
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF587B-NEK9 (FusionGDB2 ID:102469) |
Fusion Gene Summary for ZNF587B-NEK9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF587B-NEK9 | Fusion gene ID: 102469 | Hgene | Tgene | Gene symbol | ZNF587B | NEK9 | Gene ID | 100293516 | 91754 |
| Gene name | zinc finger protein 587B | NIMA related kinase 9 | |
| Synonyms | - | APUG|LCCS10|NC|NERCC|NERCC1 | |
| Cytomap | 19q13.43 | 14q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 587B | serine/threonine-protein kinase Nek9NIMA (never in mitosis gene a)- related kinase 9nercc1 kinasenimA-related protein kinase 9 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | Q8TD19 | |
| Ensembl transtripts involved in fusion gene | ENST00000316462, ENST00000442832, ENST00000594901, | ENST00000555763, ENST00000238616, | |
| Fusion gene scores | * DoF score | 12 X 7 X 7=588 | 4 X 7 X 3=84 |
| # samples | 14 | 7 | |
| ** MAII score | log2(14/588*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/84*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF587B [Title/Abstract] AND NEK9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF587B(58350517)-NEK9(75573405), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ZNF587B (5'-gene) Fusion gene breakpoints across ZNF587B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
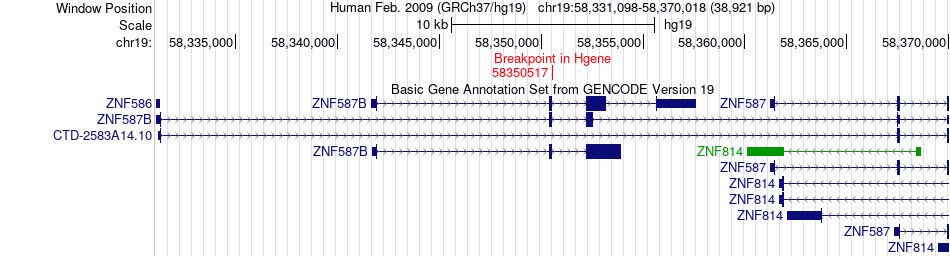 |
 Fusion gene breakpoints across NEK9 (3'-gene) Fusion gene breakpoints across NEK9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
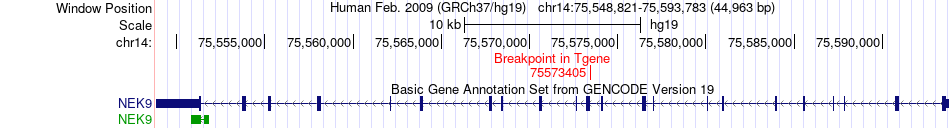 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-BH-A18S | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
Top |
Fusion Gene ORF analysis for ZNF587B-NEK9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000316462 | ENST00000555763 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
| 5CDS-intron | ENST00000442832 | ENST00000555763 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
| 5CDS-intron | ENST00000594901 | ENST00000555763 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
| In-frame | ENST00000316462 | ENST00000238616 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
| In-frame | ENST00000442832 | ENST00000238616 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
| In-frame | ENST00000594901 | ENST00000238616 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000316462 | ZNF587B | chr19 | 58350517 | + | ENST00000238616 | NEK9 | chr14 | 75573405 | - | 4400 | 342 | 125 | 1954 | 609 |
| ENST00000442832 | ZNF587B | chr19 | 58350517 | + | ENST00000238616 | NEK9 | chr14 | 75573405 | - | 4455 | 397 | 114 | 2009 | 631 |
| ENST00000594901 | ZNF587B | chr19 | 58350517 | + | ENST00000238616 | NEK9 | chr14 | 75573405 | - | 4437 | 379 | 96 | 1991 | 631 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000316462 | ENST00000238616 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - | 0.002575551 | 0.9974245 |
| ENST00000442832 | ENST00000238616 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - | 0.00253708 | 0.99746287 |
| ENST00000594901 | ENST00000238616 | ZNF587B | chr19 | 58350517 | + | NEK9 | chr14 | 75573405 | - | 0.002667868 | 0.9973321 |
Top |
Fusion Genomic Features for ZNF587B-NEK9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
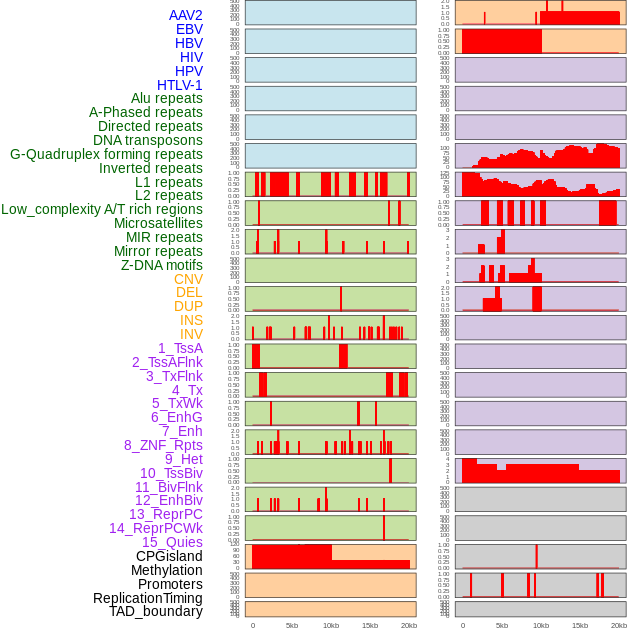 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZNF587B-NEK9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:58350517/chr14:75573405) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NEK9 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Pleiotropic regulator of mitotic progression, participating in the control of spindle dynamics and chromosome separation. Phosphorylates different histones, myelin basic protein, beta-casein, and BICD2. Phosphorylates histone H3 on serine and threonine residues and beta-casein on serine residues. Important for G1/S transition and S phase progression. Phosphorylates NEK6 and NEK7 and stimulates their activity by releasing the autoinhibitory functions of Tyr-108 and Tyr-97 respectively. {ECO:0000269|PubMed:12840024, ECO:0000269|PubMed:14660563, ECO:0000269|PubMed:19941817}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 892_939 | 442 | 980.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 752_760 | 442 | 980.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 765_888 | 442 | 980.0 | Compositional bias | Note=Pro/Ser/Thr-rich | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 445_498 | 442 | 980.0 | Repeat | Note=RCC1 2 | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 499_550 | 442 | 980.0 | Repeat | Note=RCC1 3 | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 551_615 | 442 | 980.0 | Repeat | Note=RCC1 4 | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 616_668 | 442 | 980.0 | Repeat | Note=RCC1 5 | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 669_726 | 442 | 980.0 | Repeat | Note=RCC1 6 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 15_91 | 54 | 403.0 | Domain | KRAB |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 120_142 | 54 | 403.0 | Zinc finger | C2H2-type 2%3B degenerate |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 242_264 | 54 | 403.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 270_292 | 54 | 403.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 298_320 | 54 | 403.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 326_348 | 54 | 403.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 354_383 | 54 | 403.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF587B | chr19:58350517 | chr14:75573405 | ENST00000442832 | + | 2 | 4 | 92_114 | 54 | 403.0 | Zinc finger | C2H2-type 1 |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 52_308 | 442 | 980.0 | Domain | Protein kinase | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 58_66 | 442 | 980.0 | Nucleotide binding | ATP | |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 388_444 | 442 | 980.0 | Repeat | Note=RCC1 1 |
Top |
Fusion Gene Sequence for ZNF587B-NEK9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >102469_102469_1_ZNF587B-NEK9_ZNF587B_chr19_58350517_ENST00000316462_NEK9_chr14_75573405_ENST00000238616_length(transcript)=4400nt_BP=342nt GTAGCGGCCACGTTGATCTGCGATACGCGTGTTTGCCCAGTGAGTGGCCCCGGACTGCTACGTGGGACTGCGGTGAATGAACCCAGAAGG TGGAGAGGAACCGTTCTCGGTGCACAGAGGCGGCTCTGCAGCCCGGTGAGGGCGCCTGCTGCTCCCGGGCAGTGCTTTCCCCAAGTAGTC CGATGGCTGCGGCTGCGCCGAGGCGCCCTACTCAGGGCACAGTGACTTTTGAAGACGTGGCTGTGAAATTTACCCAGGAGGAATGGAATC TCCTTAGTGAGGCTCAGAGATGCCTGTACCGTGATGTGACTCTGGAGAACCTGGCACTTATGTCCTCCCTGGATGAGGGTCAGCTCTATG CCTTCGGATCAGATTATTATGGCTGCATGGGGGTGGACAAAGTTGCTGGCCCTGAAGTGCTAGAACCCATGCAGCTGAACTTCTTCCTCA GCAATCCAGTGGAGCAGGTCTCCTGTGGAGATAATCATGTGGTGGTTCTGACACGAAACAAGGAAGTCTATTCTTGGGGCTGTGGCGAAT ATGGACGACTGGGTTTGGATTCAGAAGAGGATTATTATACACCACAAAAGGTGGATGTTCCCAAGGCCTTGATTATTGTTGCAGTTCAAT GTGGCTGTGATGGGACATTTCTGTTGACCCAGTCAGGCAAAGTGCTGGCCTGTGGACTCAATGAATTCAATAAGCTGGGTCTGAATCAGT GCATGTCGGGAATTATCAACCATGAAGCATACCATGAAGTTCCCTACACAACGTCCTTTACCTTGGCCAAACAGTTGTCCTTTTATAAGA TCCGTACCATTGCCCCAGGCAAGACTCACACAGCTGCTATTGATGAGCGAGGCCGGCTGCTGACCTTTGGCTGCAACAAGTGTGGGCAGC TGGGCGTTGGGAACTACAAGAAGCGTCTGGGAATCAACCTGTTGGGGGGACCCCTTGGTGGGAAGCAAGTGATCAGGGTCTCCTGCGGTG ATGAGTTTACCATTGCTGCCACTGATGATAATCACATTTTTGCCTGGGGCAATGGTGGTAATGGCCGCCTGGCAATGACCCCCACAGAGA GACCACATGGCTCTGATATCTGTACCTCATGGCCTCGGCCTATTTTTGGATCTCTGCATCATGTCCCGGACCTGTCTTGCCGTGGATGGC ATACCATTCTCATCGTTGAGAAAGTATTGAATTCTAAGACCATCCGTTCCAATAGCAGTGGCTTATCCATTGGAACTGTGTTTCAGAGCT CTAGCCCGGGAGGAGGCGGCGGGGGCGGCGGTGGTGAAGAAGAGGACAGTCAGCAGGAATCTGAAACTCCTGACCCAAGTGGAGGCTTCC GAGGAACAATGGAAGCAGACCGAGGAATGGAAGGTTTAATCAGTCCCACAGAGGCCATGGGGAACAGTAATGGGGCCAGCAGCTCCTGTC CTGGCTGGCTTCGAAAGGAGCTGGAAAATGCAGAATTTATCCCCATGCCTGACAGCCCATCTCCTCTCAGTGCAGCGTTTTCAGAATCTG AGAAAGATACCCTGCCCTATGAAGAGCTGCAAGGACTCAAAGTGGCCTCTGAAGCTCCTTTGGAACACAAACCCCAAGTAGAAGCCTCGT CACCTCGGCTGAATCCTGCAGTAACCTGTGCTGGGAAGGGAACACCACTGACTCCTCCTGCGTGTGCGTGCAGCTCTCTGCAGGTGGAGG TTGAGAGATTGCAGGGTCTGGTGTTAAAGTGTCTGGCTGAACAACAGAAGCTACAGCAAGAAAACCTCCAGATTTTTACCCAACTGCAGA AGTTGAACAAGAAATTAGAAGGAGGGCAGCAGGTGGGGATGCATTCCAAAGGAACTCAGACAGCAAAGGAAGAGATGGAAATGGATCCAA AGCCTGACTTAGATTCAGATTCCTGGTGCCTCCTGGGAACAGACTCCTGTAGACCCAGCCTCTAGTCTCCTGAGCCTGTAGAGCCCCCAG GAGACTGGGACCCAAAGAACTTCACAGCACACTTACCGAATGCAGAGAGCAGCTTTCCTGGCTTTGTTCACTTGCAGAAAAGGAGCGCAA GGCAGAGGCTCTGAAGCACTTTCCTTGTACATTTGGAGAGTGGCATTGCCTTTTAGATAGGATCTAGGAGTGATTTTATTGTTTTGGAGA ATGGAAGGGCCCCCATGGCCCTGGCTTTGTCATCAGTGACTGCCATAGCAACAGCAGCTCTGTACCTCATCTGTTGATCCCACCTTTGAA GAGGAGACACAGTGCTCACCTTAATTGCGCTGGTAGCAGCTTATATCCCATGTATCATTTTCACCATTGATTGGAAGCTGCCTTGGGAAT TCAGTACCAGGCATTACCCCTCTGGGTGGGAGAGGGAGAAGTGTAAAGTTGGAGTGGGCTGGAATCAGGTGTGGCCCGCCCAGTGTCCTC TGCAGAGTGGTGAAGTAGTCTGGCCCTCTTGGGAGCCCTGAGTCCAGGAAAATATGTCTGATGGAGTCAATCTAGGGCTTGTTTCGAAAA AGTTCAGTTACTCTGTGCAGCTAAATGCTTTAGGAGGAAAGGTAGGCTTAGGTTGCTTTTCCTCTGAGGGTTGATTGAAATTTCTTCAGT GAGGAATAGAGAAAGGGCAGGACCCTCCTCATCACACAGCTGGTGTTTCAGGCTGTGACCAATGCAGGGTGGGATTTCCTAACTGTGGAT GAGGGGATGAGGTGTCTCTGAGGGATGAGGTGTCTCAGAGAATTGAGTCCATGGGGCAGTCAGAATAGCCTTAAGAGAAAATCATGAAGG AGAAGAGGTCCTCCTTTAGCTGCCTCTACTTGGTATCTTAGAGAGGGCTTAGAGGGCTCTCAGTCTTCTGCCCATGAAAAGACTTCTTTG AGCCTCTGCCTTCATGGCTCTTAGGGTTCTGATCTTGATATCAGCAGCCCCAACCACTTTCTTTCTGAATGTCTAGTCAGTATTTTTCCC CTTTTGGTGTTTTATGAAGCCATGTGGTAACGAATGAATCTGTATCATTTTTCCTACCTGAGTGGCCCAAAGCCAGCACCAAACCCTGGG AGTCCCTGAAGCCTAACAGAACAGGTAGAACTTGCAAAAGGAATTTGGCTGAGAGCTCACTTCTAATCCTGTACTCACTGTGTCTTTGTA GATAGAAACAAGCTAGCTTTACCAAAGAGAAATAGTGTCATAGAAGAACAAAACTTCATATAGAAAGTTCTAGGCAAGTATTTGATGGTT TCCTTAAGGATGTGAGCTTTGTATTTCCACCTAGCCTTGTAAAATGTTCCTGTGGTATTTTGTGTCACACATCCTACCTTTGATGAGTCT CACATCCCACCTTCCTATAAACAGCTAAATTAATTTTGTTTCATCTTCCCCAGACCAAAATGTTTGATAATCTTATCAACATTGTGGGAG GTCTTGTGCAATGGAAATTTTGCCATTTCTCCAAAACTGGTGGCATAAAGGCTGATGCTTGGGGAGAACCCCATTGCTCGGGACAGGCAA CTCTGTTCAATGGGATCTTCTTTGGTTTGATGTTCCCATTGTTTTCTCAGTTCTGGGAAGCCTAGTACATTAGTACTAATGTAATCACTG AAACCTTTTCTTGAAATAAGGGAAGCAGCCAAACTTTGATTAAAGTTGCAAGTTCTGGGGACTTGCGGGGGTTGTCATAAACTGTAACAG TGGGTTTTGGTTCAGCATGTAAATGCAACTTTGATTTTCTTGAGGACCGATTGACCTGTCATGTCCCTGTATCCTCATGCTCATCATCTC AGCAGGCCTGAGAGGCTGGGTCAGTTTGGGTGTTCATCATGAGGATTGCTTCTGCCATGGAGCTGATGGACATGGGCAGGTTGCTGAGAA GGTGGGGTGAAAGTGAGTGCCGGGGGTGGGTGAGTGCCCTGGTCTTGTTCATAGGGGAGCCTTTCCCTAGCAGTGGAACGCTGTGGTCAT TTTCTCTAGCATATTCCCTTGGGAAGTCTAGATTTGCTATTAATCTGGCTGAGAATCTAAGTTCTGTGCCTTAGAGACAGTTTGCACTTT CCCATATTGTGCCTGGGACAGCCATATGATTTTTTTTCCCACCAAACAAGTATGCAAACAGAAACCAGTTCAAAGGGGGATGGAGTAAAA GATGAGGCAGTAGAAATGCCTTTGAATGGTTTTCTGTAGCTAATTCTCTTTAAATTTTGTCCTGCTTTTTTTCTTTATGCAGTGCTAGGT GTTTTAAGTTTTCTAGTAGTATTGCTTTTGAGTTACAGTATAACCTGAGTTACTCCTCTGCTCTAACATTGTTGCAGAAGAACTAGTTTA >102469_102469_1_ZNF587B-NEK9_ZNF587B_chr19_58350517_ENST00000316462_NEK9_chr14_75573405_ENST00000238616_length(amino acids)=609AA_BP=72 MQPGEGACCSRAVLSPSSPMAAAAPRRPTQGTVTFEDVAVKFTQEEWNLLSEAQRCLYRDVTLENLALMSSLDEGQLYAFGSDYYGCMGV DKVAGPEVLEPMQLNFFLSNPVEQVSCGDNHVVVLTRNKEVYSWGCGEYGRLGLDSEEDYYTPQKVDVPKALIIVAVQCGCDGTFLLTQS GKVLACGLNEFNKLGLNQCMSGIINHEAYHEVPYTTSFTLAKQLSFYKIRTIAPGKTHTAAIDERGRLLTFGCNKCGQLGVGNYKKRLGI NLLGGPLGGKQVIRVSCGDEFTIAATDDNHIFAWGNGGNGRLAMTPTERPHGSDICTSWPRPIFGSLHHVPDLSCRGWHTILIVEKVLNS KTIRSNSSGLSIGTVFQSSSPGGGGGGGGGEEEDSQQESETPDPSGGFRGTMEADRGMEGLISPTEAMGNSNGASSSCPGWLRKELENAE FIPMPDSPSPLSAAFSESEKDTLPYEELQGLKVASEAPLEHKPQVEASSPRLNPAVTCAGKGTPLTPPACACSSLQVEVERLQGLVLKCL -------------------------------------------------------------- >102469_102469_2_ZNF587B-NEK9_ZNF587B_chr19_58350517_ENST00000442832_NEK9_chr14_75573405_ENST00000238616_length(transcript)=4455nt_BP=397nt TTTTGATTGGTGTTGGGTTTATTTGTCGGAGAGGCTCCTGAGCGCTAGGTCGGCACTGCGGTGACTGAACCCAGAAGGCGGAGAACAGTT GTCCTCTGCTGCACAGAGGCGACTCTGGAGCTCTGTGACGGCGCCAAGCGTGACCCACCCCTGGGCCAGGATAGGGACCGTCATGCCCAT ATCTCCTGGCTGGTCACCCTCTCCTCCCAACCCTGCTTTAAACCACGTGGTTCGATGGCGGTGGTGGCCACGCTGAGGCTCTCTGCTCAG GGCACAGTGACTTTTGAAGACGTGGCTGTGAAATTTACCCAGGAGGAATGGAATCTCCTTAGTGAGGCTCAGAGATGCCTGTACCGTGAT GTGACTCTGGAGAACCTGGCACTTATGTCCTCCCTGGATGAGGGTCAGCTCTATGCCTTCGGATCAGATTATTATGGCTGCATGGGGGTG GACAAAGTTGCTGGCCCTGAAGTGCTAGAACCCATGCAGCTGAACTTCTTCCTCAGCAATCCAGTGGAGCAGGTCTCCTGTGGAGATAAT CATGTGGTGGTTCTGACACGAAACAAGGAAGTCTATTCTTGGGGCTGTGGCGAATATGGACGACTGGGTTTGGATTCAGAAGAGGATTAT TATACACCACAAAAGGTGGATGTTCCCAAGGCCTTGATTATTGTTGCAGTTCAATGTGGCTGTGATGGGACATTTCTGTTGACCCAGTCA GGCAAAGTGCTGGCCTGTGGACTCAATGAATTCAATAAGCTGGGTCTGAATCAGTGCATGTCGGGAATTATCAACCATGAAGCATACCAT GAAGTTCCCTACACAACGTCCTTTACCTTGGCCAAACAGTTGTCCTTTTATAAGATCCGTACCATTGCCCCAGGCAAGACTCACACAGCT GCTATTGATGAGCGAGGCCGGCTGCTGACCTTTGGCTGCAACAAGTGTGGGCAGCTGGGCGTTGGGAACTACAAGAAGCGTCTGGGAATC AACCTGTTGGGGGGACCCCTTGGTGGGAAGCAAGTGATCAGGGTCTCCTGCGGTGATGAGTTTACCATTGCTGCCACTGATGATAATCAC ATTTTTGCCTGGGGCAATGGTGGTAATGGCCGCCTGGCAATGACCCCCACAGAGAGACCACATGGCTCTGATATCTGTACCTCATGGCCT CGGCCTATTTTTGGATCTCTGCATCATGTCCCGGACCTGTCTTGCCGTGGATGGCATACCATTCTCATCGTTGAGAAAGTATTGAATTCT AAGACCATCCGTTCCAATAGCAGTGGCTTATCCATTGGAACTGTGTTTCAGAGCTCTAGCCCGGGAGGAGGCGGCGGGGGCGGCGGTGGT GAAGAAGAGGACAGTCAGCAGGAATCTGAAACTCCTGACCCAAGTGGAGGCTTCCGAGGAACAATGGAAGCAGACCGAGGAATGGAAGGT TTAATCAGTCCCACAGAGGCCATGGGGAACAGTAATGGGGCCAGCAGCTCCTGTCCTGGCTGGCTTCGAAAGGAGCTGGAAAATGCAGAA TTTATCCCCATGCCTGACAGCCCATCTCCTCTCAGTGCAGCGTTTTCAGAATCTGAGAAAGATACCCTGCCCTATGAAGAGCTGCAAGGA CTCAAAGTGGCCTCTGAAGCTCCTTTGGAACACAAACCCCAAGTAGAAGCCTCGTCACCTCGGCTGAATCCTGCAGTAACCTGTGCTGGG AAGGGAACACCACTGACTCCTCCTGCGTGTGCGTGCAGCTCTCTGCAGGTGGAGGTTGAGAGATTGCAGGGTCTGGTGTTAAAGTGTCTG GCTGAACAACAGAAGCTACAGCAAGAAAACCTCCAGATTTTTACCCAACTGCAGAAGTTGAACAAGAAATTAGAAGGAGGGCAGCAGGTG GGGATGCATTCCAAAGGAACTCAGACAGCAAAGGAAGAGATGGAAATGGATCCAAAGCCTGACTTAGATTCAGATTCCTGGTGCCTCCTG GGAACAGACTCCTGTAGACCCAGCCTCTAGTCTCCTGAGCCTGTAGAGCCCCCAGGAGACTGGGACCCAAAGAACTTCACAGCACACTTA CCGAATGCAGAGAGCAGCTTTCCTGGCTTTGTTCACTTGCAGAAAAGGAGCGCAAGGCAGAGGCTCTGAAGCACTTTCCTTGTACATTTG GAGAGTGGCATTGCCTTTTAGATAGGATCTAGGAGTGATTTTATTGTTTTGGAGAATGGAAGGGCCCCCATGGCCCTGGCTTTGTCATCA GTGACTGCCATAGCAACAGCAGCTCTGTACCTCATCTGTTGATCCCACCTTTGAAGAGGAGACACAGTGCTCACCTTAATTGCGCTGGTA GCAGCTTATATCCCATGTATCATTTTCACCATTGATTGGAAGCTGCCTTGGGAATTCAGTACCAGGCATTACCCCTCTGGGTGGGAGAGG GAGAAGTGTAAAGTTGGAGTGGGCTGGAATCAGGTGTGGCCCGCCCAGTGTCCTCTGCAGAGTGGTGAAGTAGTCTGGCCCTCTTGGGAG CCCTGAGTCCAGGAAAATATGTCTGATGGAGTCAATCTAGGGCTTGTTTCGAAAAAGTTCAGTTACTCTGTGCAGCTAAATGCTTTAGGA GGAAAGGTAGGCTTAGGTTGCTTTTCCTCTGAGGGTTGATTGAAATTTCTTCAGTGAGGAATAGAGAAAGGGCAGGACCCTCCTCATCAC ACAGCTGGTGTTTCAGGCTGTGACCAATGCAGGGTGGGATTTCCTAACTGTGGATGAGGGGATGAGGTGTCTCTGAGGGATGAGGTGTCT CAGAGAATTGAGTCCATGGGGCAGTCAGAATAGCCTTAAGAGAAAATCATGAAGGAGAAGAGGTCCTCCTTTAGCTGCCTCTACTTGGTA TCTTAGAGAGGGCTTAGAGGGCTCTCAGTCTTCTGCCCATGAAAAGACTTCTTTGAGCCTCTGCCTTCATGGCTCTTAGGGTTCTGATCT TGATATCAGCAGCCCCAACCACTTTCTTTCTGAATGTCTAGTCAGTATTTTTCCCCTTTTGGTGTTTTATGAAGCCATGTGGTAACGAAT GAATCTGTATCATTTTTCCTACCTGAGTGGCCCAAAGCCAGCACCAAACCCTGGGAGTCCCTGAAGCCTAACAGAACAGGTAGAACTTGC AAAAGGAATTTGGCTGAGAGCTCACTTCTAATCCTGTACTCACTGTGTCTTTGTAGATAGAAACAAGCTAGCTTTACCAAAGAGAAATAG TGTCATAGAAGAACAAAACTTCATATAGAAAGTTCTAGGCAAGTATTTGATGGTTTCCTTAAGGATGTGAGCTTTGTATTTCCACCTAGC CTTGTAAAATGTTCCTGTGGTATTTTGTGTCACACATCCTACCTTTGATGAGTCTCACATCCCACCTTCCTATAAACAGCTAAATTAATT TTGTTTCATCTTCCCCAGACCAAAATGTTTGATAATCTTATCAACATTGTGGGAGGTCTTGTGCAATGGAAATTTTGCCATTTCTCCAAA ACTGGTGGCATAAAGGCTGATGCTTGGGGAGAACCCCATTGCTCGGGACAGGCAACTCTGTTCAATGGGATCTTCTTTGGTTTGATGTTC CCATTGTTTTCTCAGTTCTGGGAAGCCTAGTACATTAGTACTAATGTAATCACTGAAACCTTTTCTTGAAATAAGGGAAGCAGCCAAACT TTGATTAAAGTTGCAAGTTCTGGGGACTTGCGGGGGTTGTCATAAACTGTAACAGTGGGTTTTGGTTCAGCATGTAAATGCAACTTTGAT TTTCTTGAGGACCGATTGACCTGTCATGTCCCTGTATCCTCATGCTCATCATCTCAGCAGGCCTGAGAGGCTGGGTCAGTTTGGGTGTTC ATCATGAGGATTGCTTCTGCCATGGAGCTGATGGACATGGGCAGGTTGCTGAGAAGGTGGGGTGAAAGTGAGTGCCGGGGGTGGGTGAGT GCCCTGGTCTTGTTCATAGGGGAGCCTTTCCCTAGCAGTGGAACGCTGTGGTCATTTTCTCTAGCATATTCCCTTGGGAAGTCTAGATTT GCTATTAATCTGGCTGAGAATCTAAGTTCTGTGCCTTAGAGACAGTTTGCACTTTCCCATATTGTGCCTGGGACAGCCATATGATTTTTT TTCCCACCAAACAAGTATGCAAACAGAAACCAGTTCAAAGGGGGATGGAGTAAAAGATGAGGCAGTAGAAATGCCTTTGAATGGTTTTCT GTAGCTAATTCTCTTTAAATTTTGTCCTGCTTTTTTTCTTTATGCAGTGCTAGGTGTTTTAAGTTTTCTAGTAGTATTGCTTTTGAGTTA CAGTATAACCTGAGTTACTCCTCTGCTCTAACATTGTTGCAGAAGAACTAGTTTATTGTTAGCCAGGTTGCTTGAAAGGTTGAGAGTGGA >102469_102469_2_ZNF587B-NEK9_ZNF587B_chr19_58350517_ENST00000442832_NEK9_chr14_75573405_ENST00000238616_length(amino acids)=631AA_BP=94 MELCDGAKRDPPLGQDRDRHAHISWLVTLSSQPCFKPRGSMAVVATLRLSAQGTVTFEDVAVKFTQEEWNLLSEAQRCLYRDVTLENLAL MSSLDEGQLYAFGSDYYGCMGVDKVAGPEVLEPMQLNFFLSNPVEQVSCGDNHVVVLTRNKEVYSWGCGEYGRLGLDSEEDYYTPQKVDV PKALIIVAVQCGCDGTFLLTQSGKVLACGLNEFNKLGLNQCMSGIINHEAYHEVPYTTSFTLAKQLSFYKIRTIAPGKTHTAAIDERGRL LTFGCNKCGQLGVGNYKKRLGINLLGGPLGGKQVIRVSCGDEFTIAATDDNHIFAWGNGGNGRLAMTPTERPHGSDICTSWPRPIFGSLH HVPDLSCRGWHTILIVEKVLNSKTIRSNSSGLSIGTVFQSSSPGGGGGGGGGEEEDSQQESETPDPSGGFRGTMEADRGMEGLISPTEAM GNSNGASSSCPGWLRKELENAEFIPMPDSPSPLSAAFSESEKDTLPYEELQGLKVASEAPLEHKPQVEASSPRLNPAVTCAGKGTPLTPP ACACSSLQVEVERLQGLVLKCLAEQQKLQQENLQIFTQLQKLNKKLEGGQQVGMHSKGTQTAKEEMEMDPKPDLDSDSWCLLGTDSCRPS -------------------------------------------------------------- >102469_102469_3_ZNF587B-NEK9_ZNF587B_chr19_58350517_ENST00000594901_NEK9_chr14_75573405_ENST00000238616_length(transcript)=4437nt_BP=379nt TTATTTGTCGGAGAGGCTCCTGAGCGCTAGGTCGGCACTGCGGTGACTGAACCCAGAAGGCGGAGAACAGTTGTCCTCTGCTGCACAGAG GCGACTCTGGAGCTCTGTGACGGCGCCAAGCGTGACCCACCCCTGGGCCAGGATAGGGACCGTCATGCCCATATCTCCTGGCTGGTCACC CTCTCCTCCCAACCCTGCTTTAAACCACGTGGTTCGATGGCGGTGGTGGCCACGCTGAGGCTCTCTGCTCAGGGCACAGTGACTTTTGAA GACGTGGCTGTGAAATTTACCCAGGAGGAATGGAATCTCCTTAGTGAGGCTCAGAGATGCCTGTACCGTGATGTGACTCTGGAGAACCTG GCACTTATGTCCTCCCTGGATGAGGGTCAGCTCTATGCCTTCGGATCAGATTATTATGGCTGCATGGGGGTGGACAAAGTTGCTGGCCCT GAAGTGCTAGAACCCATGCAGCTGAACTTCTTCCTCAGCAATCCAGTGGAGCAGGTCTCCTGTGGAGATAATCATGTGGTGGTTCTGACA CGAAACAAGGAAGTCTATTCTTGGGGCTGTGGCGAATATGGACGACTGGGTTTGGATTCAGAAGAGGATTATTATACACCACAAAAGGTG GATGTTCCCAAGGCCTTGATTATTGTTGCAGTTCAATGTGGCTGTGATGGGACATTTCTGTTGACCCAGTCAGGCAAAGTGCTGGCCTGT GGACTCAATGAATTCAATAAGCTGGGTCTGAATCAGTGCATGTCGGGAATTATCAACCATGAAGCATACCATGAAGTTCCCTACACAACG TCCTTTACCTTGGCCAAACAGTTGTCCTTTTATAAGATCCGTACCATTGCCCCAGGCAAGACTCACACAGCTGCTATTGATGAGCGAGGC CGGCTGCTGACCTTTGGCTGCAACAAGTGTGGGCAGCTGGGCGTTGGGAACTACAAGAAGCGTCTGGGAATCAACCTGTTGGGGGGACCC CTTGGTGGGAAGCAAGTGATCAGGGTCTCCTGCGGTGATGAGTTTACCATTGCTGCCACTGATGATAATCACATTTTTGCCTGGGGCAAT GGTGGTAATGGCCGCCTGGCAATGACCCCCACAGAGAGACCACATGGCTCTGATATCTGTACCTCATGGCCTCGGCCTATTTTTGGATCT CTGCATCATGTCCCGGACCTGTCTTGCCGTGGATGGCATACCATTCTCATCGTTGAGAAAGTATTGAATTCTAAGACCATCCGTTCCAAT AGCAGTGGCTTATCCATTGGAACTGTGTTTCAGAGCTCTAGCCCGGGAGGAGGCGGCGGGGGCGGCGGTGGTGAAGAAGAGGACAGTCAG CAGGAATCTGAAACTCCTGACCCAAGTGGAGGCTTCCGAGGAACAATGGAAGCAGACCGAGGAATGGAAGGTTTAATCAGTCCCACAGAG GCCATGGGGAACAGTAATGGGGCCAGCAGCTCCTGTCCTGGCTGGCTTCGAAAGGAGCTGGAAAATGCAGAATTTATCCCCATGCCTGAC AGCCCATCTCCTCTCAGTGCAGCGTTTTCAGAATCTGAGAAAGATACCCTGCCCTATGAAGAGCTGCAAGGACTCAAAGTGGCCTCTGAA GCTCCTTTGGAACACAAACCCCAAGTAGAAGCCTCGTCACCTCGGCTGAATCCTGCAGTAACCTGTGCTGGGAAGGGAACACCACTGACT CCTCCTGCGTGTGCGTGCAGCTCTCTGCAGGTGGAGGTTGAGAGATTGCAGGGTCTGGTGTTAAAGTGTCTGGCTGAACAACAGAAGCTA CAGCAAGAAAACCTCCAGATTTTTACCCAACTGCAGAAGTTGAACAAGAAATTAGAAGGAGGGCAGCAGGTGGGGATGCATTCCAAAGGA ACTCAGACAGCAAAGGAAGAGATGGAAATGGATCCAAAGCCTGACTTAGATTCAGATTCCTGGTGCCTCCTGGGAACAGACTCCTGTAGA CCCAGCCTCTAGTCTCCTGAGCCTGTAGAGCCCCCAGGAGACTGGGACCCAAAGAACTTCACAGCACACTTACCGAATGCAGAGAGCAGC TTTCCTGGCTTTGTTCACTTGCAGAAAAGGAGCGCAAGGCAGAGGCTCTGAAGCACTTTCCTTGTACATTTGGAGAGTGGCATTGCCTTT TAGATAGGATCTAGGAGTGATTTTATTGTTTTGGAGAATGGAAGGGCCCCCATGGCCCTGGCTTTGTCATCAGTGACTGCCATAGCAACA GCAGCTCTGTACCTCATCTGTTGATCCCACCTTTGAAGAGGAGACACAGTGCTCACCTTAATTGCGCTGGTAGCAGCTTATATCCCATGT ATCATTTTCACCATTGATTGGAAGCTGCCTTGGGAATTCAGTACCAGGCATTACCCCTCTGGGTGGGAGAGGGAGAAGTGTAAAGTTGGA GTGGGCTGGAATCAGGTGTGGCCCGCCCAGTGTCCTCTGCAGAGTGGTGAAGTAGTCTGGCCCTCTTGGGAGCCCTGAGTCCAGGAAAAT ATGTCTGATGGAGTCAATCTAGGGCTTGTTTCGAAAAAGTTCAGTTACTCTGTGCAGCTAAATGCTTTAGGAGGAAAGGTAGGCTTAGGT TGCTTTTCCTCTGAGGGTTGATTGAAATTTCTTCAGTGAGGAATAGAGAAAGGGCAGGACCCTCCTCATCACACAGCTGGTGTTTCAGGC TGTGACCAATGCAGGGTGGGATTTCCTAACTGTGGATGAGGGGATGAGGTGTCTCTGAGGGATGAGGTGTCTCAGAGAATTGAGTCCATG GGGCAGTCAGAATAGCCTTAAGAGAAAATCATGAAGGAGAAGAGGTCCTCCTTTAGCTGCCTCTACTTGGTATCTTAGAGAGGGCTTAGA GGGCTCTCAGTCTTCTGCCCATGAAAAGACTTCTTTGAGCCTCTGCCTTCATGGCTCTTAGGGTTCTGATCTTGATATCAGCAGCCCCAA CCACTTTCTTTCTGAATGTCTAGTCAGTATTTTTCCCCTTTTGGTGTTTTATGAAGCCATGTGGTAACGAATGAATCTGTATCATTTTTC CTACCTGAGTGGCCCAAAGCCAGCACCAAACCCTGGGAGTCCCTGAAGCCTAACAGAACAGGTAGAACTTGCAAAAGGAATTTGGCTGAG AGCTCACTTCTAATCCTGTACTCACTGTGTCTTTGTAGATAGAAACAAGCTAGCTTTACCAAAGAGAAATAGTGTCATAGAAGAACAAAA CTTCATATAGAAAGTTCTAGGCAAGTATTTGATGGTTTCCTTAAGGATGTGAGCTTTGTATTTCCACCTAGCCTTGTAAAATGTTCCTGT GGTATTTTGTGTCACACATCCTACCTTTGATGAGTCTCACATCCCACCTTCCTATAAACAGCTAAATTAATTTTGTTTCATCTTCCCCAG ACCAAAATGTTTGATAATCTTATCAACATTGTGGGAGGTCTTGTGCAATGGAAATTTTGCCATTTCTCCAAAACTGGTGGCATAAAGGCT GATGCTTGGGGAGAACCCCATTGCTCGGGACAGGCAACTCTGTTCAATGGGATCTTCTTTGGTTTGATGTTCCCATTGTTTTCTCAGTTC TGGGAAGCCTAGTACATTAGTACTAATGTAATCACTGAAACCTTTTCTTGAAATAAGGGAAGCAGCCAAACTTTGATTAAAGTTGCAAGT TCTGGGGACTTGCGGGGGTTGTCATAAACTGTAACAGTGGGTTTTGGTTCAGCATGTAAATGCAACTTTGATTTTCTTGAGGACCGATTG ACCTGTCATGTCCCTGTATCCTCATGCTCATCATCTCAGCAGGCCTGAGAGGCTGGGTCAGTTTGGGTGTTCATCATGAGGATTGCTTCT GCCATGGAGCTGATGGACATGGGCAGGTTGCTGAGAAGGTGGGGTGAAAGTGAGTGCCGGGGGTGGGTGAGTGCCCTGGTCTTGTTCATA GGGGAGCCTTTCCCTAGCAGTGGAACGCTGTGGTCATTTTCTCTAGCATATTCCCTTGGGAAGTCTAGATTTGCTATTAATCTGGCTGAG AATCTAAGTTCTGTGCCTTAGAGACAGTTTGCACTTTCCCATATTGTGCCTGGGACAGCCATATGATTTTTTTTCCCACCAAACAAGTAT GCAAACAGAAACCAGTTCAAAGGGGGATGGAGTAAAAGATGAGGCAGTAGAAATGCCTTTGAATGGTTTTCTGTAGCTAATTCTCTTTAA ATTTTGTCCTGCTTTTTTTCTTTATGCAGTGCTAGGTGTTTTAAGTTTTCTAGTAGTATTGCTTTTGAGTTACAGTATAACCTGAGTTAC TCCTCTGCTCTAACATTGTTGCAGAAGAACTAGTTTATTGTTAGCCAGGTTGCTTGAAAGGTTGAGAGTGGAGTGGTTTGGCATTTCTGT >102469_102469_3_ZNF587B-NEK9_ZNF587B_chr19_58350517_ENST00000594901_NEK9_chr14_75573405_ENST00000238616_length(amino acids)=631AA_BP=94 MELCDGAKRDPPLGQDRDRHAHISWLVTLSSQPCFKPRGSMAVVATLRLSAQGTVTFEDVAVKFTQEEWNLLSEAQRCLYRDVTLENLAL MSSLDEGQLYAFGSDYYGCMGVDKVAGPEVLEPMQLNFFLSNPVEQVSCGDNHVVVLTRNKEVYSWGCGEYGRLGLDSEEDYYTPQKVDV PKALIIVAVQCGCDGTFLLTQSGKVLACGLNEFNKLGLNQCMSGIINHEAYHEVPYTTSFTLAKQLSFYKIRTIAPGKTHTAAIDERGRL LTFGCNKCGQLGVGNYKKRLGINLLGGPLGGKQVIRVSCGDEFTIAATDDNHIFAWGNGGNGRLAMTPTERPHGSDICTSWPRPIFGSLH HVPDLSCRGWHTILIVEKVLNSKTIRSNSSGLSIGTVFQSSSPGGGGGGGGGEEEDSQQESETPDPSGGFRGTMEADRGMEGLISPTEAM GNSNGASSSCPGWLRKELENAEFIPMPDSPSPLSAAFSESEKDTLPYEELQGLKVASEAPLEHKPQVEASSPRLNPAVTCAGKGTPLTPP ACACSSLQVEVERLQGLVLKCLAEQQKLQQENLQIFTQLQKLNKKLEGGQQVGMHSKGTQTAKEEMEMDPKPDLDSDSWCLLGTDSCRPS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF587B-NEK9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | NEK9 | chr19:58350517 | chr14:75573405 | ENST00000238616 | 10 | 22 | 732_891 | 442.3333333333333 | 980.0 | NEK6 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF587B-NEK9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF587B-NEK9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
