCCGGTTTGATAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCAT GGAAATCTGCTCTGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCC GTTCCAGAGAGTCATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGC AATTTCTTTCCTCACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGC GGTGTCCTTGGCTCTTGCTTTGAATGGAGTCTGCACAAACACTATTAAATTAATAGTGGGAAGACCTCGCCCCGATTTCTTTTACCGCTG CTTTCCAGATGGAGTGATGAACTCGGAAATGCATTGCACAGGTGACCCCGATCTGGTGTCCGAGGGCCGCAAAAGCTTCCCCAGCATCCA TTCCTCCTTTGCCTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAG CTGGCGGCTCTGTGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGA TTCCTTTGTGGGTGGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAA ACCCTACGTTAGTCTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGAT CACCGAAGGCCCGGTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAA TGGTTTTCTGTAGTGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAA ATGTCTACTTCCAACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGG AGCTTGGCCGATTCGTCTATCTGAAATGTTTGCTGTAACAGCCACCTTCCTATGTTTTCATGGTTGTAAAACATAATAAAACCTCCCACG >102230_102230_1_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000369073_length(amino acids)=487AA_BP=234 MRPPAPGVMQLPPALCARLAAGPGAAEPLPVERDPAAGAAPFRFVARPVRFPREHQFFEDGDVQRHLYLQDVIMQVADVPEKPRVPAFAC QVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKFKTSRDRKDHMVRMHLYP ADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIWLYKNPLVQSDNIPTRLM FAISFLTPLAVICVVKIIRRTDKTEIKEAFLAVSLALALNGVCTNTIKLIVGRPRPDFFYRCFPDGVMNSEMHCTGDPDLVSEGRKSFPS IHSSFAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAMMIALSRMCDYKHHWQDSFVGGVIGLIFAYICYRQHYPPLANTAC -------------------------------------------------------------- >102230_102230_2_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000398248_length(transcript)=1811nt_BP=1149nt CCAGGGACTGCAGCGGGAGACGACAGGCGGGGCAAGGCGGACGGGAGAGGGCGTGCGAGAGGGGGACCTGCGACCCGGCTTCCCAGGGCT GGGAGGGGACACCCGGGACGACCCGGAGGCCCGCTCCGCCCCGCCCCGGCTGAGTGTCGGGCCGCGGGGCAAGACGCTGGCCTGGCCGGT GCAGGCCCGAAGGGCTTCGGCCCCAGAGTTCGGGGACCGCTGCACAGCCAGAGGGAGCGCGCGGGAAGAGCCGCGTCCCGCCAGCCCCGT CGCCTCGTGGGCCCCCCCGGGCCTGGCAGTCCCCCAACCCCCTCATCACGCCCGCGCCCGCGTTCGGCCAGGACTCACCGCAGTCCCGGA GCCACAGCCCCCGCGCAGCCCCCGACGGCGGCGGAAGTGAGCGTGACGTCACGTCCGGCTCGGCTCGCGGACGGTGGCCGACAGGCTGCG CCCGCCCGCGCCCGGGGTGATGCAGTTGCCCCCCGCGCTGTGCGCCCGCCTCGCTGCGGGGCCCGGGGCGGCGGAGCCGCTGCCTGTAGA GCGGGATCCCGCGGCTGGGGCCGCGCCCTTTCGCTTCGTTGCGCGCCCCGTGCGCTTCCCGCGGGAGCACCAGTTCTTCGAGGATGGGGA CGTGCAGCGCCACCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGT GGCCGGCTGCTGCCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCG GGCCTTCCCTTCCGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTA TCAGTGCTTGGTAGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGA CTTCCGGTTTGATAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCAT GGAAATCTGCTCTGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCC GTTCCAGAGAGTCATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTAT TCCTTTGTGGGTGGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAAA CCCTACGTTAGTCTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGATC ACCGAAGGCCCGGTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAAT GGTTTTCTGTAGTGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAAA TGTCTACTTCCAACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGGA GCTTGGCCGATTCGTCTATCTGAAATGTTTGCTGTAACAGCCACCTTCCTATGTTTTCATGGTTGTAAAACATAATAAAACCTCCCACGT >102230_102230_2_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000398248_length(amino acids)=312AA_BP=234 MRPPAPGVMQLPPALCARLAAGPGAAEPLPVERDPAAGAAPFRFVARPVRFPREHQFFEDGDVQRHLYLQDVIMQVADVPEKPRVPAFAC QVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKFKTSRDRKDHMVRMHLYP ADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIWLYKNPLVQSDNIPTRLM -------------------------------------------------------------- >102230_102230_3_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000398250_length(transcript)=2262nt_BP=1149nt CCAGGGACTGCAGCGGGAGACGACAGGCGGGGCAAGGCGGACGGGAGAGGGCGTGCGAGAGGGGGACCTGCGACCCGGCTTCCCAGGGCT GGGAGGGGACACCCGGGACGACCCGGAGGCCCGCTCCGCCCCGCCCCGGCTGAGTGTCGGGCCGCGGGGCAAGACGCTGGCCTGGCCGGT GCAGGCCCGAAGGGCTTCGGCCCCAGAGTTCGGGGACCGCTGCACAGCCAGAGGGAGCGCGCGGGAAGAGCCGCGTCCCGCCAGCCCCGT CGCCTCGTGGGCCCCCCCGGGCCTGGCAGTCCCCCAACCCCCTCATCACGCCCGCGCCCGCGTTCGGCCAGGACTCACCGCAGTCCCGGA GCCACAGCCCCCGCGCAGCCCCCGACGGCGGCGGAAGTGAGCGTGACGTCACGTCCGGCTCGGCTCGCGGACGGTGGCCGACAGGCTGCG CCCGCCCGCGCCCGGGGTGATGCAGTTGCCCCCCGCGCTGTGCGCCCGCCTCGCTGCGGGGCCCGGGGCGGCGGAGCCGCTGCCTGTAGA GCGGGATCCCGCGGCTGGGGCCGCGCCCTTTCGCTTCGTTGCGCGCCCCGTGCGCTTCCCGCGGGAGCACCAGTTCTTCGAGGATGGGGA CGTGCAGCGCCACCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGT GGCCGGCTGCTGCCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCG GGCCTTCCCTTCCGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTA TCAGTGCTTGGTAGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGA CTTCCGGTTTGATAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCAT GGAAATCTGCTCTGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCC GTTCCAGAGAGTCATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGC AATTTCTTTCCTCACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGC GGTGTCCTTGGCTCTTGCTTTGAATGGAGTCTGCACAAACACTATTAAATTAATAGTGGGAAGACCTCGCCCCGATTTCTTTTACCGCTG CTTTCCAGATGGAGTGATGAACTCGGAAATGCATTGCACAGGTGACCCCGATCTGGTGTCCGAGGGCCGCAAAAGCTTCCCCAGCATCCA TTCCTCCTTTGCCTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAG CTGGCGGCTCTGTGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGA TTCCTTTGTGGGTGGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAA ACCCTACGTTAGTCTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGAT CACCGAAGGCCCGGTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAA TGGTTTTCTGTAGTGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAA ATGTCTACTTCCAACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGG AGCTTGGCCGATTCGTCTATCTGAAATGTTTGCTGTAACAGCCACCTTCCTATGTTTTCATGGTTGTAAAACATAATAAAACCTCCCACG >102230_102230_3_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000398250_length(amino acids)=487AA_BP=234 MRPPAPGVMQLPPALCARLAAGPGAAEPLPVERDPAAGAAPFRFVARPVRFPREHQFFEDGDVQRHLYLQDVIMQVADVPEKPRVPAFAC QVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKFKTSRDRKDHMVRMHLYP ADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIWLYKNPLVQSDNIPTRLM FAISFLTPLAVICVVKIIRRTDKTEIKEAFLAVSLALALNGVCTNTIKLIVGRPRPDFFYRCFPDGVMNSEMHCTGDPDLVSEGRKSFPS IHSSFAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAMMIALSRMCDYKHHWQDSFVGGVIGLIFAYICYRQHYPPLANTAC -------------------------------------------------------------- >102230_102230_4_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000439221_length(transcript)=2073nt_BP=1149nt CCAGGGACTGCAGCGGGAGACGACAGGCGGGGCAAGGCGGACGGGAGAGGGCGTGCGAGAGGGGGACCTGCGACCCGGCTTCCCAGGGCT GGGAGGGGACACCCGGGACGACCCGGAGGCCCGCTCCGCCCCGCCCCGGCTGAGTGTCGGGCCGCGGGGCAAGACGCTGGCCTGGCCGGT GCAGGCCCGAAGGGCTTCGGCCCCAGAGTTCGGGGACCGCTGCACAGCCAGAGGGAGCGCGCGGGAAGAGCCGCGTCCCGCCAGCCCCGT CGCCTCGTGGGCCCCCCCGGGCCTGGCAGTCCCCCAACCCCCTCATCACGCCCGCGCCCGCGTTCGGCCAGGACTCACCGCAGTCCCGGA GCCACAGCCCCCGCGCAGCCCCCGACGGCGGCGGAAGTGAGCGTGACGTCACGTCCGGCTCGGCTCGCGGACGGTGGCCGACAGGCTGCG CCCGCCCGCGCCCGGGGTGATGCAGTTGCCCCCCGCGCTGTGCGCCCGCCTCGCTGCGGGGCCCGGGGCGGCGGAGCCGCTGCCTGTAGA GCGGGATCCCGCGGCTGGGGCCGCGCCCTTTCGCTTCGTTGCGCGCCCCGTGCGCTTCCCGCGGGAGCACCAGTTCTTCGAGGATGGGGA CGTGCAGCGCCACCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGT GGCCGGCTGCTGCCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCG GGCCTTCCCTTCCGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTA TCAGTGCTTGGTAGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGA CTTCCGGTTTGATAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCAT GGAAATCTGCTCTGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCC GTTCCAGAGAGTCATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGC AATTTCTTTCCTCACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGT TGCCTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAGCTGGCGGCT CTGTGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGATTCCTTTGT GGGTGGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAAACCCTACGT TAGTCTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGATCACCGAAGG CCCGGTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAATGGTTTTCT GTAGTGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAAATGTCTACT TCCAACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGGAGCTTGGCC GATTCGTCTATCTGAAATGTTTGCTGTAACAGCCACCTTCCTATGTTTTCATGGTTGTAAAACATAATAAAACCTCCCACGTGGAAGACT >102230_102230_4_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000439221_length(amino acids)=424AA_BP=234 MRPPAPGVMQLPPALCARLAAGPGAAEPLPVERDPAAGAAPFRFVARPVRFPREHQFFEDGDVQRHLYLQDVIMQVADVPEKPRVPAFAC QVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKFKTSRDRKDHMVRMHLYP ADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIWLYKNPLVQSDNIPTRLM FAISFLTPLAVICVVKIIRRTDKTEIKEAFLVAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAMMIALSRMCDYKHHWQDS -------------------------------------------------------------- >102230_102230_5_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000541332_length(transcript)=2458nt_BP=1149nt CCAGGGACTGCAGCGGGAGACGACAGGCGGGGCAAGGCGGACGGGAGAGGGCGTGCGAGAGGGGGACCTGCGACCCGGCTTCCCAGGGCT GGGAGGGGACACCCGGGACGACCCGGAGGCCCGCTCCGCCCCGCCCCGGCTGAGTGTCGGGCCGCGGGGCAAGACGCTGGCCTGGCCGGT GCAGGCCCGAAGGGCTTCGGCCCCAGAGTTCGGGGACCGCTGCACAGCCAGAGGGAGCGCGCGGGAAGAGCCGCGTCCCGCCAGCCCCGT CGCCTCGTGGGCCCCCCCGGGCCTGGCAGTCCCCCAACCCCCTCATCACGCCCGCGCCCGCGTTCGGCCAGGACTCACCGCAGTCCCGGA GCCACAGCCCCCGCGCAGCCCCCGACGGCGGCGGAAGTGAGCGTGACGTCACGTCCGGCTCGGCTCGCGGACGGTGGCCGACAGGCTGCG CCCGCCCGCGCCCGGGGTGATGCAGTTGCCCCCCGCGCTGTGCGCCCGCCTCGCTGCGGGGCCCGGGGCGGCGGAGCCGCTGCCTGTAGA GCGGGATCCCGCGGCTGGGGCCGCGCCCTTTCGCTTCGTTGCGCGCCCCGTGCGCTTCCCGCGGGAGCACCAGTTCTTCGAGGATGGGGA CGTGCAGCGCCACCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGT GGCCGGCTGCTGCCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCG GGCCTTCCCTTCCGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTA TCAGTGCTTGGTAGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGA CTTCCGGTTTGATAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCAT GGAAATCTGCTCTGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCC GTTCCAGAGAGTCATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGC AATTTCTTTCCTCACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGC GGTGTCCTTGGCTCTTGCTTTGAATGGAGTCTGCACAAACACTATTAAATTAATAGTGGGAAGACCTCGCCCCGATTTCTTTTACCGCTG CTTTCCAGATGGAGTGATGAACTCGGAAATGCATTGCACAGGTGACCCCGATCTGGTGTCCGAGGGCCGCAAAAGCTTCCCCAGCATCCA TTCCTCCTTTGCCTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAG CTGGCGGCTCTGTGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGT TGGCTGCTGTGTATGGCAGGGCCCACAGTCACATTTACCATCGGCTGCCAACCCCCGCCTCTGTCTGGGAACCTCTGAACCTTGACTGCC CATCTAAGTCCACCCGCCTCTGTGTTGGTGCCTAGGAGAGCGTAAGAAGCCTCAGCCCCTGGCTCACCCCTCCCTCCGGTCAAAGTCCAC TGCATATCCTGCTTTCAGCCTCAAGAGAGGCATTCTCAGTCAGGACTTCTGCCCTCTTCCTGCCTCCAGGATAGCCCTGGTGTTTCCCCG TGATGTAATTCCCAGGGAAGATGGACGTTGATTGCTCCCAAGTCTCACTGGCATCCTTCCTTTGTTTTACTCAAGGGAACCTGTCTCTTC TCAGCTTTCTGTGGTGTTCTGTGGTTCCTCCTGCTCAGCATATTTATAGACTCCAACTTTTCCTTCCCACTCTGGTCTAGTGATCCTCAG TTGTTGCCTGAATCATTGCAGAAGCCTCTCAGCCTGGCTCCCTCCTTCCTTCTTTGGCCCTCAACTGCATTTGGAATGTGCCTTTAGAGC CTGCGTTAGCACATGTCACTTTTGCAAAAATCCTCCAGTGATTCTCATTTCACTCAGCATCACAGCCTGTGTCCTGCCATGGTCCATAAG ATCTGCTCCCTCCGTACTTCTTTGATGCTATGTTCAAGCTACTTGTCCTCCTCGTTCACATGATTGCATCGCTTGAGCTTCTTGCTACTC >102230_102230_5_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000361518_PPAPDC1A_chr10_122263330_ENST00000541332_length(amino acids)=449AA_BP=234 MRPPAPGVMQLPPALCARLAAGPGAAEPLPVERDPAAGAAPFRFVARPVRFPREHQFFEDGDVQRHLYLQDVIMQVADVPEKPRVPAFAC QVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKFKTSRDRKDHMVRMHLYP ADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIWLYKNPLVQSDNIPTRLM FAISFLTPLAVICVVKIIRRTDKTEIKEAFLAVSLALALNGVCTNTIKLIVGRPRPDFFYRCFPDGVMNSEMHCTGDPDLVSEGRKSFPS -------------------------------------------------------------- >102230_102230_6_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000369073_length(transcript)=1616nt_BP=507nt CCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGTGGCCGGCTGCTG CCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCGGGCCTTCCCTTC CGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTATCAGTGCTTGGT AGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGACTTCCGGTTTGA TAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCATGGAAATCTGCTC TGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCCGTTCCAGAGAGT CATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGCAATTTCTTTCCT CACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGCGGTGTCCTTGGC TCTTGCTTTGAATGGAGTCTGCACAAACACTATTAAATTAATAGTGGGAAGACCTCGCCCCGATTTCTTTTACCGCTGCTTTCCAGATGG AGTGATGAACTCGGAAATGCATTGCACAGGTGACCCCGATCTGGTGTCCGAGGGCCGCAAAAGCTTCCCCAGCATCCATTCCTCCTTTGC CTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAGCTGGCGGCTCTG TGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGATTCCTTTGTGGG TGGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAAACCCTACGTTAG TCTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGATCACCGAAGGCCC GGTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAATGGTTTTCTGTA GTGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAAATGTCTACTTCC AACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGGAGCTTGGCCGAT >102230_102230_6_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000369073_length(amino acids)=414AA_BP=161 MQVADVPEKPRVPAFACQVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKF KTSRDRKDHMVRMHLYPADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIW LYKNPLVQSDNIPTRLMFAISFLTPLAVICVVKIIRRTDKTEIKEAFLAVSLALALNGVCTNTIKLIVGRPRPDFFYRCFPDGVMNSEMH CTGDPDLVSEGRKSFPSIHSSFAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAMMIALSRMCDYKHHWQDSFVGGVIGLIF -------------------------------------------------------------- >102230_102230_7_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000398248_length(transcript)=1169nt_BP=507nt CCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGTGGCCGGCTGCTG CCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCGGGCCTTCCCTTC CGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTATCAGTGCTTGGT AGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGACTTCCGGTTTGA TAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCATGGAAATCTGCTC TGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCCGTTCCAGAGAGT CATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTATTCCTTTGTGGGT GGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAAACCCTACGTTAGT CTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGATCACCGAAGGCCCG GTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAATGGTTTTCTGTAG TGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAAATGTCTACTTCCA ACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGGAGCTTGGCCGATT >102230_102230_7_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000398248_length(amino acids)=239AA_BP=161 MQVADVPEKPRVPAFACQVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKF KTSRDRKDHMVRMHLYPADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIW -------------------------------------------------------------- >102230_102230_8_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000398250_length(transcript)=1620nt_BP=507nt CCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGTGGCCGGCTGCTG CCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCGGGCCTTCCCTTC CGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTATCAGTGCTTGGT AGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGACTTCCGGTTTGA TAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCATGGAAATCTGCTC TGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCCGTTCCAGAGAGT CATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGCAATTTCTTTCCT CACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGCGGTGTCCTTGGC TCTTGCTTTGAATGGAGTCTGCACAAACACTATTAAATTAATAGTGGGAAGACCTCGCCCCGATTTCTTTTACCGCTGCTTTCCAGATGG AGTGATGAACTCGGAAATGCATTGCACAGGTGACCCCGATCTGGTGTCCGAGGGCCGCAAAAGCTTCCCCAGCATCCATTCCTCCTTTGC CTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAGCTGGCGGCTCTG TGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGATTCCTTTGTGGG TGGAGTCATCGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAAACCCTACGTTAG TCTGCGAGTCCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGATCACCGAAGGCCC GGTATGACCAGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAATGGTTTTCTGTA GTGTATTTTTCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAAATGTCTACTTCC AACATCCTTGAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGGAGCTTGGCCGAT TCGTCTATCTGAAATGTTTGCTGTAACAGCCACCTTCCTATGTTTTCATGGTTGTAAAACATAATAAAACCTCCCACGTGGAAGACTTGG >102230_102230_8_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000398250_length(amino acids)=414AA_BP=161 MQVADVPEKPRVPAFACQVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKF KTSRDRKDHMVRMHLYPADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIW LYKNPLVQSDNIPTRLMFAISFLTPLAVICVVKIIRRTDKTEIKEAFLAVSLALALNGVCTNTIKLIVGRPRPDFFYRCFPDGVMNSEMH CTGDPDLVSEGRKSFPSIHSSFAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAMMIALSRMCDYKHHWQDSFVGGVIGLIF -------------------------------------------------------------- >102230_102230_9_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000439221_length(transcript)=1431nt_BP=507nt CCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGTGGCCGGCTGCTG CCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCGGGCCTTCCCTTC CGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTATCAGTGCTTGGT AGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGACTTCCGGTTTGA TAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCATGGAAATCTGCTC TGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCCGTTCCAGAGAGT CATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGCAATTTCTTTCCT CACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGTTGCCTTTTCGGG CCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAGCTGGCGGCTCTGTGCTGCCAT CCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGATTCCTTTGTGGGTGGAGTCAT CGGCCTCATTTTTGCATACATTTGCTACAGACAGCACTATCCTCCTCTGGCCAACACAGCTTGCCATAAACCCTACGTTAGTCTGCGAGT CCCAGCCTCACTGAAGAAAGAGGAGAGGCCCACAGCTGACAGCGCACCCAGCTTGCCTCTGGAGGGGATCACCGAAGGCCCGGTATGACC AGTGTCCTGGGAGGATGGACACTAAGCCCTGGGCACATCTGCCACCCTGACATCATAACACAATAGAAATGGTTTTCTGTAGTGTATTTT TCATCAGTTGTTTCTCAAAGTCATCGTACTTCTGCTTCTGTTTCACTGATGGTGTTCCTGCTACTTTAAATGTCTACTTCCAACATCCTT GAATTTGCAAGTGAAGGACAACAATCTCTGAGAGACGTGTGGAAGAGGCTGTGAAGGTGGGGTTTGGGGAGCTTGGCCGATTCGTCTATC >102230_102230_9_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000439221_length(amino acids)=351AA_BP=161 MQVADVPEKPRVPAFACQVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKF KTSRDRKDHMVRMHLYPADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIW LYKNPLVQSDNIPTRLMFAISFLTPLAVICVVKIIRRTDKTEIKEAFLVAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAM -------------------------------------------------------------- >102230_102230_10_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000541332_length(transcript)=1816nt_BP=507nt CCTCTACCTCCAGGACGTGATCATGCAGGTGGCCGACGTGCCTGAGAAGCCCAGGGTGCCCGCGTTTGCCTGCCAGGTGGCCGGCTGCTG CCAGGTGTTCGATGCCCTGGACGACTACGAGCACCACTACCACACGCTGCACGGAAATGTTTGCTCCTTTTGCAAGCGGGCCTTCCCTTC CGGACACCTGCTGGACGCCCACATCCTGGAGTGGCACGATTCGCTCTTCCAGATCCTGTCTGAGAGGCAGGACATGTATCAGTGCTTGGT AGAAGGCTGCACAGAGAAGTTCAAGACCAGCAGAGACCGGAAGGATCACATGGTGAGGATGCACCTGTACCCCGCGGACTTCCGGTTTGA TAAGCCAAAGAAAAGCAGAAGCCCAGCCTCAGCAGAAGCCCCAGGGGACAGTGGAGAGCGGTCAGAAGGGGAGGCCATGGAAATCTGCTC TGAGCCTGTGGCAGCCTCCCCTGCACCGGCAGGTGAGAGGCGGATCTACAGACATAGTTTTACAGAGTTTTTGGATCCGTTCCAGAGAGT CATCCAGCCAGAAGAGATCTGGCTCTATAAAAATCCTTTGGTGCAATCAGATAACATACCTACCCGCCTCATGTTTGCAATTTCTTTCCT CACACCCCTGGCTGTTATTTGTGTGGTGAAAATTATCCGGCGAACAGACAAGACTGAAATTAAGGAAGCCTTCTTAGCGGTGTCCTTGGC TCTTGCTTTGAATGGAGTCTGCACAAACACTATTAAATTAATAGTGGGAAGACCTCGCCCCGATTTCTTTTACCGCTGCTTTCCAGATGG AGTGATGAACTCGGAAATGCATTGCACAGGTGACCCCGATCTGGTGTCCGAGGGCCGCAAAAGCTTCCCCAGCATCCATTCCTCCTTTGC CTTTTCGGGCCTTGGCTTCACGACGTTCTACTTGGCGGGCAAGCTGCACTGCTTCACCGAGAGTGGGCGGGGAAAGAGCTGGCGGCTCTG TGCTGCCATCCTGCCCTTGTACTGCGCCATGATGATTGCCCTGTCCCGCATGTGCGACTACAAGCATCACTGGCAAGTTGGCTGCTGTGT ATGGCAGGGCCCACAGTCACATTTACCATCGGCTGCCAACCCCCGCCTCTGTCTGGGAACCTCTGAACCTTGACTGCCCATCTAAGTCCA CCCGCCTCTGTGTTGGTGCCTAGGAGAGCGTAAGAAGCCTCAGCCCCTGGCTCACCCCTCCCTCCGGTCAAAGTCCACTGCATATCCTGC TTTCAGCCTCAAGAGAGGCATTCTCAGTCAGGACTTCTGCCCTCTTCCTGCCTCCAGGATAGCCCTGGTGTTTCCCCGTGATGTAATTCC CAGGGAAGATGGACGTTGATTGCTCCCAAGTCTCACTGGCATCCTTCCTTTGTTTTACTCAAGGGAACCTGTCTCTTCTCAGCTTTCTGT GGTGTTCTGTGGTTCCTCCTGCTCAGCATATTTATAGACTCCAACTTTTCCTTCCCACTCTGGTCTAGTGATCCTCAGTTGTTGCCTGAA TCATTGCAGAAGCCTCTCAGCCTGGCTCCCTCCTTCCTTCTTTGGCCCTCAACTGCATTTGGAATGTGCCTTTAGAGCCTGCGTTAGCAC ATGTCACTTTTGCAAAAATCCTCCAGTGATTCTCATTTCACTCAGCATCACAGCCTGTGTCCTGCCATGGTCCATAAGATCTGCTCCCTC CGTACTTCTTTGATGCTATGTTCAAGCTACTTGTCCTCCTCGTTCACATGATTGCATCGCTTGAGCTTCTTGCTACTCCTTGAACAGGCC >102230_102230_10_ZNF511-PPAPDC1A_ZNF511_chr10_135125345_ENST00000368554_PPAPDC1A_chr10_122263330_ENST00000541332_length(amino acids)=376AA_BP=161 MQVADVPEKPRVPAFACQVAGCCQVFDALDDYEHHYHTLHGNVCSFCKRAFPSGHLLDAHILEWHDSLFQILSERQDMYQCLVEGCTEKF KTSRDRKDHMVRMHLYPADFRFDKPKKSRSPASAEAPGDSGERSEGEAMEICSEPVAASPAPAGERRIYRHSFTEFLDPFQRVIQPEEIW LYKNPLVQSDNIPTRLMFAISFLTPLAVICVVKIIRRTDKTEIKEAFLAVSLALALNGVCTNTIKLIVGRPRPDFFYRCFPDGVMNSEMH CTGDPDLVSEGRKSFPSIHSSFAFSGLGFTTFYLAGKLHCFTESGRGKSWRLCAAILPLYCAMMIALSRMCDYKHHWQVGCCVWQGPQSH -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF511-PPAPDC1A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF511-PPAPDC1A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF511-PPAPDC1A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
