
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BRE-ZNF57 (FusionGDB2 ID:10245) |
Fusion Gene Summary for BRE-ZNF57 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BRE-ZNF57 | Fusion gene ID: 10245 | Hgene | Tgene | Gene symbol | BRE | ZNF57 | Gene ID | 9577 | 126295 |
| Gene name | BRISC and BRCA1 A complex member 2 | zinc finger protein 57 | |
| Synonyms | BRCC4|BRCC45|BRE | ZNF424 | |
| Cytomap | 2p23.2 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | BRISC and BRCA1-A complex member 2BRCA1-A complex subunit BREBRCA1/BRCA2-containing complex subunit 45BRCA1/BRCA2-containing complex, subunit 4brain and reproductive organ-expressed (TNFRSF1A modulator)brain and reproductive organ-expressed protein | zinc finger protein 57zinc finger protein 424 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000603461, ENST00000342045, ENST00000344773, ENST00000361704, ENST00000379624, ENST00000379632, | ENST00000306908, ENST00000523428, | |
| Fusion gene scores | * DoF score | 13 X 9 X 8=936 | 1 X 1 X 1=1 |
| # samples | 15 | 1 | |
| ** MAII score | log2(15/936*10)=-2.64154602908752 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: BRE [Title/Abstract] AND ZNF57 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BRE(28248287)-ZNF57(2915520), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BRE | GO:0043066 | negative regulation of apoptotic process | 15465831 |
 Fusion gene breakpoints across BRE (5'-gene) Fusion gene breakpoints across BRE (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
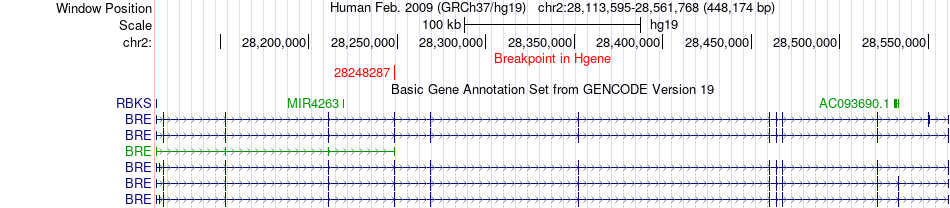 |
 Fusion gene breakpoints across ZNF57 (3'-gene) Fusion gene breakpoints across ZNF57 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
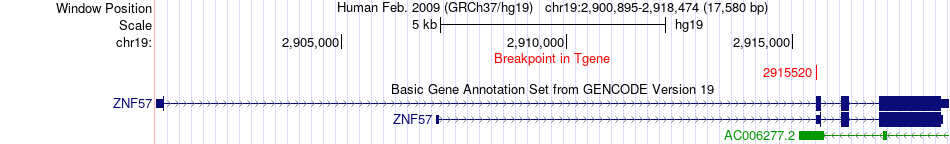 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-FS-A1Z4-06A | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
Top |
Fusion Gene ORF analysis for BRE-ZNF57 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000603461 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| 3UTR-5UTR | ENST00000603461 | ENST00000523428 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| 5CDS-5UTR | ENST00000342045 | ENST00000523428 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| 5CDS-5UTR | ENST00000344773 | ENST00000523428 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| 5CDS-5UTR | ENST00000361704 | ENST00000523428 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| 5CDS-5UTR | ENST00000379624 | ENST00000523428 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| 5CDS-5UTR | ENST00000379632 | ENST00000523428 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| In-frame | ENST00000342045 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| In-frame | ENST00000344773 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| In-frame | ENST00000361704 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| In-frame | ENST00000379624 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
| In-frame | ENST00000379632 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000344773 | BRE | chr2 | 28248287 | + | ENST00000306908 | ZNF57 | chr19 | 2915520 | + | 2485 | 633 | 138 | 2297 | 719 |
| ENST00000379624 | BRE | chr2 | 28248287 | + | ENST00000306908 | ZNF57 | chr19 | 2915520 | + | 2464 | 612 | 117 | 2276 | 719 |
| ENST00000342045 | BRE | chr2 | 28248287 | + | ENST00000306908 | ZNF57 | chr19 | 2915520 | + | 2488 | 636 | 141 | 2300 | 719 |
| ENST00000379632 | BRE | chr2 | 28248287 | + | ENST00000306908 | ZNF57 | chr19 | 2915520 | + | 2487 | 635 | 140 | 2299 | 719 |
| ENST00000361704 | BRE | chr2 | 28248287 | + | ENST00000306908 | ZNF57 | chr19 | 2915520 | + | 2421 | 569 | 74 | 2233 | 719 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000344773 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + | 0.000950093 | 0.99904984 |
| ENST00000379624 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + | 0.00093354 | 0.99906653 |
| ENST00000342045 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + | 0.000936039 | 0.999064 |
| ENST00000379632 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + | 0.000937963 | 0.999062 |
| ENST00000361704 | ENST00000306908 | BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915520 | + | 0.000956813 | 0.99904317 |
Top |
Fusion Genomic Features for BRE-ZNF57 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915519 | + | 5.95E-06 | 0.99999404 |
| BRE | chr2 | 28248287 | + | ZNF57 | chr19 | 2915519 | + | 5.95E-06 | 0.99999404 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
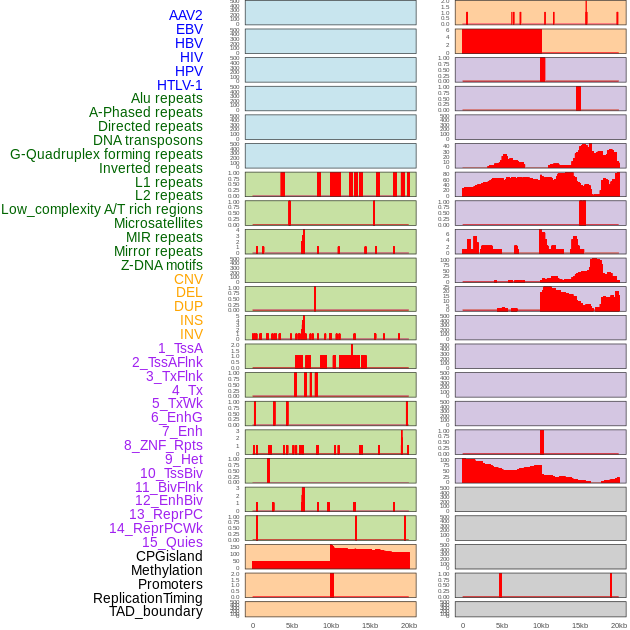 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
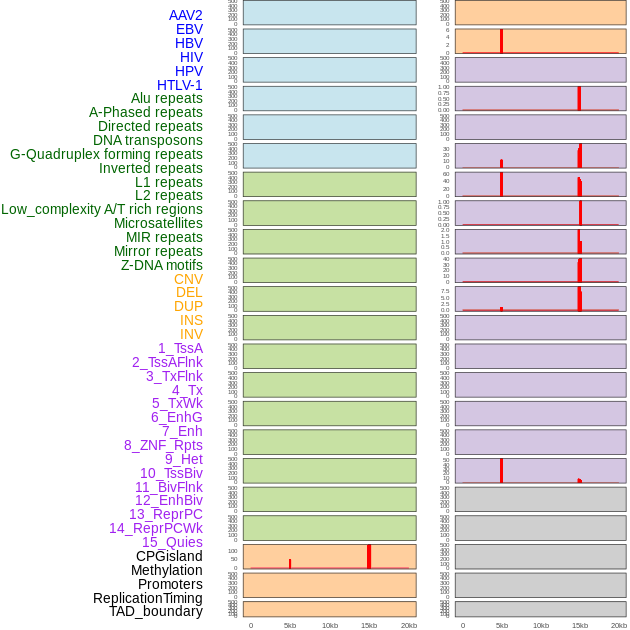 |
Top |
Fusion Protein Features for BRE-ZNF57 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:28248287/chr19:2915520) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000342045 | + | 6 | 13 | 30_147 | 165 | 384.0 | Region | Note=UEV-like 1 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000344773 | + | 5 | 13 | 30_147 | 165 | 528.6666666666666 | Region | Note=UEV-like 1 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000361704 | + | 5 | 13 | 30_147 | 165 | 511.0 | Region | Note=UEV-like 1 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000379624 | + | 5 | 12 | 30_147 | 165 | 384.0 | Region | Note=UEV-like 1 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000379632 | + | 6 | 14 | 30_147 | 165 | 511.0 | Region | Note=UEV-like 1 |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 4_76 | 1 | 556.0 | Domain | KRAB | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 140_162 | 1 | 556.0 | Zinc finger | C2H2-type 1 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 196_218 | 1 | 556.0 | Zinc finger | C2H2-type 2%3B degenerate | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 224_246 | 1 | 556.0 | Zinc finger | C2H2-type 3 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 252_274 | 1 | 556.0 | Zinc finger | C2H2-type 4 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 280_302 | 1 | 556.0 | Zinc finger | C2H2-type 5 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 308_330 | 1 | 556.0 | Zinc finger | C2H2-type 6 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 336_358 | 1 | 556.0 | Zinc finger | C2H2-type 7 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 364_386 | 1 | 556.0 | Zinc finger | C2H2-type 8 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 392_414 | 1 | 556.0 | Zinc finger | C2H2-type 9 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 420_442 | 1 | 556.0 | Zinc finger | C2H2-type 10 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 448_470 | 1 | 556.0 | Zinc finger | C2H2-type 11 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 476_498 | 1 | 556.0 | Zinc finger | C2H2-type 12 | |
| Tgene | ZNF57 | chr2:28248287 | chr19:2915520 | ENST00000306908 | 0 | 4 | 504_526 | 1 | 556.0 | Zinc finger | C2H2-type 13 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000342045 | + | 6 | 13 | 275_364 | 165 | 384.0 | Region | Note=UEV-like 2 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000344773 | + | 5 | 13 | 275_364 | 165 | 528.6666666666666 | Region | Note=UEV-like 2 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000361704 | + | 5 | 13 | 275_364 | 165 | 511.0 | Region | Note=UEV-like 2 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000379624 | + | 5 | 12 | 275_364 | 165 | 384.0 | Region | Note=UEV-like 2 |
| Hgene | BRE | chr2:28248287 | chr19:2915520 | ENST00000379632 | + | 6 | 14 | 275_364 | 165 | 511.0 | Region | Note=UEV-like 2 |
Top |
Fusion Gene Sequence for BRE-ZNF57 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >10245_10245_1_BRE-ZNF57_BRE_chr2_28248287_ENST00000342045_ZNF57_chr19_2915520_ENST00000306908_length(transcript)=2488nt_BP=636nt GGGTACCTGTACCCCACGTAGTCGCCGGTTACCGATCGGACTAAGTTCCAGAAGCAAGAGATAAAGTAATAATGGGTACTGTGGGGAAAA ACACAGAAGAACAATTCGGTAATATAGTGGTGATTTACAAGTCAAGTTAAAATGTCCCCAGAAGTGGCCTTGAACCGAATATCTCCAATG CTCTCCCCTTTCATATCTAGCGTGGTCCGGAATGGAAAAGTGGGACTGGATGCTACAAACTGTTTGAGGATAACTGACTTAAAATCTGGC TGCACATCATTGACTCCTGGGCCCAACTGTGACCGATTTAAACTGCACATACCATATGCTGGAGAGACATTAAAGTGGGATATCATTTTC AATGCCCAATACCCAGAACTGCCTCCCGATTTTATCTTTGGAGAAGATGCTGAATTCCTGCCAGACCCCTCAGCTTTGCAGAATCTTGCC TCCTGGAATCCTTCAAATCCTGAATGTCTCTTACTTGTGGTGAAGGAACTTGTGCAACAATATCACCAATTCCAATGTAGCCGCCTCCGG GAGAGCTCCCGCCTCATGTTTGAATACCAGACATTACTGGAGGAGCCACAGTATGGAGAGAACATGGAAATTTATGCTGGGAAAAAAAAC AACTGGGACTCAGTGGTCTTTGAGGATGTGGCTGTGGACTTCACCCTGGAGGAGTGGGCTTTGCTGGATTCTGCTCAGAGGGACCTCTAC AGAGATGTGATGCTGGAGACCTTCCGGAACCTGGCCTCAGTAGATGATGGGACTCAATTTAAAGCCAATGGGTCAGTTTCTCTGCAGGAT ATGTACGGGCAAGAAAAATCTAAGGAACAGACAATACCAAACTTCACAGGAAATAATTCCTGTGCCTACACTTTAGAAAAAAATTGTGAA GGCTATGGCACTGAAGACCACCACAAAAATCTGAGAAATCATATGGTGGACAGATTCTGTACACATAATGAAGGTAATCAATATGGAGAA GCCATCCATCAAATGCCAGATCTTACCCTGCACAAGAAAGTTTCTGCTGGAGAAAAACCATATGAATGCACCAAGTGCAGGACAGTCTTC ACGCATCTTTCTTCTCTTAAAAGGCACGTCAAGTCTCACTGTGGACGAAAAGCACCTCCAGGTGAGGAATGTAAGCAGGCCTGCATTTGT CCCTCACACCTACACAGTCACGGAAGAACTGACACTGAGGAGAAGCCGTATAAGTGTCAAGCATGTGGGCAAACTTTCCAACATCCTCGT TACCTCTCCCACCACGTAAAGACTCACACAGCAGAGAAAACCTACAAATGCGAGCAGTGTCGGATGGCGTTTAATGGGTTCGCAAGCTTC ACTAGACATGTGAGAACTCACACAAAAGACAGGCCATATAAATGTCAGGAATGTGGGAGAGCCTTCATTTATCCCTCGACATTTCAAAGA CACATGACAACACACACTGGAGAGAAGCCCTATAAATGTCAGCACTGTGGGAAAGCCTTCACTTACCCCCAGGCTTTTCAAAGACATGAG AAGACGCACACGGGAGAGAAGCCCTATGAATGCAAGCAGTGTGGGAAAACATTCAGTTGGTCTGAAACCTTGCGAGTCCACATGAGGATC CACACTGGGGACAAACTCTATAAATGTGAACACTGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCAAGGTCATTTGAGGACGCACACT GGAGAGAAACCTTATGAGTGTAAACAATGTGGGAAAGCCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAG CAGCTCTATAAATGTGAACAATGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCGAGGTCATTTGAGGACGCACACTGGAGAGAAGCCT TATGAGTGTAAACAATGTGGAAAAACCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGCAGCTCCATAAA TGTGAACACTGTGGGAAGGCCTTTACCTCTTCCAGAGCATTCCAAGGTCATTTGAGGATGCACACTGGAGAGAAGCCTTATGAGTGTAAA CAATGTGGAAAAACCTTCACTTGGTCCTCAACCTTACATAATCATGTGAGGATGCACACTGGAGAGAAACCTCACAAATGTAAACAATGT GGGATGTCCTTCAAGTGGCACTCCTCCTTCCGGAACCATCTGAGGATGCACACAGGACAGAAATCCCACGAATGTCAGTCATACTCAAAA GCCTTCAGTTGCCAAGTCATTCTTTCTAAAACCAGTGAGAGCACACACTAAAGAGAAATTCTATAACTTTAATGGGGTAACCTCACATTA ATTCATGTATAATGCTCCAGAAAATTCACACCAGGAGAGAAATCTTACAAGTATGATATTGTCTTTGTCAATACCTCATTTGTAAAACAG >10245_10245_1_BRE-ZNF57_BRE_chr2_28248287_ENST00000342045_ZNF57_chr19_2915520_ENST00000306908_length(amino acids)=719AA_BP=165 MSPEVALNRISPMLSPFISSVVRNGKVGLDATNCLRITDLKSGCTSLTPGPNCDRFKLHIPYAGETLKWDIIFNAQYPELPPDFIFGEDA EFLPDPSALQNLASWNPSNPECLLLVVKELVQQYHQFQCSRLRESSRLMFEYQTLLEEPQYGENMEIYAGKKNNWDSVVFEDVAVDFTLE EWALLDSAQRDLYRDVMLETFRNLASVDDGTQFKANGSVSLQDMYGQEKSKEQTIPNFTGNNSCAYTLEKNCEGYGTEDHHKNLRNHMVD RFCTHNEGNQYGEAIHQMPDLTLHKKVSAGEKPYECTKCRTVFTHLSSLKRHVKSHCGRKAPPGEECKQACICPSHLHSHGRTDTEEKPY KCQACGQTFQHPRYLSHHVKTHTAEKTYKCEQCRMAFNGFASFTRHVRTHTKDRPYKCQECGRAFIYPSTFQRHMTTHTGEKPYKCQHCG KAFTYPQAFQRHEKTHTGEKPYECKQCGKTFSWSETLRVHMRIHTGDKLYKCEHCGKAFTSSRSFQGHLRTHTGEKPYECKQCGKAFTWS STFREHVRIHTQEQLYKCEQCGKAFTSSRSFRGHLRTHTGEKPYECKQCGKTFTWSSTFREHVRIHTQEQLHKCEHCGKAFTSSRAFQGH -------------------------------------------------------------- >10245_10245_2_BRE-ZNF57_BRE_chr2_28248287_ENST00000344773_ZNF57_chr19_2915520_ENST00000306908_length(transcript)=2485nt_BP=633nt GAGTGCGCACGCATCCTGCCCGCGGCGCGCGCGCAGGTCGGTGCGTCTGTCGGGGGCGCGCTCGGGTACCTGTACCCCACGTAGTCGCCG GTTACCGATCGGACTAAGTTCCAGTGGTGATTTACAAGTCAAGTTAAAATGTCCCCAGAAGTGGCCTTGAACCGAATATCTCCAATGCTC TCCCCTTTCATATCTAGCGTGGTCCGGAATGGAAAAGTGGGACTGGATGCTACAAACTGTTTGAGGATAACTGACTTAAAATCTGGCTGC ACATCATTGACTCCTGGGCCCAACTGTGACCGATTTAAACTGCACATACCATATGCTGGAGAGACATTAAAGTGGGATATCATTTTCAAT GCCCAATACCCAGAACTGCCTCCCGATTTTATCTTTGGAGAAGATGCTGAATTCCTGCCAGACCCCTCAGCTTTGCAGAATCTTGCCTCC TGGAATCCTTCAAATCCTGAATGTCTCTTACTTGTGGTGAAGGAACTTGTGCAACAATATCACCAATTCCAATGTAGCCGCCTCCGGGAG AGCTCCCGCCTCATGTTTGAATACCAGACATTACTGGAGGAGCCACAGTATGGAGAGAACATGGAAATTTATGCTGGGAAAAAAAACAAC TGGGACTCAGTGGTCTTTGAGGATGTGGCTGTGGACTTCACCCTGGAGGAGTGGGCTTTGCTGGATTCTGCTCAGAGGGACCTCTACAGA GATGTGATGCTGGAGACCTTCCGGAACCTGGCCTCAGTAGATGATGGGACTCAATTTAAAGCCAATGGGTCAGTTTCTCTGCAGGATATG TACGGGCAAGAAAAATCTAAGGAACAGACAATACCAAACTTCACAGGAAATAATTCCTGTGCCTACACTTTAGAAAAAAATTGTGAAGGC TATGGCACTGAAGACCACCACAAAAATCTGAGAAATCATATGGTGGACAGATTCTGTACACATAATGAAGGTAATCAATATGGAGAAGCC ATCCATCAAATGCCAGATCTTACCCTGCACAAGAAAGTTTCTGCTGGAGAAAAACCATATGAATGCACCAAGTGCAGGACAGTCTTCACG CATCTTTCTTCTCTTAAAAGGCACGTCAAGTCTCACTGTGGACGAAAAGCACCTCCAGGTGAGGAATGTAAGCAGGCCTGCATTTGTCCC TCACACCTACACAGTCACGGAAGAACTGACACTGAGGAGAAGCCGTATAAGTGTCAAGCATGTGGGCAAACTTTCCAACATCCTCGTTAC CTCTCCCACCACGTAAAGACTCACACAGCAGAGAAAACCTACAAATGCGAGCAGTGTCGGATGGCGTTTAATGGGTTCGCAAGCTTCACT AGACATGTGAGAACTCACACAAAAGACAGGCCATATAAATGTCAGGAATGTGGGAGAGCCTTCATTTATCCCTCGACATTTCAAAGACAC ATGACAACACACACTGGAGAGAAGCCCTATAAATGTCAGCACTGTGGGAAAGCCTTCACTTACCCCCAGGCTTTTCAAAGACATGAGAAG ACGCACACGGGAGAGAAGCCCTATGAATGCAAGCAGTGTGGGAAAACATTCAGTTGGTCTGAAACCTTGCGAGTCCACATGAGGATCCAC ACTGGGGACAAACTCTATAAATGTGAACACTGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCAAGGTCATTTGAGGACGCACACTGGA GAGAAACCTTATGAGTGTAAACAATGTGGGAAAGCCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGCAG CTCTATAAATGTGAACAATGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCGAGGTCATTTGAGGACGCACACTGGAGAGAAGCCTTAT GAGTGTAAACAATGTGGAAAAACCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGCAGCTCCATAAATGT GAACACTGTGGGAAGGCCTTTACCTCTTCCAGAGCATTCCAAGGTCATTTGAGGATGCACACTGGAGAGAAGCCTTATGAGTGTAAACAA TGTGGAAAAACCTTCACTTGGTCCTCAACCTTACATAATCATGTGAGGATGCACACTGGAGAGAAACCTCACAAATGTAAACAATGTGGG ATGTCCTTCAAGTGGCACTCCTCCTTCCGGAACCATCTGAGGATGCACACAGGACAGAAATCCCACGAATGTCAGTCATACTCAAAAGCC TTCAGTTGCCAAGTCATTCTTTCTAAAACCAGTGAGAGCACACACTAAAGAGAAATTCTATAACTTTAATGGGGTAACCTCACATTAATT CATGTATAATGCTCCAGAAAATTCACACCAGGAGAGAAATCTTACAAGTATGATATTGTCTTTGTCAATACCTCATTTGTAAAACAGACC >10245_10245_2_BRE-ZNF57_BRE_chr2_28248287_ENST00000344773_ZNF57_chr19_2915520_ENST00000306908_length(amino acids)=719AA_BP=165 MSPEVALNRISPMLSPFISSVVRNGKVGLDATNCLRITDLKSGCTSLTPGPNCDRFKLHIPYAGETLKWDIIFNAQYPELPPDFIFGEDA EFLPDPSALQNLASWNPSNPECLLLVVKELVQQYHQFQCSRLRESSRLMFEYQTLLEEPQYGENMEIYAGKKNNWDSVVFEDVAVDFTLE EWALLDSAQRDLYRDVMLETFRNLASVDDGTQFKANGSVSLQDMYGQEKSKEQTIPNFTGNNSCAYTLEKNCEGYGTEDHHKNLRNHMVD RFCTHNEGNQYGEAIHQMPDLTLHKKVSAGEKPYECTKCRTVFTHLSSLKRHVKSHCGRKAPPGEECKQACICPSHLHSHGRTDTEEKPY KCQACGQTFQHPRYLSHHVKTHTAEKTYKCEQCRMAFNGFASFTRHVRTHTKDRPYKCQECGRAFIYPSTFQRHMTTHTGEKPYKCQHCG KAFTYPQAFQRHEKTHTGEKPYECKQCGKTFSWSETLRVHMRIHTGDKLYKCEHCGKAFTSSRSFQGHLRTHTGEKPYECKQCGKAFTWS STFREHVRIHTQEQLYKCEQCGKAFTSSRSFRGHLRTHTGEKPYECKQCGKTFTWSSTFREHVRIHTQEQLHKCEHCGKAFTSSRAFQGH -------------------------------------------------------------- >10245_10245_3_BRE-ZNF57_BRE_chr2_28248287_ENST00000361704_ZNF57_chr19_2915520_ENST00000306908_length(transcript)=2421nt_BP=569nt GGTACCTGTACCCCACGTAGTCGCCGGTTACCGATCGGACTAAGTTCCAGTGGTGATTTACAAGTCAAGTTAAAATGTCCCCAGAAGTGG CCTTGAACCGAATATCTCCAATGCTCTCCCCTTTCATATCTAGCGTGGTCCGGAATGGAAAAGTGGGACTGGATGCTACAAACTGTTTGA GGATAACTGACTTAAAATCTGGCTGCACATCATTGACTCCTGGGCCCAACTGTGACCGATTTAAACTGCACATACCATATGCTGGAGAGA CATTAAAGTGGGATATCATTTTCAATGCCCAATACCCAGAACTGCCTCCCGATTTTATCTTTGGAGAAGATGCTGAATTCCTGCCAGACC CCTCAGCTTTGCAGAATCTTGCCTCCTGGAATCCTTCAAATCCTGAATGTCTCTTACTTGTGGTGAAGGAACTTGTGCAACAATATCACC AATTCCAATGTAGCCGCCTCCGGGAGAGCTCCCGCCTCATGTTTGAATACCAGACATTACTGGAGGAGCCACAGTATGGAGAGAACATGG AAATTTATGCTGGGAAAAAAAACAACTGGGACTCAGTGGTCTTTGAGGATGTGGCTGTGGACTTCACCCTGGAGGAGTGGGCTTTGCTGG ATTCTGCTCAGAGGGACCTCTACAGAGATGTGATGCTGGAGACCTTCCGGAACCTGGCCTCAGTAGATGATGGGACTCAATTTAAAGCCA ATGGGTCAGTTTCTCTGCAGGATATGTACGGGCAAGAAAAATCTAAGGAACAGACAATACCAAACTTCACAGGAAATAATTCCTGTGCCT ACACTTTAGAAAAAAATTGTGAAGGCTATGGCACTGAAGACCACCACAAAAATCTGAGAAATCATATGGTGGACAGATTCTGTACACATA ATGAAGGTAATCAATATGGAGAAGCCATCCATCAAATGCCAGATCTTACCCTGCACAAGAAAGTTTCTGCTGGAGAAAAACCATATGAAT GCACCAAGTGCAGGACAGTCTTCACGCATCTTTCTTCTCTTAAAAGGCACGTCAAGTCTCACTGTGGACGAAAAGCACCTCCAGGTGAGG AATGTAAGCAGGCCTGCATTTGTCCCTCACACCTACACAGTCACGGAAGAACTGACACTGAGGAGAAGCCGTATAAGTGTCAAGCATGTG GGCAAACTTTCCAACATCCTCGTTACCTCTCCCACCACGTAAAGACTCACACAGCAGAGAAAACCTACAAATGCGAGCAGTGTCGGATGG CGTTTAATGGGTTCGCAAGCTTCACTAGACATGTGAGAACTCACACAAAAGACAGGCCATATAAATGTCAGGAATGTGGGAGAGCCTTCA TTTATCCCTCGACATTTCAAAGACACATGACAACACACACTGGAGAGAAGCCCTATAAATGTCAGCACTGTGGGAAAGCCTTCACTTACC CCCAGGCTTTTCAAAGACATGAGAAGACGCACACGGGAGAGAAGCCCTATGAATGCAAGCAGTGTGGGAAAACATTCAGTTGGTCTGAAA CCTTGCGAGTCCACATGAGGATCCACACTGGGGACAAACTCTATAAATGTGAACACTGTGGGAAGGCTTTTACCTCTTCCAGATCATTCC AAGGTCATTTGAGGACGCACACTGGAGAGAAACCTTATGAGTGTAAACAATGTGGGAAAGCCTTCACTTGGTCCTCAACGTTTAGAGAAC ATGTGAGAATTCACACGCAAGAGCAGCTCTATAAATGTGAACAATGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCGAGGTCATTTGA GGACGCACACTGGAGAGAAGCCTTATGAGTGTAAACAATGTGGAAAAACCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTC ACACGCAAGAGCAGCTCCATAAATGTGAACACTGTGGGAAGGCCTTTACCTCTTCCAGAGCATTCCAAGGTCATTTGAGGATGCACACTG GAGAGAAGCCTTATGAGTGTAAACAATGTGGAAAAACCTTCACTTGGTCCTCAACCTTACATAATCATGTGAGGATGCACACTGGAGAGA AACCTCACAAATGTAAACAATGTGGGATGTCCTTCAAGTGGCACTCCTCCTTCCGGAACCATCTGAGGATGCACACAGGACAGAAATCCC ACGAATGTCAGTCATACTCAAAAGCCTTCAGTTGCCAAGTCATTCTTTCTAAAACCAGTGAGAGCACACACTAAAGAGAAATTCTATAAC TTTAATGGGGTAACCTCACATTAATTCATGTATAATGCTCCAGAAAATTCACACCAGGAGAGAAATCTTACAAGTATGATATTGTCTTTG >10245_10245_3_BRE-ZNF57_BRE_chr2_28248287_ENST00000361704_ZNF57_chr19_2915520_ENST00000306908_length(amino acids)=719AA_BP=165 MSPEVALNRISPMLSPFISSVVRNGKVGLDATNCLRITDLKSGCTSLTPGPNCDRFKLHIPYAGETLKWDIIFNAQYPELPPDFIFGEDA EFLPDPSALQNLASWNPSNPECLLLVVKELVQQYHQFQCSRLRESSRLMFEYQTLLEEPQYGENMEIYAGKKNNWDSVVFEDVAVDFTLE EWALLDSAQRDLYRDVMLETFRNLASVDDGTQFKANGSVSLQDMYGQEKSKEQTIPNFTGNNSCAYTLEKNCEGYGTEDHHKNLRNHMVD RFCTHNEGNQYGEAIHQMPDLTLHKKVSAGEKPYECTKCRTVFTHLSSLKRHVKSHCGRKAPPGEECKQACICPSHLHSHGRTDTEEKPY KCQACGQTFQHPRYLSHHVKTHTAEKTYKCEQCRMAFNGFASFTRHVRTHTKDRPYKCQECGRAFIYPSTFQRHMTTHTGEKPYKCQHCG KAFTYPQAFQRHEKTHTGEKPYECKQCGKTFSWSETLRVHMRIHTGDKLYKCEHCGKAFTSSRSFQGHLRTHTGEKPYECKQCGKAFTWS STFREHVRIHTQEQLYKCEQCGKAFTSSRSFRGHLRTHTGEKPYECKQCGKTFTWSSTFREHVRIHTQEQLHKCEHCGKAFTSSRAFQGH -------------------------------------------------------------- >10245_10245_4_BRE-ZNF57_BRE_chr2_28248287_ENST00000379624_ZNF57_chr19_2915520_ENST00000306908_length(transcript)=2464nt_BP=612nt GCGGCGCGCGCGCAGGTCGGTGCGTCTGTCGGGGGCGCGCTCGGGTACCTGTACCCCACGTAGTCGCCGGTTACCGATCGGACTAAGTTC CAGTGGTGATTTACAAGTCAAGTTAAAATGTCCCCAGAAGTGGCCTTGAACCGAATATCTCCAATGCTCTCCCCTTTCATATCTAGCGTG GTCCGGAATGGAAAAGTGGGACTGGATGCTACAAACTGTTTGAGGATAACTGACTTAAAATCTGGCTGCACATCATTGACTCCTGGGCCC AACTGTGACCGATTTAAACTGCACATACCATATGCTGGAGAGACATTAAAGTGGGATATCATTTTCAATGCCCAATACCCAGAACTGCCT CCCGATTTTATCTTTGGAGAAGATGCTGAATTCCTGCCAGACCCCTCAGCTTTGCAGAATCTTGCCTCCTGGAATCCTTCAAATCCTGAA TGTCTCTTACTTGTGGTGAAGGAACTTGTGCAACAATATCACCAATTCCAATGTAGCCGCCTCCGGGAGAGCTCCCGCCTCATGTTTGAA TACCAGACATTACTGGAGGAGCCACAGTATGGAGAGAACATGGAAATTTATGCTGGGAAAAAAAACAACTGGGACTCAGTGGTCTTTGAG GATGTGGCTGTGGACTTCACCCTGGAGGAGTGGGCTTTGCTGGATTCTGCTCAGAGGGACCTCTACAGAGATGTGATGCTGGAGACCTTC CGGAACCTGGCCTCAGTAGATGATGGGACTCAATTTAAAGCCAATGGGTCAGTTTCTCTGCAGGATATGTACGGGCAAGAAAAATCTAAG GAACAGACAATACCAAACTTCACAGGAAATAATTCCTGTGCCTACACTTTAGAAAAAAATTGTGAAGGCTATGGCACTGAAGACCACCAC AAAAATCTGAGAAATCATATGGTGGACAGATTCTGTACACATAATGAAGGTAATCAATATGGAGAAGCCATCCATCAAATGCCAGATCTT ACCCTGCACAAGAAAGTTTCTGCTGGAGAAAAACCATATGAATGCACCAAGTGCAGGACAGTCTTCACGCATCTTTCTTCTCTTAAAAGG CACGTCAAGTCTCACTGTGGACGAAAAGCACCTCCAGGTGAGGAATGTAAGCAGGCCTGCATTTGTCCCTCACACCTACACAGTCACGGA AGAACTGACACTGAGGAGAAGCCGTATAAGTGTCAAGCATGTGGGCAAACTTTCCAACATCCTCGTTACCTCTCCCACCACGTAAAGACT CACACAGCAGAGAAAACCTACAAATGCGAGCAGTGTCGGATGGCGTTTAATGGGTTCGCAAGCTTCACTAGACATGTGAGAACTCACACA AAAGACAGGCCATATAAATGTCAGGAATGTGGGAGAGCCTTCATTTATCCCTCGACATTTCAAAGACACATGACAACACACACTGGAGAG AAGCCCTATAAATGTCAGCACTGTGGGAAAGCCTTCACTTACCCCCAGGCTTTTCAAAGACATGAGAAGACGCACACGGGAGAGAAGCCC TATGAATGCAAGCAGTGTGGGAAAACATTCAGTTGGTCTGAAACCTTGCGAGTCCACATGAGGATCCACACTGGGGACAAACTCTATAAA TGTGAACACTGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCAAGGTCATTTGAGGACGCACACTGGAGAGAAACCTTATGAGTGTAAA CAATGTGGGAAAGCCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGCAGCTCTATAAATGTGAACAATGT GGGAAGGCTTTTACCTCTTCCAGATCATTCCGAGGTCATTTGAGGACGCACACTGGAGAGAAGCCTTATGAGTGTAAACAATGTGGAAAA ACCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGCAGCTCCATAAATGTGAACACTGTGGGAAGGCCTTT ACCTCTTCCAGAGCATTCCAAGGTCATTTGAGGATGCACACTGGAGAGAAGCCTTATGAGTGTAAACAATGTGGAAAAACCTTCACTTGG TCCTCAACCTTACATAATCATGTGAGGATGCACACTGGAGAGAAACCTCACAAATGTAAACAATGTGGGATGTCCTTCAAGTGGCACTCC TCCTTCCGGAACCATCTGAGGATGCACACAGGACAGAAATCCCACGAATGTCAGTCATACTCAAAAGCCTTCAGTTGCCAAGTCATTCTT TCTAAAACCAGTGAGAGCACACACTAAAGAGAAATTCTATAACTTTAATGGGGTAACCTCACATTAATTCATGTATAATGCTCCAGAAAA TTCACACCAGGAGAGAAATCTTACAAGTATGATATTGTCTTTGTCAATACCTCATTTGTAAAACAGACCCATTAGTCGTGAATTTCCAGC >10245_10245_4_BRE-ZNF57_BRE_chr2_28248287_ENST00000379624_ZNF57_chr19_2915520_ENST00000306908_length(amino acids)=719AA_BP=165 MSPEVALNRISPMLSPFISSVVRNGKVGLDATNCLRITDLKSGCTSLTPGPNCDRFKLHIPYAGETLKWDIIFNAQYPELPPDFIFGEDA EFLPDPSALQNLASWNPSNPECLLLVVKELVQQYHQFQCSRLRESSRLMFEYQTLLEEPQYGENMEIYAGKKNNWDSVVFEDVAVDFTLE EWALLDSAQRDLYRDVMLETFRNLASVDDGTQFKANGSVSLQDMYGQEKSKEQTIPNFTGNNSCAYTLEKNCEGYGTEDHHKNLRNHMVD RFCTHNEGNQYGEAIHQMPDLTLHKKVSAGEKPYECTKCRTVFTHLSSLKRHVKSHCGRKAPPGEECKQACICPSHLHSHGRTDTEEKPY KCQACGQTFQHPRYLSHHVKTHTAEKTYKCEQCRMAFNGFASFTRHVRTHTKDRPYKCQECGRAFIYPSTFQRHMTTHTGEKPYKCQHCG KAFTYPQAFQRHEKTHTGEKPYECKQCGKTFSWSETLRVHMRIHTGDKLYKCEHCGKAFTSSRSFQGHLRTHTGEKPYECKQCGKAFTWS STFREHVRIHTQEQLYKCEQCGKAFTSSRSFRGHLRTHTGEKPYECKQCGKTFTWSSTFREHVRIHTQEQLHKCEHCGKAFTSSRAFQGH -------------------------------------------------------------- >10245_10245_5_BRE-ZNF57_BRE_chr2_28248287_ENST00000379632_ZNF57_chr19_2915520_ENST00000306908_length(transcript)=2487nt_BP=635nt GGTACCTGTACCCCACGTAGTCGCCGGTTACCGATCGGACTAAGTTCCAGAAGCAAGAGATAAAGTAATAATGGGTACTGTGGGGAAAAA CACAGAAGAACAATTCGGTAATATAGTGGTGATTTACAAGTCAAGTTAAAATGTCCCCAGAAGTGGCCTTGAACCGAATATCTCCAATGC TCTCCCCTTTCATATCTAGCGTGGTCCGGAATGGAAAAGTGGGACTGGATGCTACAAACTGTTTGAGGATAACTGACTTAAAATCTGGCT GCACATCATTGACTCCTGGGCCCAACTGTGACCGATTTAAACTGCACATACCATATGCTGGAGAGACATTAAAGTGGGATATCATTTTCA ATGCCCAATACCCAGAACTGCCTCCCGATTTTATCTTTGGAGAAGATGCTGAATTCCTGCCAGACCCCTCAGCTTTGCAGAATCTTGCCT CCTGGAATCCTTCAAATCCTGAATGTCTCTTACTTGTGGTGAAGGAACTTGTGCAACAATATCACCAATTCCAATGTAGCCGCCTCCGGG AGAGCTCCCGCCTCATGTTTGAATACCAGACATTACTGGAGGAGCCACAGTATGGAGAGAACATGGAAATTTATGCTGGGAAAAAAAACA ACTGGGACTCAGTGGTCTTTGAGGATGTGGCTGTGGACTTCACCCTGGAGGAGTGGGCTTTGCTGGATTCTGCTCAGAGGGACCTCTACA GAGATGTGATGCTGGAGACCTTCCGGAACCTGGCCTCAGTAGATGATGGGACTCAATTTAAAGCCAATGGGTCAGTTTCTCTGCAGGATA TGTACGGGCAAGAAAAATCTAAGGAACAGACAATACCAAACTTCACAGGAAATAATTCCTGTGCCTACACTTTAGAAAAAAATTGTGAAG GCTATGGCACTGAAGACCACCACAAAAATCTGAGAAATCATATGGTGGACAGATTCTGTACACATAATGAAGGTAATCAATATGGAGAAG CCATCCATCAAATGCCAGATCTTACCCTGCACAAGAAAGTTTCTGCTGGAGAAAAACCATATGAATGCACCAAGTGCAGGACAGTCTTCA CGCATCTTTCTTCTCTTAAAAGGCACGTCAAGTCTCACTGTGGACGAAAAGCACCTCCAGGTGAGGAATGTAAGCAGGCCTGCATTTGTC CCTCACACCTACACAGTCACGGAAGAACTGACACTGAGGAGAAGCCGTATAAGTGTCAAGCATGTGGGCAAACTTTCCAACATCCTCGTT ACCTCTCCCACCACGTAAAGACTCACACAGCAGAGAAAACCTACAAATGCGAGCAGTGTCGGATGGCGTTTAATGGGTTCGCAAGCTTCA CTAGACATGTGAGAACTCACACAAAAGACAGGCCATATAAATGTCAGGAATGTGGGAGAGCCTTCATTTATCCCTCGACATTTCAAAGAC ACATGACAACACACACTGGAGAGAAGCCCTATAAATGTCAGCACTGTGGGAAAGCCTTCACTTACCCCCAGGCTTTTCAAAGACATGAGA AGACGCACACGGGAGAGAAGCCCTATGAATGCAAGCAGTGTGGGAAAACATTCAGTTGGTCTGAAACCTTGCGAGTCCACATGAGGATCC ACACTGGGGACAAACTCTATAAATGTGAACACTGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCAAGGTCATTTGAGGACGCACACTG GAGAGAAACCTTATGAGTGTAAACAATGTGGGAAAGCCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGC AGCTCTATAAATGTGAACAATGTGGGAAGGCTTTTACCTCTTCCAGATCATTCCGAGGTCATTTGAGGACGCACACTGGAGAGAAGCCTT ATGAGTGTAAACAATGTGGAAAAACCTTCACTTGGTCCTCAACGTTTAGAGAACATGTGAGAATTCACACGCAAGAGCAGCTCCATAAAT GTGAACACTGTGGGAAGGCCTTTACCTCTTCCAGAGCATTCCAAGGTCATTTGAGGATGCACACTGGAGAGAAGCCTTATGAGTGTAAAC AATGTGGAAAAACCTTCACTTGGTCCTCAACCTTACATAATCATGTGAGGATGCACACTGGAGAGAAACCTCACAAATGTAAACAATGTG GGATGTCCTTCAAGTGGCACTCCTCCTTCCGGAACCATCTGAGGATGCACACAGGACAGAAATCCCACGAATGTCAGTCATACTCAAAAG CCTTCAGTTGCCAAGTCATTCTTTCTAAAACCAGTGAGAGCACACACTAAAGAGAAATTCTATAACTTTAATGGGGTAACCTCACATTAA TTCATGTATAATGCTCCAGAAAATTCACACCAGGAGAGAAATCTTACAAGTATGATATTGTCTTTGTCAATACCTCATTTGTAAAACAGA >10245_10245_5_BRE-ZNF57_BRE_chr2_28248287_ENST00000379632_ZNF57_chr19_2915520_ENST00000306908_length(amino acids)=719AA_BP=165 MSPEVALNRISPMLSPFISSVVRNGKVGLDATNCLRITDLKSGCTSLTPGPNCDRFKLHIPYAGETLKWDIIFNAQYPELPPDFIFGEDA EFLPDPSALQNLASWNPSNPECLLLVVKELVQQYHQFQCSRLRESSRLMFEYQTLLEEPQYGENMEIYAGKKNNWDSVVFEDVAVDFTLE EWALLDSAQRDLYRDVMLETFRNLASVDDGTQFKANGSVSLQDMYGQEKSKEQTIPNFTGNNSCAYTLEKNCEGYGTEDHHKNLRNHMVD RFCTHNEGNQYGEAIHQMPDLTLHKKVSAGEKPYECTKCRTVFTHLSSLKRHVKSHCGRKAPPGEECKQACICPSHLHSHGRTDTEEKPY KCQACGQTFQHPRYLSHHVKTHTAEKTYKCEQCRMAFNGFASFTRHVRTHTKDRPYKCQECGRAFIYPSTFQRHMTTHTGEKPYKCQHCG KAFTYPQAFQRHEKTHTGEKPYECKQCGKTFSWSETLRVHMRIHTGDKLYKCEHCGKAFTSSRSFQGHLRTHTGEKPYECKQCGKAFTWS STFREHVRIHTQEQLYKCEQCGKAFTSSRSFRGHLRTHTGEKPYECKQCGKTFTWSSTFREHVRIHTQEQLHKCEHCGKAFTSSRAFQGH -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BRE-ZNF57 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BRE-ZNF57 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BRE-ZNF57 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
