
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BTBD1-ETFA (FusionGDB2 ID:10441) |
Fusion Gene Summary for BTBD1-ETFA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BTBD1-ETFA | Fusion gene ID: 10441 | Hgene | Tgene | Gene symbol | BTBD1 | ETFA | Gene ID | 53339 | 2108 |
| Gene name | BTB domain containing 1 | electron transfer flavoprotein subunit alpha | |
| Synonyms | C15orf1|NS5ATP8 | EMA|GA2|MADD | |
| Cytomap | 15q25.2 | 15q24.2-q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | BTB/POZ domain-containing protein 1BTB (POZ) domain containing 1HCV NS5A-transactivated protein 8hepatitis C virus NS5A-transactivated protein 8 | electron transfer flavoprotein subunit alpha, mitochondrialalpha-ETFelectron transfer flavoprotein alpha subunitelectron transfer flavoprotein, alpha polypeptideepididymis secretory sperm binding proteinglutaric aciduria II | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q9H0C5 | P13804 | |
| Ensembl transtripts involved in fusion gene | ENST00000261721, ENST00000379403, ENST00000560015, | ENST00000433983, ENST00000559602, ENST00000560726, ENST00000560816, ENST00000557943, | |
| Fusion gene scores | * DoF score | 11 X 9 X 9=891 | 8 X 7 X 6=336 |
| # samples | 13 | 8 | |
| ** MAII score | log2(13/891*10)=-2.77691380849347 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/336*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: BTBD1 [Title/Abstract] AND ETFA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BTBD1(83725140)-ETFA(76588078), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ETFA | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 25416781 |
 Fusion gene breakpoints across BTBD1 (5'-gene) Fusion gene breakpoints across BTBD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
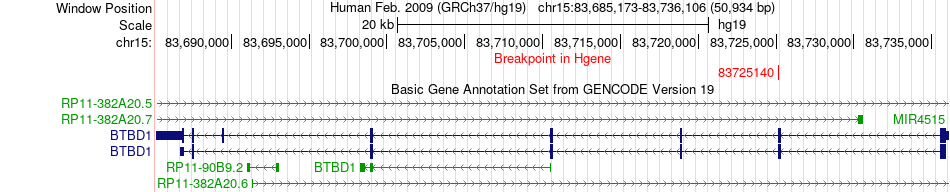 |
 Fusion gene breakpoints across ETFA (3'-gene) Fusion gene breakpoints across ETFA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
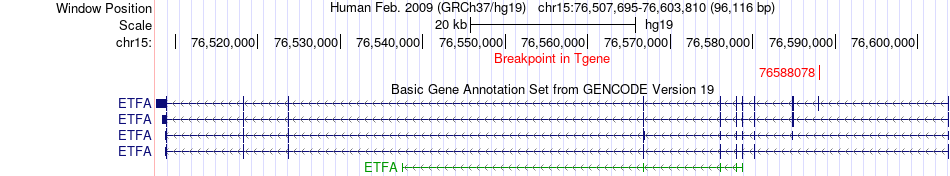 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 2397N | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
Top |
Fusion Gene ORF analysis for BTBD1-ETFA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000261721 | ENST00000433983 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000261721 | ENST00000559602 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000261721 | ENST00000560726 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000261721 | ENST00000560816 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000379403 | ENST00000433983 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000379403 | ENST00000559602 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000379403 | ENST00000560726 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| 5CDS-intron | ENST00000379403 | ENST00000560816 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| In-frame | ENST00000261721 | ENST00000557943 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| In-frame | ENST00000379403 | ENST00000557943 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| intron-3CDS | ENST00000560015 | ENST00000557943 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| intron-intron | ENST00000560015 | ENST00000433983 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| intron-intron | ENST00000560015 | ENST00000559602 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| intron-intron | ENST00000560015 | ENST00000560726 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
| intron-intron | ENST00000560015 | ENST00000560816 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000261721 | BTBD1 | chr15 | 83725140 | - | ENST00000557943 | ETFA | chr15 | 76588078 | - | 2928 | 761 | 203 | 1723 | 506 |
| ENST00000379403 | BTBD1 | chr15 | 83725140 | - | ENST00000557943 | ETFA | chr15 | 76588078 | - | 2725 | 558 | 0 | 1520 | 506 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000261721 | ENST00000557943 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - | 0.001177459 | 0.9988225 |
| ENST00000379403 | ENST00000557943 | BTBD1 | chr15 | 83725140 | - | ETFA | chr15 | 76588078 | - | 0.001185361 | 0.9988147 |
Top |
Fusion Genomic Features for BTBD1-ETFA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
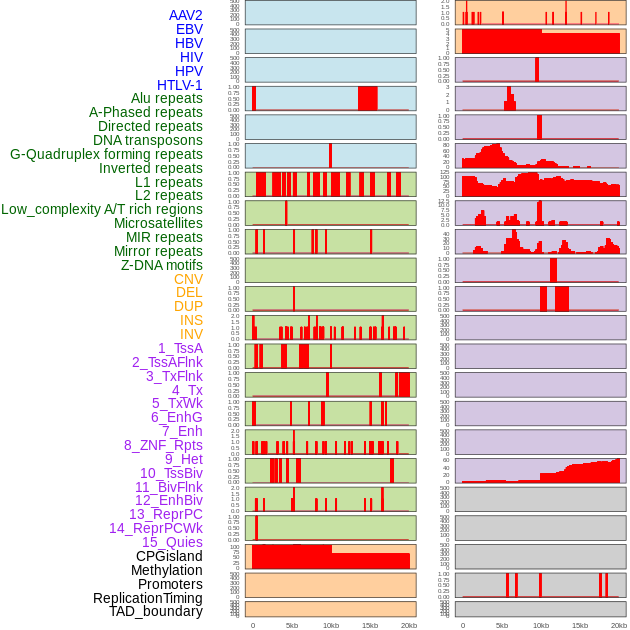 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for BTBD1-ETFA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:83725140/chr15:76588078) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BTBD1 | ETFA |
| FUNCTION: Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins (PubMed:14528312). Seems to regulate expression levels and/or subnuclear distribution of TOP1, via an unknown mechanism (By similarity). May play a role in mesenchymal differentiation where it promotes myogenic differentiation and suppresses adipogenesis (By similarity). {ECO:0000250|UniProtKB:P58544, ECO:0000269|PubMed:14528312}. | FUNCTION: Heterodimeric electron transfer flavoprotein that accepts electrons from several mitochondrial dehydrogenases, including acyl-CoA dehydrogenases, glutaryl-CoA and sarcosine dehydrogenase (PubMed:27499296, PubMed:15159392, PubMed:15975918, PubMed:9334218, PubMed:10356313). It transfers the electrons to the main mitochondrial respiratory chain via ETF-ubiquinone oxidoreductase (ETF dehydrogenase) (PubMed:9334218). Required for normal mitochondrial fatty acid oxidation and normal amino acid metabolism (PubMed:12815589, PubMed:1882842, PubMed:1430199). {ECO:0000269|PubMed:10356313, ECO:0000269|PubMed:12815589, ECO:0000269|PubMed:1430199, ECO:0000269|PubMed:15159392, ECO:0000269|PubMed:15975918, ECO:0000269|PubMed:27499296, ECO:0000269|PubMed:9334218, ECO:0000303|PubMed:17941859, ECO:0000305|PubMed:1882842}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BTBD1 | chr15:83725140 | chr15:76588078 | ENST00000261721 | - | 2 | 8 | 25_31 | 186 | 483.0 | Compositional bias | Poly-Pro |
| Hgene | BTBD1 | chr15:83725140 | chr15:76588078 | ENST00000379403 | - | 2 | 7 | 25_31 | 186 | 337.3333333333333 | Compositional bias | Poly-Pro |
| Hgene | BTBD1 | chr15:83725140 | chr15:76588078 | ENST00000261721 | - | 2 | 8 | 69_145 | 186 | 483.0 | Domain | BTB |
| Hgene | BTBD1 | chr15:83725140 | chr15:76588078 | ENST00000379403 | - | 2 | 7 | 69_145 | 186 | 337.3333333333333 | Domain | BTB |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000433983 | 0 | 11 | 263_266 | 0 | 285.0 | Nucleotide binding | FAD | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000433983 | 0 | 11 | 281_286 | 0 | 285.0 | Nucleotide binding | FAD | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000433983 | 0 | 11 | 318_319 | 0 | 285.0 | Nucleotide binding | FAD | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000557943 | 0 | 12 | 263_266 | 13 | 334.0 | Nucleotide binding | FAD | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000557943 | 0 | 12 | 281_286 | 13 | 334.0 | Nucleotide binding | FAD | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000557943 | 0 | 12 | 318_319 | 13 | 334.0 | Nucleotide binding | FAD | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000433983 | 0 | 11 | 205_333 | 0 | 285.0 | Region | Domain II | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000433983 | 0 | 11 | 20_204 | 0 | 285.0 | Region | Domain I | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000557943 | 0 | 12 | 205_333 | 13 | 334.0 | Region | Domain II | |
| Tgene | ETFA | chr15:83725140 | chr15:76588078 | ENST00000557943 | 0 | 12 | 20_204 | 13 | 334.0 | Region | Domain I |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BTBD1 | chr15:83725140 | chr15:76588078 | ENST00000261721 | - | 2 | 8 | 184_284 | 186 | 483.0 | Domain | BACK |
| Hgene | BTBD1 | chr15:83725140 | chr15:76588078 | ENST00000379403 | - | 2 | 7 | 184_284 | 186 | 337.3333333333333 | Domain | BACK |
Top |
Fusion Gene Sequence for BTBD1-ETFA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >10441_10441_1_BTBD1-ETFA_BTBD1_chr15_83725140_ENST00000261721_ETFA_chr15_76588078_ENST00000557943_length(transcript)=2928nt_BP=761nt AGTCCTGTTACACCTCCCGCGGCCGCGCACAGCGGCAACCCGGGCGTGCGGGGGCGCGCGCGGTCAGGGAGGGGCCTGCGGAGGCGCGAG GCCCCCGGAAGCGCCTTTCCCGGAAGGCAGGGAGGCAGCGCCGGCCTCCGGAGGCGGCCTGGGCGATGGCGGCGGAGTTTTGTCCATAAC GTGGGCAACCGCGCAGCTGGAGGATGGCCTCACTCGGGCCTGCCGCAGCTGGGGAGCAGGCGTCGGGGGCTGAGGCGGAGCCGGGCCCCG CGGGGCCGCCGCCGCCGCCCTCACCGTCCTCTCTGGGGCCCCTGCTCCCCCTGCAGCGGGAACCTCTCTACAACTGGCAGGCGACCAAGG CGTCGCTGAAGGAGCGCTTCGCCTTCCTCTTCAACTCGGAGCTGCTGAGCGATGTGCGCTTCGTACTGGGCAAGGGTCGCGGCGCCGCCG CCGCTGGGGGCCCGCAGCGCATCCCCGCCCACCGCTTCGTGCTGGCGGCCGGCAGCGCCGTCTTTGACGCCATGTTCAACGGCGGCATGG CCACCACGTCGGCCGAGATCGAGCTGCCGGACGTGGAGCCCGCAGCCTTCCTGGCGCTGCTGAGATTTCTATATTCAGATGAAGTTCAAA TTGGTCCAGAAACAGTTATGACCACTCTTTATACTGCCAAGAAATACGCAGTCCCAGCCTTGGAAGCACACTGTGTAGAATTTCTCACCA AACATCTTAGGGCAGATAATGCCTTTATGTTACTTACTCAGGCCTCATTGCTACGATTTCAGAGTACCCTGGTAATAGCTGAGCATGCAA ATGATTCCCTAGCACCCATTACTTTAAATACCATTACTGCAGCCACACGCCTTGGAGGTGAAGTGTCCTGCTTAGTAGCTGGAACCAAAT GTGACAAGGTGGCACAAGATCTCTGTAAAGTAGCAGGCATAGCAAAAGTTCTGGTGGCTCAGCATGATGTGTACAAAGGCCTACTTCCAG AGGAACTGACACCATTGATTTTGGCAACTCAGAAGCAGTTCAATTACACACACATCTGTGCTGGAGCATCTGCCTTCGGAAAGAACCTTT TGCCCAGAGTAGCAGCCAAACTTGAGGTTGCCCCGATTTCTGACATCATTGCAATCAAGTCACCTGACACATTTGTGAGAACTATTTATG CAGGAAATGCTCTATGTACAGTGAAGTGTGATGAGAAAGTGAAAGTGTTTTCTGTCCGTGGAACATCCTTTGATGCTGCAGCAACAAGTG GCGGTAGTGCCAGTTCAGAAAAGGCATCAAGTACTTCACCAGTGGAAATATCAGAGTGGCTTGACCAGAAATTAACAAAAAGTGATCGAC CAGAGCTAACAGGTGCCAAAGTGGTGGTATCTGGTGGTCGAGGCTTGAAGAGTGGAGAGAACTTTAAGTTGTTATATGACTTGGCAGATC AACTACATGCTGCAGTTGGTGCTTCCCGTGCTGCTGTTGATGCTGGCTTTGTTCCCAATGACATGCAAGTTGGACAGACGGGAAAAATAG TAGCACCAGAACTTTATATTGCTGTTGGAATATCTGGAGCCATCCAACATTTAGCTGGGATGAAAGACAGCAAGACAATTGTGGCAATTA ATAAAGACCCAGAAGCTCCAATTTTCCAAGTGGCAGATTATGGAATAGTTGCAGATTTATTTAAGGTAGTTCCTGAAATGACTGAGATAT TGAAGAAAAAATGAATCAGGATCATGCCTTAAAAAGAAAACTTTTGTTAAAGTATTCCACTGAAATCACAGATATTTGTGGGTATTATAA CAATCATTGGAAAGCATGGAGAGCTACATTTCATAATTTGAGGGAAAATTTCTAACAGATGCCAGAATGCTTGTTTATGGGATTGCTGTG TTTCCTTTTAATTATTTGTGGTTCCAAACAATTATTGTTTGAACTTTTTAAATTCTGTACTAAAATCTATAATAAAGCTTTTCCACAGCT TTAAAACTATCAGAAATATGCCTAGCCTCTAGAGAAAATTGAAGAAAAGGAGTAAAAATTTTGTTTATTTATTTATTTTTTTGCCTTTCT TGTTCATGGAGAGCTCTATGTTTTAGGAAGATTGTAAACATGATTGTCTCCACCAGCCTAATCAACCTACTAATCTCATTGTTGAAGGGC AAACTTCATAGGAGAAGGTAACTGAATTGGCAAGTGCAGAGTTAAAAAAGCACGGCTATAAAGCATGAATATTATCTTAAACATACTCCA GTCTTCAGAAGACCATAGCTAATGTGACAAAAGCAGACCTGACTTAAACCTGACCCAGCTCCATGTATTTAGTTGGCTTAAGGTAAATTT CATTTTCCGTTTAATTTTAAGGTTTTGTCAGTTTATAGGACTTTTGTCTGTAAAGGAGGGAGAGTGGAGGAGGGGCTGCTTTGAGACAGC ATGGCCTCTGATGCAAGCAGATAGTTTGATGTTATTATAAACATTCAAAATCCTGTGCAGAGAATGCTCTAGCCTTTCTTTTGCCTTCTG TTGGCTTCTTTTGGGCTCTCTTAGGAAGCACCCAGGCCTGTGTAGTCACCTCTGGCACTCTCATTCTGCTGCCCAGGGCACAGCTGAGGT CCAAGAGTGTCACCAGGGCTGCAGCTGGTGTGGATGCAGGCCCAAGAGCAGAGGCTGGTGAGGCTTTGAACAGGCCTGCAGGAAGGCGTG GGTAGGTAGGTGAGTAGAACAAAGAGGGCTGCCCTCCTGAATCAGATCTGCAGAAGTAGGAGTAGTGACCTGGATCAAGCAGACAGTAGG GAAGAGAAGGGAAGTGATAAGTGATGGAGGTGGCTAGAGTGCCTTTGTGTGACCAGCCTGGGGCCCTTCCCCAGCTGACTAGTGAACCCT >10441_10441_1_BTBD1-ETFA_BTBD1_chr15_83725140_ENST00000261721_ETFA_chr15_76588078_ENST00000557943_length(amino acids)=506AA_BP=186 MASLGPAAAGEQASGAEAEPGPAGPPPPPSPSSLGPLLPLQREPLYNWQATKASLKERFAFLFNSELLSDVRFVLGKGRGAAAAGGPQRI PAHRFVLAAGSAVFDAMFNGGMATTSAEIELPDVEPAAFLALLRFLYSDEVQIGPETVMTTLYTAKKYAVPALEAHCVEFLTKHLRADNA FMLLTQASLLRFQSTLVIAEHANDSLAPITLNTITAATRLGGEVSCLVAGTKCDKVAQDLCKVAGIAKVLVAQHDVYKGLLPEELTPLIL ATQKQFNYTHICAGASAFGKNLLPRVAAKLEVAPISDIIAIKSPDTFVRTIYAGNALCTVKCDEKVKVFSVRGTSFDAAATSGGSASSEK ASSTSPVEISEWLDQKLTKSDRPELTGAKVVVSGGRGLKSGENFKLLYDLADQLHAAVGASRAAVDAGFVPNDMQVGQTGKIVAPELYIA -------------------------------------------------------------- >10441_10441_2_BTBD1-ETFA_BTBD1_chr15_83725140_ENST00000379403_ETFA_chr15_76588078_ENST00000557943_length(transcript)=2725nt_BP=558nt ATGGCCTCACTCGGGCCTGCCGCAGCTGGGGAGCAGGCGTCGGGGGCTGAGGCGGAGCCGGGCCCCGCGGGGCCGCCGCCGCCGCCCTCA CCGTCCTCTCTGGGGCCCCTGCTCCCCCTGCAGCGGGAACCTCTCTACAACTGGCAGGCGACCAAGGCGTCGCTGAAGGAGCGCTTCGCC TTCCTCTTCAACTCGGAGCTGCTGAGCGATGTGCGCTTCGTACTGGGCAAGGGTCGCGGCGCCGCCGCCGCTGGGGGCCCGCAGCGCATC CCCGCCCACCGCTTCGTGCTGGCGGCCGGCAGCGCCGTCTTTGACGCCATGTTCAACGGCGGCATGGCCACCACGTCGGCCGAGATCGAG CTGCCGGACGTGGAGCCCGCAGCCTTCCTGGCGCTGCTGAGATTTCTATATTCAGATGAAGTTCAAATTGGTCCAGAAACAGTTATGACC ACTCTTTATACTGCCAAGAAATACGCAGTCCCAGCCTTGGAAGCACACTGTGTAGAATTTCTCACCAAACATCTTAGGGCAGATAATGCC TTTATGTTACTTACTCAGGCCTCATTGCTACGATTTCAGAGTACCCTGGTAATAGCTGAGCATGCAAATGATTCCCTAGCACCCATTACT TTAAATACCATTACTGCAGCCACACGCCTTGGAGGTGAAGTGTCCTGCTTAGTAGCTGGAACCAAATGTGACAAGGTGGCACAAGATCTC TGTAAAGTAGCAGGCATAGCAAAAGTTCTGGTGGCTCAGCATGATGTGTACAAAGGCCTACTTCCAGAGGAACTGACACCATTGATTTTG GCAACTCAGAAGCAGTTCAATTACACACACATCTGTGCTGGAGCATCTGCCTTCGGAAAGAACCTTTTGCCCAGAGTAGCAGCCAAACTT GAGGTTGCCCCGATTTCTGACATCATTGCAATCAAGTCACCTGACACATTTGTGAGAACTATTTATGCAGGAAATGCTCTATGTACAGTG AAGTGTGATGAGAAAGTGAAAGTGTTTTCTGTCCGTGGAACATCCTTTGATGCTGCAGCAACAAGTGGCGGTAGTGCCAGTTCAGAAAAG GCATCAAGTACTTCACCAGTGGAAATATCAGAGTGGCTTGACCAGAAATTAACAAAAAGTGATCGACCAGAGCTAACAGGTGCCAAAGTG GTGGTATCTGGTGGTCGAGGCTTGAAGAGTGGAGAGAACTTTAAGTTGTTATATGACTTGGCAGATCAACTACATGCTGCAGTTGGTGCT TCCCGTGCTGCTGTTGATGCTGGCTTTGTTCCCAATGACATGCAAGTTGGACAGACGGGAAAAATAGTAGCACCAGAACTTTATATTGCT GTTGGAATATCTGGAGCCATCCAACATTTAGCTGGGATGAAAGACAGCAAGACAATTGTGGCAATTAATAAAGACCCAGAAGCTCCAATT TTCCAAGTGGCAGATTATGGAATAGTTGCAGATTTATTTAAGGTAGTTCCTGAAATGACTGAGATATTGAAGAAAAAATGAATCAGGATC ATGCCTTAAAAAGAAAACTTTTGTTAAAGTATTCCACTGAAATCACAGATATTTGTGGGTATTATAACAATCATTGGAAAGCATGGAGAG CTACATTTCATAATTTGAGGGAAAATTTCTAACAGATGCCAGAATGCTTGTTTATGGGATTGCTGTGTTTCCTTTTAATTATTTGTGGTT CCAAACAATTATTGTTTGAACTTTTTAAATTCTGTACTAAAATCTATAATAAAGCTTTTCCACAGCTTTAAAACTATCAGAAATATGCCT AGCCTCTAGAGAAAATTGAAGAAAAGGAGTAAAAATTTTGTTTATTTATTTATTTTTTTGCCTTTCTTGTTCATGGAGAGCTCTATGTTT TAGGAAGATTGTAAACATGATTGTCTCCACCAGCCTAATCAACCTACTAATCTCATTGTTGAAGGGCAAACTTCATAGGAGAAGGTAACT GAATTGGCAAGTGCAGAGTTAAAAAAGCACGGCTATAAAGCATGAATATTATCTTAAACATACTCCAGTCTTCAGAAGACCATAGCTAAT GTGACAAAAGCAGACCTGACTTAAACCTGACCCAGCTCCATGTATTTAGTTGGCTTAAGGTAAATTTCATTTTCCGTTTAATTTTAAGGT TTTGTCAGTTTATAGGACTTTTGTCTGTAAAGGAGGGAGAGTGGAGGAGGGGCTGCTTTGAGACAGCATGGCCTCTGATGCAAGCAGATA GTTTGATGTTATTATAAACATTCAAAATCCTGTGCAGAGAATGCTCTAGCCTTTCTTTTGCCTTCTGTTGGCTTCTTTTGGGCTCTCTTA GGAAGCACCCAGGCCTGTGTAGTCACCTCTGGCACTCTCATTCTGCTGCCCAGGGCACAGCTGAGGTCCAAGAGTGTCACCAGGGCTGCA GCTGGTGTGGATGCAGGCCCAAGAGCAGAGGCTGGTGAGGCTTTGAACAGGCCTGCAGGAAGGCGTGGGTAGGTAGGTGAGTAGAACAAA GAGGGCTGCCCTCCTGAATCAGATCTGCAGAAGTAGGAGTAGTGACCTGGATCAAGCAGACAGTAGGGAAGAGAAGGGAAGTGATAAGTG ATGGAGGTGGCTAGAGTGCCTTTGTGTGACCAGCCTGGGGCCCTTCCCCAGCTGACTAGTGAACCCTTTCTGATTTGTGTCACAATCAGT >10441_10441_2_BTBD1-ETFA_BTBD1_chr15_83725140_ENST00000379403_ETFA_chr15_76588078_ENST00000557943_length(amino acids)=506AA_BP=186 MASLGPAAAGEQASGAEAEPGPAGPPPPPSPSSLGPLLPLQREPLYNWQATKASLKERFAFLFNSELLSDVRFVLGKGRGAAAAGGPQRI PAHRFVLAAGSAVFDAMFNGGMATTSAEIELPDVEPAAFLALLRFLYSDEVQIGPETVMTTLYTAKKYAVPALEAHCVEFLTKHLRADNA FMLLTQASLLRFQSTLVIAEHANDSLAPITLNTITAATRLGGEVSCLVAGTKCDKVAQDLCKVAGIAKVLVAQHDVYKGLLPEELTPLIL ATQKQFNYTHICAGASAFGKNLLPRVAAKLEVAPISDIIAIKSPDTFVRTIYAGNALCTVKCDEKVKVFSVRGTSFDAAATSGGSASSEK ASSTSPVEISEWLDQKLTKSDRPELTGAKVVVSGGRGLKSGENFKLLYDLADQLHAAVGASRAAVDAGFVPNDMQVGQTGKIVAPELYIA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BTBD1-ETFA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BTBD1-ETFA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BTBD1-ETFA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
