
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF571-TSHZ3 (FusionGDB2 ID:102419) |
Fusion Gene Summary for ZNF571-TSHZ3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF571-TSHZ3 | Fusion gene ID: 102419 | Hgene | Tgene | Gene symbol | ZNF571 | TSHZ3 | Gene ID | 51276 | 57616 |
| Gene name | zinc finger protein 571 | teashirt zinc finger homeobox 3 | |
| Synonyms | HSPC059 | TSH3|ZNF537 | |
| Cytomap | 19q13.12 | 19q12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 571 | teashirt homolog 3teashirt family zinc finger 3zinc finger protein 537 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000328550, ENST00000358744, ENST00000451802, ENST00000590751, ENST00000593133, | ENST00000558569, ENST00000240587, | |
| Fusion gene scores | * DoF score | 3 X 2 X 2=12 | 5 X 4 X 4=80 |
| # samples | 3 | 5 | |
| ** MAII score | log2(3/12*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(5/80*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF571 [Title/Abstract] AND TSHZ3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF571(38077569)-TSHZ3(31770658), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ZNF571-TSHZ3 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. ZNF571-TSHZ3 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. ZNF571-TSHZ3 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | TSHZ3 | GO:0045892 | negative regulation of transcription, DNA-templated | 19343227 |
 Fusion gene breakpoints across ZNF571 (5'-gene) Fusion gene breakpoints across ZNF571 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
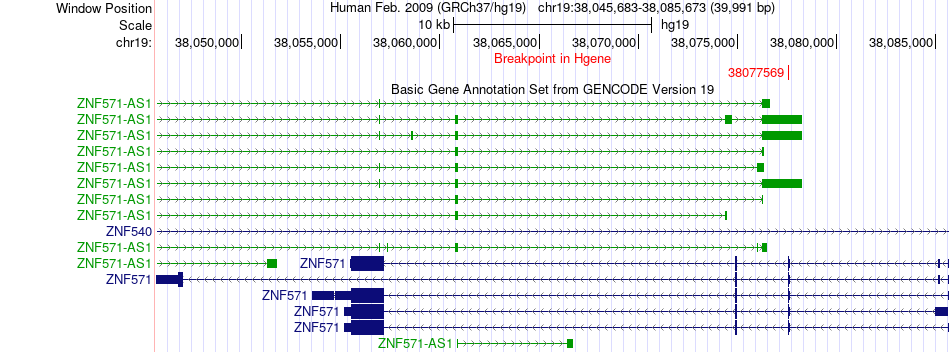 |
 Fusion gene breakpoints across TSHZ3 (3'-gene) Fusion gene breakpoints across TSHZ3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
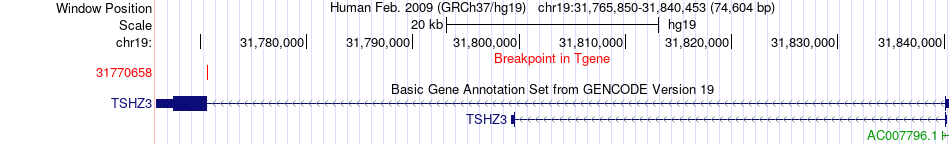 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-37-A5EN-01A | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
Top |
Fusion Gene ORF analysis for ZNF571-TSHZ3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000328550 | ENST00000558569 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| 5CDS-intron | ENST00000358744 | ENST00000558569 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| 5CDS-intron | ENST00000451802 | ENST00000558569 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| 5CDS-intron | ENST00000590751 | ENST00000558569 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| 5CDS-intron | ENST00000593133 | ENST00000558569 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| Frame-shift | ENST00000328550 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| Frame-shift | ENST00000451802 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| Frame-shift | ENST00000593133 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| In-frame | ENST00000358744 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
| In-frame | ENST00000590751 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000590751 | ZNF571 | chr19 | 38077569 | - | ENST00000240587 | TSHZ3 | chr19 | 31770658 | - | 5001 | 193 | 225 | 3398 | 1057 |
| ENST00000358744 | ZNF571 | chr19 | 38077569 | - | ENST00000240587 | TSHZ3 | chr19 | 31770658 | - | 5007 | 199 | 231 | 3404 | 1057 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000590751 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - | 0.001389182 | 0.99861073 |
| ENST00000358744 | ENST00000240587 | ZNF571 | chr19 | 38077569 | - | TSHZ3 | chr19 | 31770658 | - | 0.001398707 | 0.99860126 |
Top |
Fusion Genomic Features for ZNF571-TSHZ3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
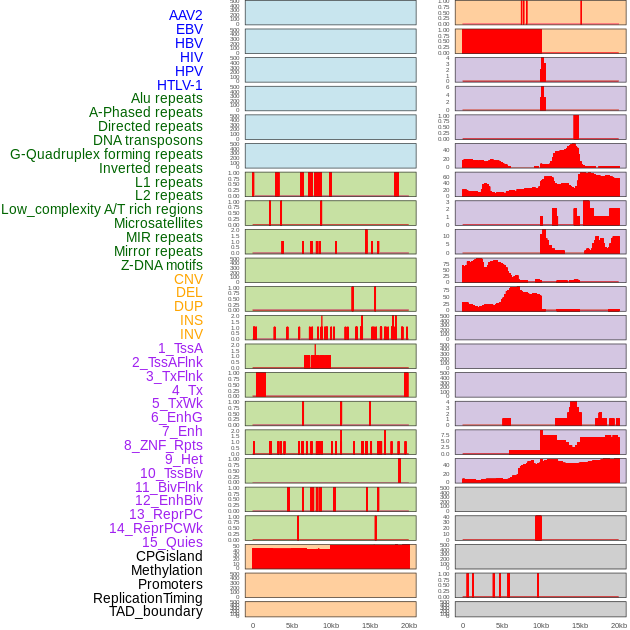 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZNF571-TSHZ3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:38077569/chr19:31770658) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 606_630 | 13 | 1082.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 142_164 | 13 | 1082.0 | Compositional bias | Note=Ser-rich | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 493_496 | 13 | 1082.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 891_961 | 13 | 1082.0 | DNA binding | Homeobox%3B atypical | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 1041_1064 | 13 | 1082.0 | Zinc finger | C2H2-type 5 | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 214_238 | 13 | 1082.0 | Zinc finger | C2H2-type 1 | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 275_299 | 13 | 1082.0 | Zinc finger | C2H2-type 2 | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 386_404 | 13 | 1082.0 | Zinc finger | C2H2-type 3%3B atypical | |
| Tgene | TSHZ3 | chr19:38077569 | chr19:31770658 | ENST00000240587 | 0 | 2 | 976_998 | 13 | 1082.0 | Zinc finger | C2H2-type 4 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 6_84 | 3 | 1776.3333333333333 | Domain | KRAB |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 6_84 | 3 | 610.0 | Domain | KRAB |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 6_84 | 3 | 610.0 | Domain | KRAB |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 6_84 | 3 | 610.0 | Domain | KRAB |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 139_161 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 164_189 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 2%3B atypical |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 195_217 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 223_245 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 251_273 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 279_301 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 307_329 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 335_357 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 363_385 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 391_413 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 419_441 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 447_469 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 474_496 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 502_524 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 14 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 530_552 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 15 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 558_580 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 16 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000328550 | - | 2 | 5 | 586_608 | 3 | 1776.3333333333333 | Zinc finger | C2H2-type 17 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 139_161 | 3 | 610.0 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 164_189 | 3 | 610.0 | Zinc finger | C2H2-type 2%3B atypical |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 195_217 | 3 | 610.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 223_245 | 3 | 610.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 251_273 | 3 | 610.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 279_301 | 3 | 610.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 307_329 | 3 | 610.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 335_357 | 3 | 610.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 363_385 | 3 | 610.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 391_413 | 3 | 610.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 419_441 | 3 | 610.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 447_469 | 3 | 610.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 474_496 | 3 | 610.0 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 502_524 | 3 | 610.0 | Zinc finger | C2H2-type 14 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 530_552 | 3 | 610.0 | Zinc finger | C2H2-type 15 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 558_580 | 3 | 610.0 | Zinc finger | C2H2-type 16 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000358744 | - | 3 | 5 | 586_608 | 3 | 610.0 | Zinc finger | C2H2-type 17 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 139_161 | 3 | 610.0 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 164_189 | 3 | 610.0 | Zinc finger | C2H2-type 2%3B atypical |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 195_217 | 3 | 610.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 223_245 | 3 | 610.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 251_273 | 3 | 610.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 279_301 | 3 | 610.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 307_329 | 3 | 610.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 335_357 | 3 | 610.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 363_385 | 3 | 610.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 391_413 | 3 | 610.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 419_441 | 3 | 610.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 447_469 | 3 | 610.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 474_496 | 3 | 610.0 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 502_524 | 3 | 610.0 | Zinc finger | C2H2-type 14 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 530_552 | 3 | 610.0 | Zinc finger | C2H2-type 15 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 558_580 | 3 | 610.0 | Zinc finger | C2H2-type 16 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000451802 | - | 2 | 4 | 586_608 | 3 | 610.0 | Zinc finger | C2H2-type 17 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 139_161 | 3 | 610.0 | Zinc finger | C2H2-type 1 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 164_189 | 3 | 610.0 | Zinc finger | C2H2-type 2%3B atypical |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 195_217 | 3 | 610.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 223_245 | 3 | 610.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 251_273 | 3 | 610.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 279_301 | 3 | 610.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 307_329 | 3 | 610.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 335_357 | 3 | 610.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 363_385 | 3 | 610.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 391_413 | 3 | 610.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 419_441 | 3 | 610.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 447_469 | 3 | 610.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 474_496 | 3 | 610.0 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 502_524 | 3 | 610.0 | Zinc finger | C2H2-type 14 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 530_552 | 3 | 610.0 | Zinc finger | C2H2-type 15 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 558_580 | 3 | 610.0 | Zinc finger | C2H2-type 16 |
| Hgene | ZNF571 | chr19:38077569 | chr19:31770658 | ENST00000593133 | - | 2 | 4 | 586_608 | 3 | 610.0 | Zinc finger | C2H2-type 17 |
Top |
Fusion Gene Sequence for ZNF571-TSHZ3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >102419_102419_1_ZNF571-TSHZ3_ZNF571_chr19_38077569_ENST00000358744_TSHZ3_chr19_31770658_ENST00000240587_length(transcript)=5007nt_BP=199nt GAGCTTGTCTGCGCCGGCCAGCGCTTGGCCCCTCCCGGGCCGCAGTACTCCTCGTGTGACACGGACATCATTAAGCCAGCTGAGAATCGT GCGGTTACAGCTGCTGCAGTTTCTTTGCAGACACTGGCTTCTCTGGACTTTGCACACCTCCAGGAGAAGAACCCAGAAAATTGATCAGTT CTAAAAAACCATGCCCCACCCTATGTTTCCGAAGAGTTAAAGGCTGCTGCCCTGGTGGACGAAGGTTTAGACCCAGAGGAGCATACGGCA GATGGAGAGCCCTCGGCCAAGTACATGTGCCCGGAGAAGGAGCTCGCCAGGGCCTGCCCCAGCTACCAGAACTCCCCGGCCGCCGAGTTT TCCTGCCATGAAATGGACAGCGAGTCACACATCAGTGAGACCAGTGACCGAATGGCTGACTTTGAAAGCGGCTCCATCAAGAACGAAGAG GAGACCAAGGAGGTCACGGTCCCACTGGAAGACACGACTGTGTCGGATAGCCTGGAGCAGATGAAGGCCGTGTACAACAACTTCCTCTCC AACTCCTACTGGTCCAACCTCAACCTCAACCTGCACCAGCCCTCCTCGGAGAAGAACAACGGCAGCAGCAGCAGCAGCAGTAGCAGCAGC AGCAGCTGTGGCAGCGGGAGCTTCGACTGGCACCAGAGCGCCATGGCTAAGACGCTGCAGCAGGTGTCACAGAGCCGCATGCTCCCGGAG CCCAGCCTCTTCAGCACCGTGCAGCTGTACCGGCAGAGCAGCAAGCTCTATGGCTCCATCTTCACGGGGGCCAGCAAGTTCCGCTGTAAG GACTGCAGCGCTGCCTACGACACCCTGGTGGAGTTGACGGTGCACATGAACGAGACGGGGCATTACCGCGACGACAACCATGAGACCGAT AACAACAACCCCAAGCGCTGGTCCAAGCCTCGCAAACGCTCCTTGCTGGAAATGGAAGGGAAGGAAGACGCCCAGAAGGTGCTGAAGTGC ATGTACTGTGGCCACTCCTTTGAGTCCCTGCAGGATTTGAGTGTCCATATGATCAAAACAAAACACTACCAAAAAGTGCCTCTGAAGGAA CCCGTCACTCCTGTCGCCGCCAAAATCATCCCTGCCACTCGGAAGAAAGCTTCCCTGGAGCTGGAGCTCCCCAGCTCCCCAGATTCCACA GGTGGAACCCCCAAAGCCACCATCTCAGACACCAACGATGCACTTCAGAAGAACTCCAACCCTTACATCACGCCAAATAATCGGTACGGC CACCAGAATGGGGCCAGCTATGCATGGCACTTTGAGGCCCGGAAGTCGCAGATCCTGAAGTGCATGGAGTGTGGGAGCTCGCATGACACC CTGCAGGAGCTCACTGCCCACATGATGGTCACTGGCCACTTCATCAAGGTCACCAACTCTGCTATGAAAAAGGGGAAGCCCATTGTGGAG ACGCCTGTCACACCTACCATCACAACCCTGCTGGATGAGAAGGTCCAGTCCGTGCCCCTGGCAGCCACCACCTTCACGTCCCCCTCCAAT ACACCTGCCAGCATCTCCCCAAAACTGAATGTGGAGGTCAAGAAGGAAGTCGACAAGGAGAAAGCGGTCACTGACGAGAAACCTAAGCAA AAAGACAAGCCTGGCGAAGAAGAGGAGAAGTGTGACATCTCTTCCAAATACCATTACTTGACTGAAAATGACTTAGAAGAGAGTCCCAAG GGGGGGCTTGATATCCTCAAATCCTTGGAAAACACAGTGACATCCGCAATCAACAAGGCCCAGAACGGCACTCCTAGCTGGGGGGGCTAT CCCAGCATCCATGCCGCCTACCAACTTCCCAACATGATGAAGTTGTCCCTGGGCTCGTCGGGGAAGAGCACGCCCCTGAAACCCATGTTT GGCAACAGTGAGATTGTCTCCCCGACGAAAAACCAGACCCTGGTCTCTCCACCCAGCAGCCAGACGTCCCCCATGCCCAAGACAAACTTT CATGCCATGGAGGAGCTGGTGAAAAAGGTCACTGAGAAAGTTGCCAAAGTGGAGGAGAAGATGAAGGAGCCGGATGGGAAGCTTTCCCCG CCCAAGCGGGCCACTCCCTCCCCATGTAGCAGCGAAGTCGGGGAACCCATCAAGATGGAGGCATCCAGCGATGGGGGCTTCCGCAGCCAG GAGAACAGCCCCAGCCCCCCGCGGGATGGGTGCAAGGATGGGAGCCCCCTCGCTGAGCCGGTGGAGAATGGCAAGGAGCTGGTGAAGCCC CTAGCCAGCAGTTTGAGTGGCAGCACGGCCATCATCACCGACCACCCGCCTGAACAGCCTTTTGTTAACCCTTTGAGCGCCCTGCAGTCA GTCATGAACATTCACCTGGGCAAGGCCGCCAAGCCCTCCCTGCCTGCCCTGGACCCCATGAGCATGCTTTTCAAGATGAGCAACAGCCTG GCGGAGAAGGCTGCTGTGGCCACCCCGCCGCCCCTGCAGTCCAAGAAGGCAGACCACCTCGACCGCTATTTCTACCACGTCAACAACGAC CAGCCCATAGACTTGACAAAAGGGAAGAGTGACAAAGGCTGCTCCTTGGGTTCAGTGCTTCTGTCACCCACGTCCACAGCCCCGGCAACC TCCTCATCCACGGTGACAACGGCAAAGACATCTGCCGTCGTATCATTCATGTCAAACTCGCCGCTACGCGAGAATGCCTTGTCAGATATA TCCGATATGCTGAAGAACTTGACAGAGAGCCACACGTCAAAATCCTCCACTCCTTCCAGCATCTCCGAGAAGTCTGACATTGACGGGGCC ACTCTGGAGGAGGCTGAGGAGTCGACGCCCGCCCAGAAGAGGAAGGGCCGCCAGTCAAACTGGAACCCCCAGCACCTCCTGATCCTCCAG GCCCAGTTTGCCGCCAGCCTCCGGCAGACCTCAGAAGGGAAGTACATCATGTCAGACCTGAGCCCCCAGGAGCGGATGCATATCTCCAGG TTCACCGGGCTGTCCATGACCACCATCAGCCACTGGCTGGCCAACGTGAAATACCAGCTTCGAAGGACAGGTGGAACAAAGTTCCTCAAA AACTTGGACACTGGCCACCCCGTCTTCTTTTGTAACGATTGTGCGTCCCAAATCAGGACTCCTTCCACGTACATCAGTCACCTAGAGTCA CACTTAGGCTTCCGGCTACGGGACTTATCCAAACTGTCCACCGAACAGATTAACAGTCAGATAGCACAAACCAAGTCACCGTCAGAAAAA ATGGTGACGTCCTCCCCCGAGGAAGACCTGGGGACTTCCTATCAGTGCAAACTTTGCAATCGGACCTTTGCCAGCAAGCACGCTGTTAAA CTTCACCTTAGCAAAACACACGGGAAATCTCCGGAAGACCACCTTCTGTATGTCTCTGAGTTAGAGAAGCAGTAGCATTTGCTTTTGATA GAAAGGACTGCAGTTTGCTTTGAGGGAAACTGTGGAAGGCACCTTCAGGCCCCCTCTGACTTGTTGTTCTTGGCACATGTTCTTATTTTA ACTGCAGAGAATCACTCTGGGCTGGACTGTTTTGTATAACTGTACAGTGTTTAATAGAGGTGCATAATCAGCTGTTGTTACTGGTAAAAT ATGAAGGTTAAAATGCAGTGGTAAGTGTTTGGAACTTTGTGTAAACGGGATTTAGTTGTGAGCATCCTCCCGATGCTTCAAGCTGCATGC ATTAACAGACAGTTTAATTAAGCATTTATAACGGAATCAGGCACACCTTTTCCACGAGACTCGAGTGTGCTGGCATTTCTCACCCTTTCA TCTTTAGCCCTCTGAGTACTTTGAAGCACTTTTGCATTAATTTGGTTAAAAAATAAAATAAAATAATAATAATGTATGAAGCTCTGTTTT TTAAACTCCTTACCAGCTTAGTTATAATGAATAATATGAACCTCCATTTATGCAGGTCTGCAGGGGTATAACACGCCTTGAAATTTAAAA GAATATTATTTTCACATTGAAACATAGATGTATATATTGTATAGATTTCAGACTCTCTTATGAAAAAAAATGTGATTGTGGTTAAATGAC CTTTTTCTTGCATTTATAGCAACAGTGTTTTATGCACCTGCTATGCTCTGGGCATAAGCTGTGCCTATGTATAGTGTATATTTCTTTTTT TCTTTTTTTTAAGGTCTATGGGTTTTGTTTTTTACATGCAAACATTGTAAATTATACAGAAGATACCACAGATAGCATTTATAAAGTATA CAGAAACATTATCTGAAAGCAAAGTATGATAGTTTGTTTTGCTATACAGTACATCTATATTGATAGAGGTTCATGTTTAAATTATACATA TTTATTAGCATCATATTGTCATTTGTTTTGAGCAGTCTGAATAAACGAGACCGGGAAAGACATCCCTGGCAGGCATCAGAACTATTTTGC ACATGATTTTTAAAGGTATTTATTAGAAATCAAAGAACACTCAAAATAAACTCAGTGCTCAAAGGGTTAAGTCTATTTGAAAAGGTTAAA AAAAAGAACAAAAAAAAAAAAAGAACTTGTACTGTATTTCCTAAACATTGATAAAGCCTTTAAAATGTTTGTACTGTAATACTTTGCTTA AAAGTCATGAGGCATTCTGTGATCCAACCTCTTTCACTTATTTATAAGCCCTCTTGGTTGCTATTCCATATTGTAGGATGCCTTTCTATT TCAATTGGTAACTTTCTGTTTTGTTCTTCCTAATTATTCTCCCAAGATCCCACACTGCAGCTTTATCTTTAGGCTTATGAAAGGTAACCC GTGGTTACCGGCTCTCCAAGTGATTCTGTTCTTCTCCATTTTTGGCAGTTAATTTGCAGAAGTAACTGACAGCTGACACCATATGAGAAC CTTTGTATAAAATATTGGCATGTAAACAGCACAGACACCGTAACACACTCTGTGCCCTGTTTGGTTGTTGACAATGAAGCACCATTATGT >102419_102419_1_ZNF571-TSHZ3_ZNF571_chr19_38077569_ENST00000358744_TSHZ3_chr19_31770658_ENST00000240587_length(amino acids)=1057AA_BP= MVDEGLDPEEHTADGEPSAKYMCPEKELARACPSYQNSPAAEFSCHEMDSESHISETSDRMADFESGSIKNEEETKEVTVPLEDTTVSDS LEQMKAVYNNFLSNSYWSNLNLNLHQPSSEKNNGSSSSSSSSSSSCGSGSFDWHQSAMAKTLQQVSQSRMLPEPSLFSTVQLYRQSSKLY GSIFTGASKFRCKDCSAAYDTLVELTVHMNETGHYRDDNHETDNNNPKRWSKPRKRSLLEMEGKEDAQKVLKCMYCGHSFESLQDLSVHM IKTKHYQKVPLKEPVTPVAAKIIPATRKKASLELELPSSPDSTGGTPKATISDTNDALQKNSNPYITPNNRYGHQNGASYAWHFEARKSQ ILKCMECGSSHDTLQELTAHMMVTGHFIKVTNSAMKKGKPIVETPVTPTITTLLDEKVQSVPLAATTFTSPSNTPASISPKLNVEVKKEV DKEKAVTDEKPKQKDKPGEEEEKCDISSKYHYLTENDLEESPKGGLDILKSLENTVTSAINKAQNGTPSWGGYPSIHAAYQLPNMMKLSL GSSGKSTPLKPMFGNSEIVSPTKNQTLVSPPSSQTSPMPKTNFHAMEELVKKVTEKVAKVEEKMKEPDGKLSPPKRATPSPCSSEVGEPI KMEASSDGGFRSQENSPSPPRDGCKDGSPLAEPVENGKELVKPLASSLSGSTAIITDHPPEQPFVNPLSALQSVMNIHLGKAAKPSLPAL DPMSMLFKMSNSLAEKAAVATPPPLQSKKADHLDRYFYHVNNDQPIDLTKGKSDKGCSLGSVLLSPTSTAPATSSSTVTTAKTSAVVSFM SNSPLRENALSDISDMLKNLTESHTSKSSTPSSISEKSDIDGATLEEAEESTPAQKRKGRQSNWNPQHLLILQAQFAASLRQTSEGKYIM SDLSPQERMHISRFTGLSMTTISHWLANVKYQLRRTGGTKFLKNLDTGHPVFFCNDCASQIRTPSTYISHLESHLGFRLRDLSKLSTEQI -------------------------------------------------------------- >102419_102419_2_ZNF571-TSHZ3_ZNF571_chr19_38077569_ENST00000590751_TSHZ3_chr19_31770658_ENST00000240587_length(transcript)=5001nt_BP=193nt GTCTGCGCCGGCCAGCGCTTGGCCCCTCCCGGGCCGCAGTACTCCTCGTGTGACACGGACATCATTAAGCCAGCTGAGAATCGTGCGGTT ACAGCTGCTGCAGTTTCTTTGCAGACACTGGCTTCTCTGGACTTTGCACACCTCCAGGAGAAGAACCCAGAAAATTGATCAGTTCTAAAA AACCATGCCCCACCCTATGTTTCCGAAGAGTTAAAGGCTGCTGCCCTGGTGGACGAAGGTTTAGACCCAGAGGAGCATACGGCAGATGGA GAGCCCTCGGCCAAGTACATGTGCCCGGAGAAGGAGCTCGCCAGGGCCTGCCCCAGCTACCAGAACTCCCCGGCCGCCGAGTTTTCCTGC CATGAAATGGACAGCGAGTCACACATCAGTGAGACCAGTGACCGAATGGCTGACTTTGAAAGCGGCTCCATCAAGAACGAAGAGGAGACC AAGGAGGTCACGGTCCCACTGGAAGACACGACTGTGTCGGATAGCCTGGAGCAGATGAAGGCCGTGTACAACAACTTCCTCTCCAACTCC TACTGGTCCAACCTCAACCTCAACCTGCACCAGCCCTCCTCGGAGAAGAACAACGGCAGCAGCAGCAGCAGCAGTAGCAGCAGCAGCAGC TGTGGCAGCGGGAGCTTCGACTGGCACCAGAGCGCCATGGCTAAGACGCTGCAGCAGGTGTCACAGAGCCGCATGCTCCCGGAGCCCAGC CTCTTCAGCACCGTGCAGCTGTACCGGCAGAGCAGCAAGCTCTATGGCTCCATCTTCACGGGGGCCAGCAAGTTCCGCTGTAAGGACTGC AGCGCTGCCTACGACACCCTGGTGGAGTTGACGGTGCACATGAACGAGACGGGGCATTACCGCGACGACAACCATGAGACCGATAACAAC AACCCCAAGCGCTGGTCCAAGCCTCGCAAACGCTCCTTGCTGGAAATGGAAGGGAAGGAAGACGCCCAGAAGGTGCTGAAGTGCATGTAC TGTGGCCACTCCTTTGAGTCCCTGCAGGATTTGAGTGTCCATATGATCAAAACAAAACACTACCAAAAAGTGCCTCTGAAGGAACCCGTC ACTCCTGTCGCCGCCAAAATCATCCCTGCCACTCGGAAGAAAGCTTCCCTGGAGCTGGAGCTCCCCAGCTCCCCAGATTCCACAGGTGGA ACCCCCAAAGCCACCATCTCAGACACCAACGATGCACTTCAGAAGAACTCCAACCCTTACATCACGCCAAATAATCGGTACGGCCACCAG AATGGGGCCAGCTATGCATGGCACTTTGAGGCCCGGAAGTCGCAGATCCTGAAGTGCATGGAGTGTGGGAGCTCGCATGACACCCTGCAG GAGCTCACTGCCCACATGATGGTCACTGGCCACTTCATCAAGGTCACCAACTCTGCTATGAAAAAGGGGAAGCCCATTGTGGAGACGCCT GTCACACCTACCATCACAACCCTGCTGGATGAGAAGGTCCAGTCCGTGCCCCTGGCAGCCACCACCTTCACGTCCCCCTCCAATACACCT GCCAGCATCTCCCCAAAACTGAATGTGGAGGTCAAGAAGGAAGTCGACAAGGAGAAAGCGGTCACTGACGAGAAACCTAAGCAAAAAGAC AAGCCTGGCGAAGAAGAGGAGAAGTGTGACATCTCTTCCAAATACCATTACTTGACTGAAAATGACTTAGAAGAGAGTCCCAAGGGGGGG CTTGATATCCTCAAATCCTTGGAAAACACAGTGACATCCGCAATCAACAAGGCCCAGAACGGCACTCCTAGCTGGGGGGGCTATCCCAGC ATCCATGCCGCCTACCAACTTCCCAACATGATGAAGTTGTCCCTGGGCTCGTCGGGGAAGAGCACGCCCCTGAAACCCATGTTTGGCAAC AGTGAGATTGTCTCCCCGACGAAAAACCAGACCCTGGTCTCTCCACCCAGCAGCCAGACGTCCCCCATGCCCAAGACAAACTTTCATGCC ATGGAGGAGCTGGTGAAAAAGGTCACTGAGAAAGTTGCCAAAGTGGAGGAGAAGATGAAGGAGCCGGATGGGAAGCTTTCCCCGCCCAAG CGGGCCACTCCCTCCCCATGTAGCAGCGAAGTCGGGGAACCCATCAAGATGGAGGCATCCAGCGATGGGGGCTTCCGCAGCCAGGAGAAC AGCCCCAGCCCCCCGCGGGATGGGTGCAAGGATGGGAGCCCCCTCGCTGAGCCGGTGGAGAATGGCAAGGAGCTGGTGAAGCCCCTAGCC AGCAGTTTGAGTGGCAGCACGGCCATCATCACCGACCACCCGCCTGAACAGCCTTTTGTTAACCCTTTGAGCGCCCTGCAGTCAGTCATG AACATTCACCTGGGCAAGGCCGCCAAGCCCTCCCTGCCTGCCCTGGACCCCATGAGCATGCTTTTCAAGATGAGCAACAGCCTGGCGGAG AAGGCTGCTGTGGCCACCCCGCCGCCCCTGCAGTCCAAGAAGGCAGACCACCTCGACCGCTATTTCTACCACGTCAACAACGACCAGCCC ATAGACTTGACAAAAGGGAAGAGTGACAAAGGCTGCTCCTTGGGTTCAGTGCTTCTGTCACCCACGTCCACAGCCCCGGCAACCTCCTCA TCCACGGTGACAACGGCAAAGACATCTGCCGTCGTATCATTCATGTCAAACTCGCCGCTACGCGAGAATGCCTTGTCAGATATATCCGAT ATGCTGAAGAACTTGACAGAGAGCCACACGTCAAAATCCTCCACTCCTTCCAGCATCTCCGAGAAGTCTGACATTGACGGGGCCACTCTG GAGGAGGCTGAGGAGTCGACGCCCGCCCAGAAGAGGAAGGGCCGCCAGTCAAACTGGAACCCCCAGCACCTCCTGATCCTCCAGGCCCAG TTTGCCGCCAGCCTCCGGCAGACCTCAGAAGGGAAGTACATCATGTCAGACCTGAGCCCCCAGGAGCGGATGCATATCTCCAGGTTCACC GGGCTGTCCATGACCACCATCAGCCACTGGCTGGCCAACGTGAAATACCAGCTTCGAAGGACAGGTGGAACAAAGTTCCTCAAAAACTTG GACACTGGCCACCCCGTCTTCTTTTGTAACGATTGTGCGTCCCAAATCAGGACTCCTTCCACGTACATCAGTCACCTAGAGTCACACTTA GGCTTCCGGCTACGGGACTTATCCAAACTGTCCACCGAACAGATTAACAGTCAGATAGCACAAACCAAGTCACCGTCAGAAAAAATGGTG ACGTCCTCCCCCGAGGAAGACCTGGGGACTTCCTATCAGTGCAAACTTTGCAATCGGACCTTTGCCAGCAAGCACGCTGTTAAACTTCAC CTTAGCAAAACACACGGGAAATCTCCGGAAGACCACCTTCTGTATGTCTCTGAGTTAGAGAAGCAGTAGCATTTGCTTTTGATAGAAAGG ACTGCAGTTTGCTTTGAGGGAAACTGTGGAAGGCACCTTCAGGCCCCCTCTGACTTGTTGTTCTTGGCACATGTTCTTATTTTAACTGCA GAGAATCACTCTGGGCTGGACTGTTTTGTATAACTGTACAGTGTTTAATAGAGGTGCATAATCAGCTGTTGTTACTGGTAAAATATGAAG GTTAAAATGCAGTGGTAAGTGTTTGGAACTTTGTGTAAACGGGATTTAGTTGTGAGCATCCTCCCGATGCTTCAAGCTGCATGCATTAAC AGACAGTTTAATTAAGCATTTATAACGGAATCAGGCACACCTTTTCCACGAGACTCGAGTGTGCTGGCATTTCTCACCCTTTCATCTTTA GCCCTCTGAGTACTTTGAAGCACTTTTGCATTAATTTGGTTAAAAAATAAAATAAAATAATAATAATGTATGAAGCTCTGTTTTTTAAAC TCCTTACCAGCTTAGTTATAATGAATAATATGAACCTCCATTTATGCAGGTCTGCAGGGGTATAACACGCCTTGAAATTTAAAAGAATAT TATTTTCACATTGAAACATAGATGTATATATTGTATAGATTTCAGACTCTCTTATGAAAAAAAATGTGATTGTGGTTAAATGACCTTTTT CTTGCATTTATAGCAACAGTGTTTTATGCACCTGCTATGCTCTGGGCATAAGCTGTGCCTATGTATAGTGTATATTTCTTTTTTTCTTTT TTTTAAGGTCTATGGGTTTTGTTTTTTACATGCAAACATTGTAAATTATACAGAAGATACCACAGATAGCATTTATAAAGTATACAGAAA CATTATCTGAAAGCAAAGTATGATAGTTTGTTTTGCTATACAGTACATCTATATTGATAGAGGTTCATGTTTAAATTATACATATTTATT AGCATCATATTGTCATTTGTTTTGAGCAGTCTGAATAAACGAGACCGGGAAAGACATCCCTGGCAGGCATCAGAACTATTTTGCACATGA TTTTTAAAGGTATTTATTAGAAATCAAAGAACACTCAAAATAAACTCAGTGCTCAAAGGGTTAAGTCTATTTGAAAAGGTTAAAAAAAAG AACAAAAAAAAAAAAAGAACTTGTACTGTATTTCCTAAACATTGATAAAGCCTTTAAAATGTTTGTACTGTAATACTTTGCTTAAAAGTC ATGAGGCATTCTGTGATCCAACCTCTTTCACTTATTTATAAGCCCTCTTGGTTGCTATTCCATATTGTAGGATGCCTTTCTATTTCAATT GGTAACTTTCTGTTTTGTTCTTCCTAATTATTCTCCCAAGATCCCACACTGCAGCTTTATCTTTAGGCTTATGAAAGGTAACCCGTGGTT ACCGGCTCTCCAAGTGATTCTGTTCTTCTCCATTTTTGGCAGTTAATTTGCAGAAGTAACTGACAGCTGACACCATATGAGAACCTTTGT ATAAAATATTGGCATGTAAACAGCACAGACACCGTAACACACTCTGTGCCCTGTTTGGTTGTTGACAATGAAGCACCATTATGTGACTCT >102419_102419_2_ZNF571-TSHZ3_ZNF571_chr19_38077569_ENST00000590751_TSHZ3_chr19_31770658_ENST00000240587_length(amino acids)=1057AA_BP= MVDEGLDPEEHTADGEPSAKYMCPEKELARACPSYQNSPAAEFSCHEMDSESHISETSDRMADFESGSIKNEEETKEVTVPLEDTTVSDS LEQMKAVYNNFLSNSYWSNLNLNLHQPSSEKNNGSSSSSSSSSSSCGSGSFDWHQSAMAKTLQQVSQSRMLPEPSLFSTVQLYRQSSKLY GSIFTGASKFRCKDCSAAYDTLVELTVHMNETGHYRDDNHETDNNNPKRWSKPRKRSLLEMEGKEDAQKVLKCMYCGHSFESLQDLSVHM IKTKHYQKVPLKEPVTPVAAKIIPATRKKASLELELPSSPDSTGGTPKATISDTNDALQKNSNPYITPNNRYGHQNGASYAWHFEARKSQ ILKCMECGSSHDTLQELTAHMMVTGHFIKVTNSAMKKGKPIVETPVTPTITTLLDEKVQSVPLAATTFTSPSNTPASISPKLNVEVKKEV DKEKAVTDEKPKQKDKPGEEEEKCDISSKYHYLTENDLEESPKGGLDILKSLENTVTSAINKAQNGTPSWGGYPSIHAAYQLPNMMKLSL GSSGKSTPLKPMFGNSEIVSPTKNQTLVSPPSSQTSPMPKTNFHAMEELVKKVTEKVAKVEEKMKEPDGKLSPPKRATPSPCSSEVGEPI KMEASSDGGFRSQENSPSPPRDGCKDGSPLAEPVENGKELVKPLASSLSGSTAIITDHPPEQPFVNPLSALQSVMNIHLGKAAKPSLPAL DPMSMLFKMSNSLAEKAAVATPPPLQSKKADHLDRYFYHVNNDQPIDLTKGKSDKGCSLGSVLLSPTSTAPATSSSTVTTAKTSAVVSFM SNSPLRENALSDISDMLKNLTESHTSKSSTPSSISEKSDIDGATLEEAEESTPAQKRKGRQSNWNPQHLLILQAQFAASLRQTSEGKYIM SDLSPQERMHISRFTGLSMTTISHWLANVKYQLRRTGGTKFLKNLDTGHPVFFCNDCASQIRTPSTYISHLESHLGFRLRDLSKLSTEQI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF571-TSHZ3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF571-TSHZ3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF571-TSHZ3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
