
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF66-MLLT3 (FusionGDB2 ID:102680) |
Fusion Gene Summary for ZNF66-MLLT3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF66-MLLT3 | Fusion gene ID: 102680 | Hgene | Tgene | Gene symbol | ZNF66 | MLLT3 | Gene ID | 7617 | 4300 |
| Gene name | zinc finger protein 66 | MLLT3 super elongation complex subunit | |
| Synonyms | ZNF66P | AF9|YEATS3 | |
| Cytomap | 19p12 | 9p21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | putative zinc finger protein 66 | protein AF-9ALL1-fused gene from chromosome 9 proteinKMT2A/MLLT3 fusionKMT2A/MLLT3 fusion proteinYEATS domain-containing protein 3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog); translocated to, 3myeloid/lymphoid or mixed-lineage leu | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | P42568 | |
| Ensembl transtripts involved in fusion gene | ENST00000344519, ENST00000360204, ENST00000425625, ENST00000594534, | ENST00000355930, ENST00000429426, ENST00000380321, ENST00000475957, ENST00000380338, | |
| Fusion gene scores | * DoF score | 1 X 1 X 1=1 | 18 X 37 X 9=5994 |
| # samples | 1 | 35 | |
| ** MAII score | log2(1/1*10)=3.32192809488736 | log2(35/5994*10)=-4.09809225668125 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF66 [Title/Abstract] AND MLLT3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF66(20976684)-MLLT3(20448264), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ZNF66-MLLT3 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. ZNF66-MLLT3 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MLLT3 | GO:0045893 | positive regulation of transcription, DNA-templated | 25417107|27105114 |
| Tgene | MLLT3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 19591803 |
 Fusion gene breakpoints across ZNF66 (5'-gene) Fusion gene breakpoints across ZNF66 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
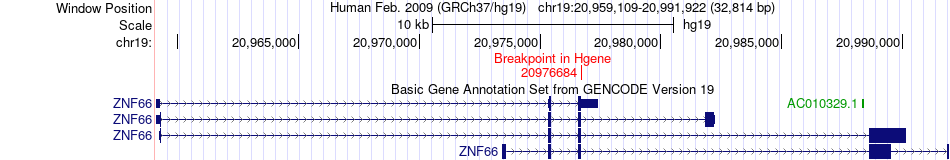 |
 Fusion gene breakpoints across MLLT3 (3'-gene) Fusion gene breakpoints across MLLT3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
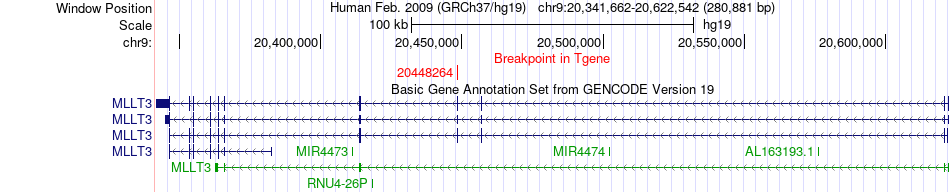 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-69-7980-01A | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
Top |
Fusion Gene ORF analysis for ZNF66-MLLT3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000344519 | ENST00000355930 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000344519 | ENST00000429426 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000360204 | ENST00000355930 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000360204 | ENST00000429426 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000425625 | ENST00000355930 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000425625 | ENST00000429426 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000594534 | ENST00000355930 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-5UTR | ENST00000594534 | ENST00000429426 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000344519 | ENST00000380321 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000344519 | ENST00000475957 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000360204 | ENST00000380321 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000360204 | ENST00000475957 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000425625 | ENST00000380321 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000425625 | ENST00000475957 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000594534 | ENST00000380321 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| 5CDS-intron | ENST00000594534 | ENST00000475957 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| Frame-shift | ENST00000344519 | ENST00000380338 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| Frame-shift | ENST00000425625 | ENST00000380338 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| Frame-shift | ENST00000594534 | ENST00000380338 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
| In-frame | ENST00000360204 | ENST00000380338 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000360204 | ZNF66 | chr19 | 20976684 | + | ENST00000380338 | MLLT3 | chr9 | 20448264 | - | 7233 | 1024 | 1063 | 2454 | 463 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000360204 | ENST00000380338 | ZNF66 | chr19 | 20976684 | + | MLLT3 | chr9 | 20448264 | - | 0.000171084 | 0.99982893 |
Top |
Fusion Genomic Features for ZNF66-MLLT3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
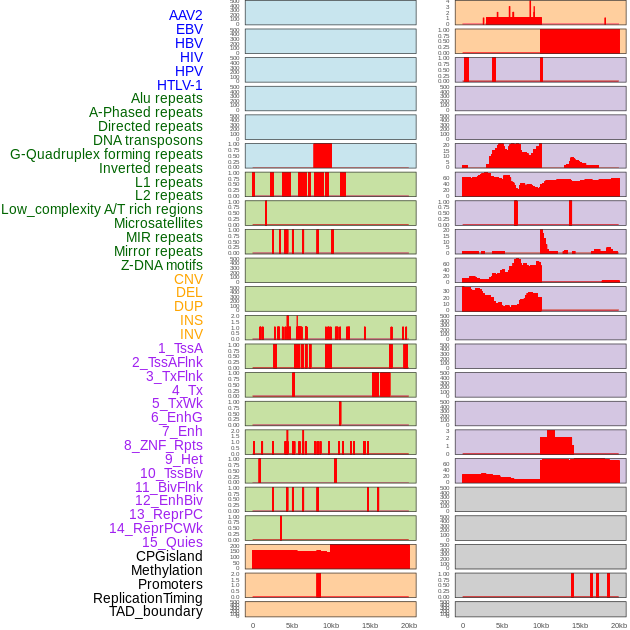 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ZNF66-MLLT3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:20976684/chr9:20448264) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MLLT3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Chromatin reader component of the super elongation complex (SEC), a complex required to increase the catalytic rate of RNA polymerase II transcription by suppressing transient pausing by the polymerase at multiple sites along the DNA (PubMed:20159561, PubMed:20471948, PubMed:25417107, PubMed:27105114, PubMed:27545619). Specifically recognizes and binds acylated histone H3, with a preference for histone H3 that is crotonylated (PubMed:25417107, PubMed:27105114, PubMed:27545619, PubMed:30374167, PubMed:30385749). Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors (PubMed:25417107, PubMed:27105114, PubMed:27545619). Recognizes and binds histone H3 crotonylated at 'Lys-9' (H3K9cr), and with slightly lower affinity histone H3 crotonylated at 'Lys-18' (H3K18cr) (PubMed:27105114). Also recognizes and binds histone H3 acetylated and butyrylated at 'Lys-9' (H3K9ac and H3K9bu, respectively), but with lower affinity than crotonylated histone H3 (PubMed:25417107, PubMed:27105114, PubMed:30385749). In the SEC complex, MLLT3 is required to recruit the complex to crotonylated histones (PubMed:27105114, PubMed:27545619). Recruitment of the SEC complex to crotonylated histones promotes recruitment of DOT1L on active chromatin to deposit histone H3 'Lys-79' methylation (H3K79me) (PubMed:25417107). Plays a key role in hematopoietic stem cell (HSC) maintenance by preserving, rather than confering, HSC stemness (PubMed:31776511). Acts by binding to the transcription start site of active genes in HSCs and sustaining level of H3K79me2, probably by recruiting DOT1L (PubMed:31776511). {ECO:0000269|PubMed:20159561, ECO:0000269|PubMed:20471948, ECO:0000269|PubMed:25417107, ECO:0000269|PubMed:27105114, ECO:0000269|PubMed:27545619, ECO:0000269|PubMed:30374167, ECO:0000269|PubMed:30385749, ECO:0000269|PubMed:31776511}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 4_75 | 75 | 574.0 | Domain | KRAB |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 149_194 | 92 | 569.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 383_391 | 92 | 569.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 466_469 | 92 | 569.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 295_300 | 92 | 569.0 | Motif | Nuclear localization signal | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 106_108 | 92 | 569.0 | Region | Histone H3K9cr binding | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 106_109 | 92 | 569.0 | Region | Inhibitor XL-07i binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 145_167 | 75 | 574.0 | Zinc finger | C2H2-type 1%3B atypical |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 171_195 | 75 | 574.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 201_223 | 75 | 574.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 229_251 | 75 | 574.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 257_279 | 75 | 574.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 285_307 | 75 | 574.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 313_335 | 75 | 574.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 341_363 | 75 | 574.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 369_391 | 75 | 574.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 397_419 | 75 | 574.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 425_447 | 75 | 574.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 453_475 | 75 | 574.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 481_503 | 75 | 574.0 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 509_531 | 75 | 574.0 | Zinc finger | C2H2-type 14 |
| Hgene | ZNF66 | chr19:20976684 | chr9:20448264 | ENST00000344519 | + | 3 | 4 | 537_559 | 75 | 574.0 | Zinc finger | C2H2-type 15 |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 1_138 | 92 | 569.0 | Domain | YEATS | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 56_58 | 92 | 569.0 | Region | Inhibitor XL-07i binding | |
| Tgene | MLLT3 | chr19:20976684 | chr9:20448264 | ENST00000380338 | 2 | 11 | 78_80 | 92 | 569.0 | Region | Histone H3K9cr binding |
Top |
Fusion Gene Sequence for ZNF66-MLLT3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >102680_102680_1_ZNF66-MLLT3_ZNF66_chr19_20976684_ENST00000360204_MLLT3_chr9_20448264_ENST00000380338_length(transcript)=7233nt_BP=1024nt GTCTCTCCCTGCAGCTGGAGCTCCAGGTCGTCTGTTCACTGCTCTCTGTCTTCTTCTCCTAGAGGCCCAGCCTCTGTGGCCCTGTGTCCT GCAGGTATTGGGAGATCCACAGCTAAGACGCCAGGACCCCCTGGAAGCCTAGAAATGGTGAGAGTGCCGGTCCAGCATCCCGAGAGAGGG GCCATTGCAATTTAGAGATGTGGCCATAGAATTCTCTCTGGAGGAGTGGCATTGCCTGGACATGGCACAGCGGAATTTATATAGGGATGT GATGTTAGAGAACTACAGAAACCTGGTCTTTCTTGGTAGGTGTGAGTGAAAATGAATACAACAGACAACACAGTAAGAAGTCCAAGGTCA AAAAGAAAGCCAGTCCTTAAGGTGTGATTCTGGAAGCTGTGTTCCAAAGGAACTTCTGGGCAGCTGTTTTTTTTGTTTTTTGTTTTTGTT TTTGTTGTTTTAAATTTTGCTGTCACAAAGGGGCATATTGTCTTATGCTTCTAAATTCTCTAAAAATTCTACTTTTCTCTCAGTGAGCTT CCTTCACATTCACAGTGAGAGCCAAAGTCCTCTTCATGACATATAAGAGACAGCACAATCCAGCTGCTTTTTCATTGTTTTGGGGACACA CAAATATCTTCCTAATTTTGAGAAACTGAAACTATTTTTTAGTTTTCTTTTTGTATCCGGTCTGAAATTTGTGAGAGTAGTAGTTTCTGT TGCATTTTTTTTTGTTCATTTTTGTGCACAGTCCATTCTATTTTTATTACTATATAGTCTTGAAATATAGTTTGAAATTGTAAGTACGAT ATTCTGTTTTCTTCTTTTTCCTCAAGATTGCTTTGGCTATTCAAAGTTTATTTTAGTTTCATGTAAATTTTAGAATTGTATTTTCCATTA TTGTAAAAAAAAACACTGCAATTTTAATAGGAAGATTATTGAGTCTGTAAATCACTTTGGATAATATGATTCTTTAATAATATTTATTCT TTTAATTCATTGACATAAAATATTTTAAAATTAAGAAGAACCTAGGAAAGTCCGCTTTGATTATGACTTATTCCTGCATCTTGAAGGCCA TCCACCAGTGAATCACCTCCGCTGTGAAAAGCTAACTTTCAACAACCCCACAGAGGACTTTAGGAGAAAGTTGCTGAAGGCAGGAGGGGA CCCTAATAGGAGTATTCATACCAGCAGCAGCAGCAGCAGCAGCAGTAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGTAGCAGCAGCAG CAGCAGCAGCAGCAGCAGCAGTAGCAGCAGCAGTAGCAGCAGCAGCAGCAGCAGTAGTACCAGTTTTTCAAAGCCTCACAAATTAATGAA GGAGCACAAGGAAAAACCTTCTAAAGACTCCAGAGAACATAAAAGTGCCTTCAAAGAACCTTCCAGGGATCACAACAAATCTTCCAAAGA ATCCTCTAAGAAACCCAAAGAAAATAAACCACTGAAAGAAGAGAAAATAGTTCCTAAGATGGCCTTCAAGGAACCTAAACCCATGTCAAA AGAGCCAAAACCAGATAGTAACTTACTCACCATCACCAGTGGACAAGATAAGAAGGCTCCTAGTAAAAGGCCGCCCATTTCAGATTCTGA AGAACTCTCAGCCAAAAAAAGGAAAAAGAGTAGCTCAGAGGCTTTATTTAAAAGTTTTTCTAGCGCACCACCACTGATACTCACTTGTTC TGCTGACAAAAAACAGATAAAAGATAAATCTCATGTCAAGATGGGAAAGGTCAAAATTGAAAGTGAGACATCAGAGAAGAAGAAATCAAC GTTACCGCCATTTGATGATATTGTGGATCCCAATGATTCAGATGTGGAGGAGAATATATCCTCTAAATCTGATTCTGAACAACCCAGTCC TGCCAGCTCCAGCTCCAGCTCCAGCTCCAGCTTCACACCATCCCAGACCAGGCAACAAGGTCCTTTGAGGTCTATAATGAAAGATCTGCA TTCTGATGACAATGAGGAGGAATCAGATGAAGTGGAGGATAACGACAATGACTCTGAAATGGAGAGGCCTGTAAATAGAGGAGGCAGCCG AAGTCGCAGAGTTAGCTTAAGTGATGGCAGCGATAGTGAAAGCAGTTCTGCTTCTTCACCCCTACATCACGAACCTCCACCACCCTTACT AAAAACCAACAACAACCAGATTCTTGAAGTGAAAAGTCCAATAAAGCAAAGCAAATCAGATAAGCAAATAAAGAATGGTGAATGTGACAA GGCATACCTAGATGAACTGGTAGAGCTTCACAGAAGGTTAATGACATTGAGAGAAAGACACATTCTGCAGCAGATCGTGAACCTTATAGA AGAAACTGGACACTTTCATATCACAAACACAACATTTGATTTTGATCTTTGCTCGCTGGACAAAACCACAGTCCGTAAACTACAGAGTTA CCTGGAAACATCTGGAACATCCTGAGGATATAACAACTGGATGCATCAAGAACTATTGTGTTTTTTTTTTTTGGTTTTTTTTTTTTTTGG TTGTGATTTTTTGTTCTTGTTGTTTATATGAAAACACTCAAAATGATGCAACCAAAAGGGAAAAAATAAAAATCAAACAACCTTCAGCTT TATTTTTCTTTAAAGCCAGTCATCATCTCTTGATAAAGGAGAGGTTAAAGCAAACCAGCCTCAGCGGACCACTCTTCTCTCCAAGGAAAT CCCCGGGAAGAGTTAGCCTGGATAGCCTTGAAAACAAACAAATCAAACACAACACAAGAAAACTCAAAGAATGTGTATGGTATCATGTAT CTCTCTGTGGTGGTTCATTCCACAGGACGAATGCATATTCAACACACTGCCTTATTACATAACTGATCTATTTATTATCGCATACAGATA TTCTAAGTCGTTGAGGGAATGACACCATCAGACATTATAAGTACTTGGTCCCGTGGATGCTCTTTCAATGCAGCACCCTTGCCATCCCAA GCCCAGTGACCTTACTCGTATACCGTGCCACTTTCCACCAACTTTTTCCAAGTCCTTTAACTCGTTGCAGTCTGTATTTTCCACCTTTTG TTTTTCCAGTTCCAGGACACAGATTATCAACTGGGGGGACCAAATAGCCACCTTGATTTTCTTCTTTGTGGTCTTTTTCCTGAAAGTTGG GGCCCAGTCCTTGGCTGTATCCATGTAATGATCTTGGACCATGGTAGAAAATGCACCAAATAGGATCATATGAATTGCTGTCTAGCCTTA GTCAATAAACTTGTAGGACTTTTAAACAAAAGTGTACCTGTAAATGTCCTGAATCCAGCATTGTTGAGCTGTCATCAACATTCTTGTGTC TGTTTTACTGTTACAATATTAGGTGAATATGGAAGTAAAGGCATTCCACAGGATCATCATTTAAAAAAAAAGAATTCTGGTCCTGTTTTC TAAAAAAAAAAACTGTTGTAGAAATTCTTAATTTGGATCTATTTATTAGTCAGAGTTTCAGCTTTCTTCAGCTGCCAGTGTGTTACTCAT CTTTATCCTAAAAATCTGGAATCAGAGATTTTTGTTTGTTCACATATGATTCTCTTAGACACTTTTATATTTGAAAAAATTAAAATCTTT CTTTGGGGAAAAATTCTTGGTTATTCTGCCATAACAGATTATGTATTAACTTGTAGATTCAGTGGTTCAATACCTGTTTAGTTGCTTGCT AATATTTCCAGAAGGATTTCTTGTATTGGTGAAAGACGGTTGGGGATGGGGGGATTTTTTTGTTCTTGTTGTACCCTTGTTTTGAAACTA GAAATCTGTCCTGTGGCATGCAAAAGAAAGCAAATTATTTTTAAAAGAAAAAAACCAAAGTACTTTTGGTGTCATTATTCCATCTTCTCC ATAAGTGGAGAAATGAAAAGTAAGAACAGCTCATCTTCAAAGTTTTTACTAGAAATTCAGCCGAAAAGAAGAGAAATGAAGAAATACTTC TGGATCCAAAGGTTCGTCACTGGATCAGCCTTAAGAAAGTCTCTATGTGTGCTAATAGACCAGTATCTCCCTTGTTTGTTCCCATGCCAG ATGCTGTCTTTTCATACAGAGATTAACTAGTGTCTGTACAGAAAATGGTCTTTCCAGACCCATTCTTTTGGGCAGTTATATAAATGCAGA TTGATAATGGAGTTATTTCTGCATTTTAAAAAGGGGATTTTTTACACTGTTTCTTTCTATCCAAAGAAACTATTTTTCCTTTAAAAAATT ATAATACAGCCATGCTCCTTGTAAATTATTTCTCTGAAGCGTCCTCCCAGAGGACAAGTGTACTTTCTCAATAATGTTATGATTCTTAAT GACGATTGGCATGAACCGTACTTGAAGCATTTGTTGTATTGTCCACTTCTTGATGTGCAGAGAAACTTTGAGTCATTTTGGACATTAGGT GATTGATGAATTGTGTGGTTTCAGTTGCGTTTTGTTTTTAACTTTATAGCTGGAAGGGGAGGGTAATGTTTATATGGAAATGTTGCAAAT TTTTAAAATATTTTAAAATTTCTAACTGCTAACACATATTCTGATCAAGCCAGAAGTTTATGTGATTTAATATATCTGTGTAGTTTAACT AGAAACGTATATATGTAGTTTGGGCCATTTTTATTTCTGTAACTAAATTGTGGGGCAAAAAATATGATTTGCCAATGTAAAAATTTTTTC CTTTAAGTTACTAACCAAGAAGCTAGGTAAGATAAGAAATGACCAAACAAAAATAATGATAAGAAGGACAGCTATTATATGATAAACTTT AGACTTGAAAATTAGGTAATAAAGTTAATTTCTTCAGAATATCCTAAAATGTAAGTTTGTAAATTGATAAAGTTCAGAAAAACTTATCCA TCTTCTACTCATCAAGATAGCAAAGTAAGTTTTTCACAAAATTAAGACTGAGAGTTACGTTATTTTTTTTGAAGTGGTAATCTTGCTCTC ATCTAATTTTTAAATACAGAAATAAAAAGATCCCTTTGAATTAATTTCAAGTAGTTTTAATACCTCTCTTAAAGTAACCCCTTCAGTTCT GTCACATTTATATACACCCAACTTTGAAGCCTATTAAGAATATCTTTGACATGTGAGTACCCTAACAGTGGCTCTGGTTTCTTAAGCACA AAGGGAAAATGGCAGGGTTTAATTTCTTTGGTGTACATATATAAATAGATGTTTCAAATAAAATTAATTGGGTAAATGTGAATATGGTTT GAACCAGCCAGAGTATGATAAACTTTTAAAAAATTTTTCCTCAGTGATCAGCCAGGGGAAAAAATATATGTACAGAGCTCCTGCCCCTCT GCCCCCTTATAACTTAGCTTTTGCCTCTGGTTTTGCAGCAGGATGTGTGTTGGTGGGTTTGGGGTTCCTTCTGTCTTCGGATGTAACAGT CATGTTAGCAACAATATTTTGCAGTTACTTCACCATCCTCAGCATAATCTGCCTCTCTCTCAATCTTAATACAGACAAATAGGTATGTCT ATAAATGCGTTCATATAGTGTCAAGGTAGGGAAGACTATAAAAACTTCAGATCAGGAAAAAAATATCTCTTAAGAGAGTGTCTTAAACAT TTTAAATTGCTAGGCAATTTAAATCTCTCTAATCTGAATATTCACCTTTTGGTGGAAAGAGCCAGTAATATTGGTTTTCTTCCACTAAAA AGTTTCTTCAAAATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCCGATATCTTTTACAGGCCCACCTAATCTTTAAAAAAAAGAGAAG AAACAATTTTAGGCCTGTATAACATCCTATTTACTACTTCAAGCTCCACAAATGTAGTTAATATATTTGTTTAAGAGAGACTTTACTCTT AAATGTTTTAACTTTGCCTACTTAATGGAGTTGGATGTCCAGTGTTGGAAACAGTCAGTGTCTGTTTGGTGGCTCGTGTTTATTTTTGTC ACTGTAGAAGTGATCTGCTCTGGAGTAATTTGCTGTATGCTTATTACTTTCCACCTTTAGCGTTTTGTGTACTAATGCTATGTACTGCTC CTATATTTATATATACATATATATATATATATACACATATATATTATCAGAGTAATCTTGCTTTGGACTGTTGCTCTAAAACTATGAATA TGATGTAGAGACTACTACAGCTGCAGTGAATGTTAAGGAGTCAATTCCCATGGTTTAGTTTAGTCTCGGGCTGGAAGTCTACATCTTTTG ACCTCTTTTGTTTTATATGATGCTTTTTATTATGTAGGCACTGCCATCCCTGAATATTCCTTGTAAAGATGCAGTGAAGAAGTCAATTTA AAAGGAAGACAAAGCCACCGCTGTCTGTAAACTGTAAATGTGTATTTTGGCACTTGTACTCCAGTATTCTGAAAATAGAGTTATGATGGA AATGCTGACAGATCCTTAATGGTGGCCAGAGCTACCATGCAGTGCAAAAATAAATTAAGTGAAAATGTATTATTGAGATTAACAGTGCAA GGCCAATACATTTTATAAATCTGTCAGTATCTTTTTAATGGACTGTGTATGTGTAGTTATGTTGAAAGATACATTGTGTATGTGTACATG CTTTTGTGTGAATTCAAGGCACACTGTAAGATGTATTTCCCTTCTCTAAAATCCATGTTGAAATGTAAGTACAGATTGATAGAAATAGGT GTTTTACTTAAACTCATATAATCTGTTTCAGCACCTTTTTTAAAATGTATAATTGTACTGATTTGCTGAGAGTCATTTGACCAAAGTTTG GCTTTATTTTGCATACAGGAAAAATTAGCCCCTAGCCTTTTCTCAAGAAGTGATGCTGTTTCTAAAGAAGAATAGATGGGGAGATCTGAG ACATCCAGAGAGAGGACAATGTGTCCAAAACCAGCTGAAGGCAGAGAGTCCATGCCTCATTCATTTTAGGAGTCCCTCATGTCTAACACC ATCTTCTGTGTCCTAAAGCCAGTATATATATAATATTTTTAAAACTCTGCAAAGATTGGAACAAGAAGTGGTGGGATACATTTTTTGGCA TAAACTGGGCTATGTGTGCCCGGTATTTTTTCTTTCCCCCCTATTTCTTTGGAGGGTTTTGTTCAGAAACAATGTGCTATAGCCAGAGAG AGTCGATCTTACAAAGTCCCTGCATTTACTTTCACAGCAAGAACTGCTACCTTGATTAGACAGGAAGGAGAACAACTACTACAACAAAGA >102680_102680_1_ZNF66-MLLT3_ZNF66_chr19_20976684_ENST00000360204_MLLT3_chr9_20448264_ENST00000380338_length(amino acids)=463AA_BP= MHLEGHPPVNHLRCEKLTFNNPTEDFRRKLLKAGGDPNRSIHTSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSTSFSK PHKLMKEHKEKPSKDSREHKSAFKEPSRDHNKSSKESSKKPKENKPLKEEKIVPKMAFKEPKPMSKEPKPDSNLLTITSGQDKKAPSKRP PISDSEELSAKKRKKSSSEALFKSFSSAPPLILTCSADKKQIKDKSHVKMGKVKIESETSEKKKSTLPPFDDIVDPNDSDVEENISSKSD SEQPSPASSSSSSSSSFTPSQTRQQGPLRSIMKDLHSDDNEEESDEVEDNDNDSEMERPVNRGGSRSRRVSLSDGSDSESSSASSPLHHE PPPPLLKTNNNQILEVKSPIKQSKSDKQIKNGECDKAYLDELVELHRRLMTLRERHILQQIVNLIEETGHFHITNTTFDFDLCSLDKTTV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF66-MLLT3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF66-MLLT3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF66-MLLT3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
