
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ZNF814-EIF3B (FusionGDB2 ID:103004) |
Fusion Gene Summary for ZNF814-EIF3B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ZNF814-EIF3B | Fusion gene ID: 103004 | Hgene | Tgene | Gene symbol | ZNF814 | EIF3B | Gene ID | 730051 | 8662 |
| Gene name | zinc finger protein 814 | eukaryotic translation initiation factor 3 subunit B | |
| Synonyms | - | EIF3-ETA|EIF3-P110|EIF3-P116|EIF3S9|PRT1 | |
| Cytomap | 19q13.43 | 7p22.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein 814 | eukaryotic translation initiation factor 3 subunit Beukaryotic translation initiation factor 3 subunit 9eukaryotic translation initiation factor 3, subunit 9 (eta, 116kD)eukaryotic translation initiation factor 3, subunit 9 eta, 116kDaprt1 homolog | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | . | P55884 | |
| Ensembl transtripts involved in fusion gene | ENST00000435989, ENST00000595295, ENST00000596604, ENST00000597342, ENST00000597807, ENST00000597832, ENST00000600634, ENST00000597652, | ENST00000360876, ENST00000397011, | |
| Fusion gene scores | * DoF score | 6 X 5 X 4=120 | 16 X 22 X 8=2816 |
| # samples | 7 | 22 | |
| ** MAII score | log2(7/120*10)=-0.777607578663552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/2816*10)=-3.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ZNF814 [Title/Abstract] AND EIF3B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ZNF814(58400135)-EIF3B(2416585), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | EIF3B | GO:0006413 | translational initiation | 8995410|17581632 |
| Tgene | EIF3B | GO:0006446 | regulation of translational initiation | 8995410 |
| Tgene | EIF3B | GO:0075522 | IRES-dependent viral translational initiation | 9573242 |
| Tgene | EIF3B | GO:0075525 | viral translational termination-reinitiation | 21347434 |
 Fusion gene breakpoints across ZNF814 (5'-gene) Fusion gene breakpoints across ZNF814 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
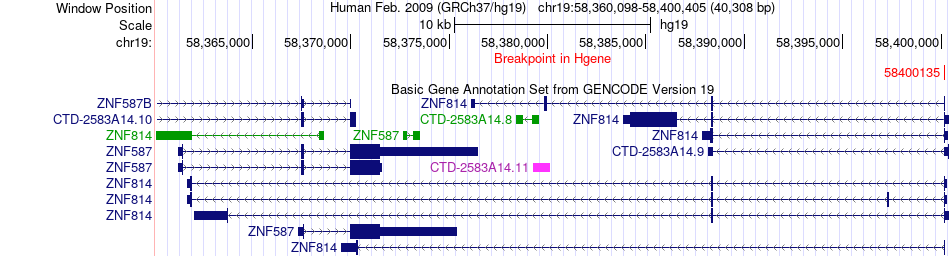 |
 Fusion gene breakpoints across EIF3B (3'-gene) Fusion gene breakpoints across EIF3B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
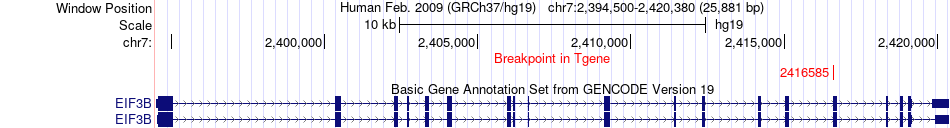 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-FS-A1ZK-06A | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
Top |
Fusion Gene ORF analysis for ZNF814-EIF3B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000435989 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000435989 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000595295 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000595295 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000596604 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000596604 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000597342 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000597342 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000597807 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000597807 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000597832 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000597832 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000600634 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| In-frame | ENST00000600634 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| intron-3CDS | ENST00000597652 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
| intron-3CDS | ENST00000597652 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000596604 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1125 | 154 | 34 | 570 | 178 |
| ENST00000596604 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 1052 | 154 | 34 | 570 | 178 |
| ENST00000597342 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1163 | 192 | 36 | 608 | 190 |
| ENST00000597342 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 1090 | 192 | 36 | 608 | 190 |
| ENST00000597832 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1224 | 253 | 97 | 669 | 190 |
| ENST00000597832 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 1151 | 253 | 97 | 669 | 190 |
| ENST00000595295 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1026 | 55 | 967 | 446 | 173 |
| ENST00000595295 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 953 | 55 | 19 | 471 | 150 |
| ENST00000600634 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1022 | 51 | 963 | 442 | 173 |
| ENST00000600634 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 949 | 51 | 15 | 467 | 150 |
| ENST00000435989 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1242 | 271 | 115 | 687 | 190 |
| ENST00000435989 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 1169 | 271 | 115 | 687 | 190 |
| ENST00000597807 | ZNF814 | chr19 | 58400135 | - | ENST00000360876 | EIF3B | chr7 | 2416585 | + | 1174 | 203 | 47 | 619 | 190 |
| ENST00000597807 | ZNF814 | chr19 | 58400135 | - | ENST00000397011 | EIF3B | chr7 | 2416585 | + | 1101 | 203 | 47 | 619 | 190 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000596604 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.005236101 | 0.99476385 |
| ENST00000596604 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.00573713 | 0.9942629 |
| ENST00000597342 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.005800227 | 0.99419975 |
| ENST00000597342 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.005192303 | 0.99480766 |
| ENST00000597832 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004901311 | 0.99509865 |
| ENST00000597832 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004659319 | 0.9953407 |
| ENST00000595295 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004332333 | 0.9956677 |
| ENST00000595295 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004142241 | 0.9958578 |
| ENST00000600634 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004423915 | 0.99557614 |
| ENST00000600634 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004521477 | 0.9954786 |
| ENST00000435989 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.003848108 | 0.99615186 |
| ENST00000435989 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.004692748 | 0.99530727 |
| ENST00000597807 | ENST00000360876 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.00602846 | 0.9939715 |
| ENST00000597807 | ENST00000397011 | ZNF814 | chr19 | 58400135 | - | EIF3B | chr7 | 2416585 | + | 0.006987726 | 0.99301225 |
Top |
Fusion Genomic Features for ZNF814-EIF3B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ZNF814 | chr19 | 58400134 | - | EIF3B | chr7 | 2416584 | + | 6.01E-09 | 1 |
| ZNF814 | chr19 | 58400134 | - | EIF3B | chr7 | 2416584 | + | 6.01E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
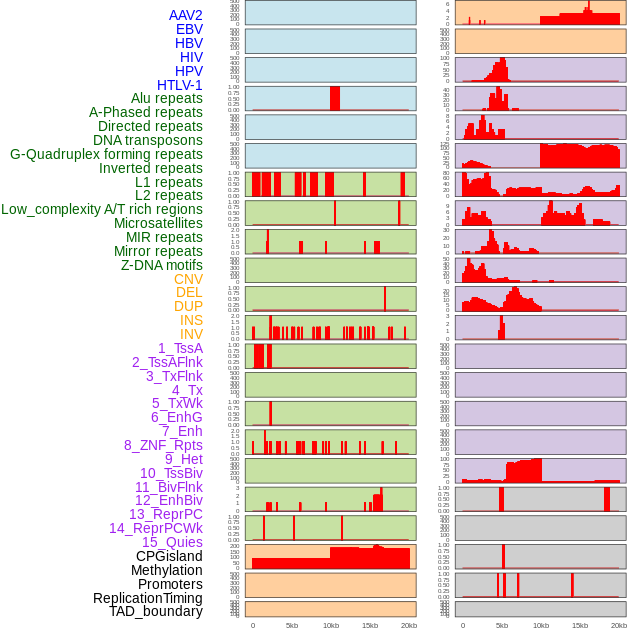 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
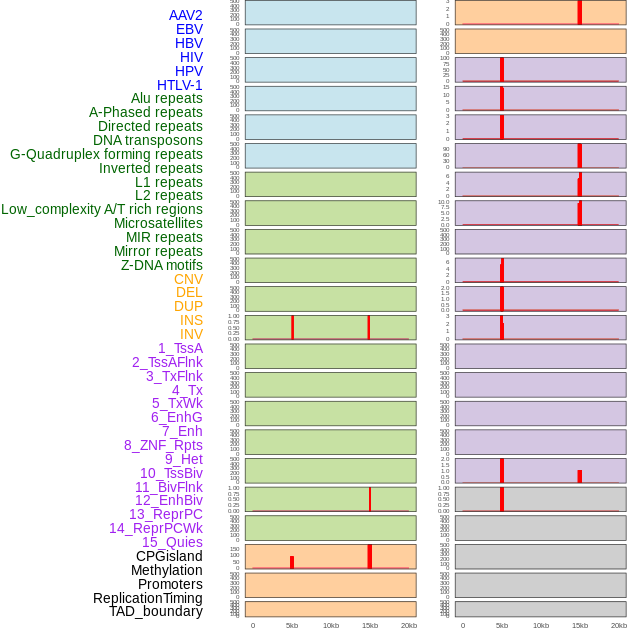 |
Top |
Fusion Protein Features for ZNF814-EIF3B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:58400135/chr7:2416585) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | EIF3B |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: RNA-binding component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis (PubMed:9388245, PubMed:17581632, PubMed:25849773, PubMed:27462815). The eIF-3 complex associates with the 40S ribosome and facilitates the recruitment of eIF-1, eIF-1A, eIF-2:GTP:methionyl-tRNAi and eIF-5 to form the 43S pre-initiation complex (43S PIC). The eIF-3 complex stimulates mRNA recruitment to the 43S PIC and scanning of the mRNA for AUG recognition. The eIF-3 complex is also required for disassembly and recycling of post-termination ribosomal complexes and subsequently prevents premature joining of the 40S and 60S ribosomal subunits prior to initiation (PubMed:9388245, PubMed:17581632). The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation, including cell cycling, differentiation and apoptosis, and uses different modes of RNA stem-loop binding to exert either translational activation or repression (PubMed:25849773). {ECO:0000255|HAMAP-Rule:MF_03001, ECO:0000269|PubMed:17581632, ECO:0000269|PubMed:25849773, ECO:0000269|PubMed:27462815, ECO:0000269|PubMed:9388245}.; FUNCTION: (Microbial infection) In case of FCV infection, plays a role in the ribosomal termination-reinitiation event leading to the translation of VP2 (PubMed:18056426). {ECO:0000269|PubMed:18056426}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 15_91 | 12 | 856.0 | Domain | KRAB |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 120_142 | 12 | 856.0 | Zinc finger | C2H2-type 1%3B degenerate |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 242_264 | 12 | 856.0 | Zinc finger | C2H2-type 2 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 269_291 | 12 | 856.0 | Zinc finger | C2H2-type 3 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 296_318 | 12 | 856.0 | Zinc finger | C2H2-type 4 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 324_346 | 12 | 856.0 | Zinc finger | C2H2-type 5 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 352_374 | 12 | 856.0 | Zinc finger | C2H2-type 6 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 380_402 | 12 | 856.0 | Zinc finger | C2H2-type 7 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 408_430 | 12 | 856.0 | Zinc finger | C2H2-type 8 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 436_458 | 12 | 856.0 | Zinc finger | C2H2-type 9 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 464_486 | 12 | 856.0 | Zinc finger | C2H2-type 10 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 492_514 | 12 | 856.0 | Zinc finger | C2H2-type 11 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 520_542 | 12 | 856.0 | Zinc finger | C2H2-type 12 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 548_570 | 12 | 856.0 | Zinc finger | C2H2-type 13 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 576_598 | 12 | 856.0 | Zinc finger | C2H2-type 14 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 604_626 | 12 | 856.0 | Zinc finger | C2H2-type 15 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 632_654 | 12 | 856.0 | Zinc finger | C2H2-type 16 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 660_682 | 12 | 856.0 | Zinc finger | C2H2-type 17 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 688_710 | 12 | 856.0 | Zinc finger | C2H2-type 18 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 716_738 | 12 | 856.0 | Zinc finger | C2H2-type 19 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 744_766 | 12 | 856.0 | Zinc finger | C2H2-type 20 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 772_794 | 12 | 856.0 | Zinc finger | C2H2-type 21 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 800_822 | 12 | 856.0 | Zinc finger | C2H2-type 22 |
| Hgene | ZNF814 | chr19:58400135 | chr7:2416585 | ENST00000435989 | - | 1 | 3 | 828_850 | 12 | 856.0 | Zinc finger | C2H2-type 23 |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 185_268 | 676 | 810.0 | Domain | RRM | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 185_268 | 676 | 791.0 | Domain | RRM | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 285_324 | 676 | 810.0 | Repeat | Note=WD 1 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 325_365 | 676 | 810.0 | Repeat | Note=WD 2 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 366_425 | 676 | 810.0 | Repeat | Note=WD 3 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 426_489 | 676 | 810.0 | Repeat | Note=WD 4 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 490_553 | 676 | 810.0 | Repeat | Note=WD 5 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 554_598 | 676 | 810.0 | Repeat | Note=WD 6 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 599_642 | 676 | 810.0 | Repeat | Note=WD 7 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000360876 | 13 | 19 | 643_685 | 676 | 810.0 | Repeat | Note=WD 8 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 285_324 | 676 | 791.0 | Repeat | Note=WD 1 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 325_365 | 676 | 791.0 | Repeat | Note=WD 2 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 366_425 | 676 | 791.0 | Repeat | Note=WD 3 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 426_489 | 676 | 791.0 | Repeat | Note=WD 4 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 490_553 | 676 | 791.0 | Repeat | Note=WD 5 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 554_598 | 676 | 791.0 | Repeat | Note=WD 6 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 599_642 | 676 | 791.0 | Repeat | Note=WD 7 | |
| Tgene | EIF3B | chr19:58400135 | chr7:2416585 | ENST00000397011 | 13 | 19 | 643_685 | 676 | 791.0 | Repeat | Note=WD 8 |
Top |
Fusion Gene Sequence for ZNF814-EIF3B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >103004_103004_1_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000435989_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1242nt_BP=271nt ATTTTGATTGGTGTTGGATTTATTTGTAGGAGAGGCTCCTGAGCGCTAGGTCCGCACTGTGGTGACTGAACCCAGAAGTCGGGGAGCAGT TGTCCTCCGCTGCACAGAGGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGACCGTCGTGCCCA TATCTCCTGGCTGGTCGCCCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCA GGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCC CCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCG TTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGA GCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGA GGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGGGGACGGACTCCGCCTGC TGTTCCCGCGCTGAGCTACAGGACTCCCGAGTGTGAGCCGCGGTTCCTCTGTTGCAGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTC CTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTT TTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGC CGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGC AGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTA >103004_103004_1_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000435989_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_2_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000435989_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=1169nt_BP=271nt ATTTTGATTGGTGTTGGATTTATTTGTAGGAGAGGCTCCTGAGCGCTAGGTCCGCACTGTGGTGACTGAACCCAGAAGTCGGGGAGCAGT TGTCCTCCGCTGCACAGAGGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGACCGTCGTGCCCA TATCTCCTGGCTGGTCGCCCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCA GGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCC CCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCG TTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGA GCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGA GGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGCGCAGCCGTGTGTGCTGT GGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGC CATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTT TTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCC TGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCG >103004_103004_2_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000435989_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_3_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000595295_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1026nt_BP=55nt CTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGG ACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAA GCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGA GAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTT GCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCAT TCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGGGGACGGACTCCGCCTGCTGTTCCCGCGCTGAGCTACAGGACTCCCGAGTGTGA GCCGCGGTTCCTCTGTTGCAGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGT GCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGA TTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGC GGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGG GCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGGTTTTCTCCGCA >103004_103004_3_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000595295_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=173AA_BP= MHGGGTRHGGALPVGLRDTSSTARNGDASPSTEMLLGCVQAPWKHHWAQEKVPPVASLSLRSQRRRFQQKTPNKKEVEAGALNPPLQTPG -------------------------------------------------------------- >103004_103004_4_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000595295_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=953nt_BP=55nt CTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGG ACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAA GCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGA GAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTT GCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCAT TCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTC CCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGT GTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTT AGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTT GCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGG >103004_103004_4_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000595295_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=150AA_BP=10 MAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMM -------------------------------------------------------------- >103004_103004_5_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000596604_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1125nt_BP=154nt GGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGACCGTCGTGCCCATATCTCCTGGCTGGTCGCCCTATCCTC CCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGAC TTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGA ACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGA ATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCG CCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGA AGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGGGGACGGACTCCGCCTGCTGTTCCCGCGCTGAGCTACAGGACTCC CGAGTGTGAGCCGCGGTTCCTCTGTTGCAGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCT CCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGC AGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGAC GCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCC CCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGGTT >103004_103004_5_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000596604_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=178AA_BP=38 MGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQEQIKQIKKDLKK -------------------------------------------------------------- >103004_103004_6_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000596604_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=1052nt_BP=154nt GGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGACCGTCGTGCCCATATCTCCTGGCTGGTCGCCCTATCCTC CCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGAC TTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGA ACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGA ATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCG CCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGA AGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCC GCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCA GCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTC CGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTT GAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGC >103004_103004_6_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000596604_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=178AA_BP=38 MGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQEQIKQIKKDLKK -------------------------------------------------------------- >103004_103004_7_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597342_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1163nt_BP=192nt CGGGGAGCAGTTGTCCTCCGCTGCACAGAGGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGAC CGTCGTGCCCATATCTCCTGGCTGGTCGCCCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGG CTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTG CTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAA CAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAA ATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGAC GACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGGGGACGG ACTCCGCCTGCTGTTCCCGCGCTGAGCTACAGGACTCCCGAGTGTGAGCCGCGGTTCCTCTGTTGCAGCGCAGCCGTGTGTGCTGTGGAG CCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATT GCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCT GTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTA CACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGC >103004_103004_7_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597342_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_8_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597342_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=1090nt_BP=192nt CGGGGAGCAGTTGTCCTCCGCTGCACAGAGGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGAC CGTCGTGCCCATATCTCCTGGCTGGTCGCCCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGG CTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTG CTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAA CAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAA ATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGAC GACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGCGCAGCC GTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGC CTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCT TGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTT TCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCA CCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGGTTTTCTCCGCAGGTTGAACATGGAAATAAAAGCAAACTT >103004_103004_8_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597342_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_9_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597807_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1174nt_BP=203nt AACCCAGAAGTCGGGGAGCAGTTGTCCTCCGCTGCACAGAGGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCC AGGATACGGACCGTCGTGCCCATATCTCCTGGCTGGTCGCCCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGG CTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCT TCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTA AGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGA AGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACA GCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCAC TGTGGGGACGGACTCCGCCTGCTGTTCCCGCGCTGAGCTACAGGACTCCCGAGTGTGAGCCGCGGTTCCTCTGTTGCAGCGCAGCCGTGT GTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGG ATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTT TGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCA CGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGG GAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGGTTTTCTCCGCAGGTTGAACATGGAAATAAAAGCAAACTTGTAT >103004_103004_9_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597807_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_10_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597807_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=1101nt_BP=203nt AACCCAGAAGTCGGGGAGCAGTTGTCCTCCGCTGCACAGAGGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCC AGGATACGGACCGTCGTGCCCATATCTCCTGGCTGGTCGCCCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGG CTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCT TCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTA AGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGA AGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACA GCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCAC TGTGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGAC TGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTC CACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGC CCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTC CCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGGTTTTCTCCGCAGGTTGAACATGGAAATA >103004_103004_10_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597807_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_11_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597832_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1224nt_BP=253nt TTTATTTGTAGGAGAGGCTCCTGAGCGCTAGGTCCGCACTGTGGTGACTGAACCCAGAAGTCGGGGAGCAGTTGTCCTCCGCTGCACAGA GGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGACCGTCGTGCCCATATCTCCTGGCTGGTCGC CCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTG GCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCT GAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGC CTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAA AAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTT CGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGGGGACGGACTCCGCCTGCTGTTCCCGCGCTGAGCTA CAGGACTCCCGAGTGTGAGCCGCGGTTCCTCTGTTGCAGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGA CTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCT GGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGA CTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGA CTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTG >103004_103004_11_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597832_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_12_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597832_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=1151nt_BP=253nt TTTATTTGTAGGAGAGGCTCCTGAGCGCTAGGTCCGCACTGTGGTGACTGAACCCAGAAGTCGGGGAGCAGTTGTCCTCCGCTGCACAGA GGCTACTCTGGAGCTCTGTGACGGCGCCCAGCGTGACCCACTCCTGGGCCAGGATACGGACCGTCGTGCCCATATCTCCTGGCTGGTCGC CCTATCCTCCCGACTCTGCTTAAAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTG GCTGTGGACTTTCCAGGGACGCCTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCT GAGCCAGGAACAGATCAAGCAAATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGC CTCAAAGGAATTGGTGGAGAGAAGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAA AAACGAGCGCCTGGAGTTGCGAGGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTT CGTCACTGAAGAAATCATTCCCCTCGGGAATCAGGAGTGACCTGGAGCACTGTGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTG CAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTG TGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGG CGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGC ATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCC >103004_103004_12_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000597832_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=190AA_BP=50 MELCDGAQRDPLLGQDTDRRAHISWLVALSSRLCLKPRGSMAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQ EQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVT -------------------------------------------------------------- >103004_103004_13_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000600634_EIF3B_chr7_2416585_ENST00000360876_length(transcript)=1022nt_BP=51nt AAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGC CTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAA ATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGA AGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGA GGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCC CTCGGGAATCAGGAGTGACCTGGAGCACTGTGGGGACGGACTCCGCCTGCTGTTCCCGCGCTGAGCTACAGGACTCCCGAGTGTGAGCCG CGGTTCCTCTGTTGCAGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGCCTCCTCCCTGTGCTC TCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCTGCAGTGGGGGATTTA AGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCGACGCCACTGGCGGCA CCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCATCCCCATTGCGGGCAG TGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGGTTTTCTCCGCAGGTT >103004_103004_13_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000600634_EIF3B_chr7_2416585_ENST00000360876_length(amino acids)=173AA_BP= MHGGGTRHGGALPVGLRDTSSTARNGDASPSTEMLLGCVQAPWKHHWAQEKVPPVASLSLRSQRRRFQQKTPNKKEVEAGALNPPLQTPG -------------------------------------------------------------- >103004_103004_14_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000600634_EIF3B_chr7_2416585_ENST00000397011_length(transcript)=949nt_BP=51nt AAACCACGTGGTTCGATGGCTGCCGCGGCTACGCTGAGGCTCTCCGCTCAGGTGGACAACGCGTACTGGCTGTGGACTTTCCAGGGACGC CTCCTGCAGAAGAACAACAAGGACCGCTTCTGCCAGCTGCTGTGGCGGCCCCGGCCTCCCACACTCCTGAGCCAGGAACAGATCAAGCAA ATTAAAAAGGATCTGAAGAAATACTCTAAGATCTTTGAACAGAAGGATCGTTTGAGTCAGTCCAAAGCCTCAAAGGAATTGGTGGAGAGA AGGCGCACCATGATGGAAGATTTCCGGAAGTACCGGAAAATGGCCCAGGAGCTCTATATGGAGCAGAAAAACGAGCGCCTGGAGTTGCGA GGAGGGGTGGACACTGACGAGCTGGACAGCAACGTGGACGACTGGGAAGAGGAGACCATTGAGTTCTTCGTCACTGAAGAAATCATTCCC CTCGGGAATCAGGAGTGACCTGGAGCACTGTGCGCAGCCGTGTGTGCTGTGGAGCCGAGGCCGTCCTGCAGGAAGCCGCGTGACTCCCGC CTCCTCCCTGTGCTCTCTGGCTCTGGACTGTGACTGCGCCTGGATTCTGCCATTGCGACACATTTTTGTGCCTTTCAGCCCCTGGTGTCT GCAGTGGGGGATTTAAGGCACCCGCTTCCACTTCTTTCTTGTTTGGAGTTTTCTGTTGGAACCGCCGGCGTTGGCTCCGAAGACTTAGCG ACGCCACTGGCGGCACCTTCTCCTGCGCCCAGTGATGTTTCCACGGTGCCTGTACACAGCCGAGCAGCATTTCCGTTGAAGGACTTGCAT CCCCATTGCGGGCAGTGCTGGACGTGTCCCGGAGACCCACCGGGAGGGCGCCGCCATGCCTTGTACCCCCACCGTGCAGGTTGTGGCCGG >103004_103004_14_ZNF814-EIF3B_ZNF814_chr19_58400135_ENST00000600634_EIF3B_chr7_2416585_ENST00000397011_length(amino acids)=150AA_BP=10 MAAAATLRLSAQVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ZNF814-EIF3B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ZNF814-EIF3B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ZNF814-EIF3B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
