
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:C9orf96-SQSTM1 (FusionGDB2 ID:12165) |
Fusion Gene Summary for C9orf96-SQSTM1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: C9orf96-SQSTM1 | Fusion gene ID: 12165 | Hgene | Tgene | Gene symbol | C9orf96 | SQSTM1 | Gene ID | 169436 | 8878 |
| Gene name | serine/threonine kinase like domain containing 1 | sequestosome 1 | |
| Synonyms | C9orf96|SgK071|Sk521 | A170|DMRV|FTDALS3|NADGP|OSIL|PDB3|ZIP3|p60|p62|p62B | |
| Cytomap | 9q34.2 | 5q35.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine kinase-like domain-containing protein STKLD1probable inactive protein kinase-like protein SgK071serine/threonine kinase-like domain-containing protein 1sugen kinase 071 | sequestosome-1EBI3-associated protein of 60 kDaEBI3-associated protein p60EBIAPautophagy receptor p62oxidative stress induced likephosphotyrosine independent ligand for the Lck SH2 domain p62phosphotyrosine-independent ligand for the Lck SH2 domain | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | Q13501 | |
| Ensembl transtripts involved in fusion gene | ENST00000371957, ENST00000426926, ENST00000371955, ENST00000468046, | ENST00000360718, ENST00000389805, ENST00000402874, ENST00000506690, ENST00000510187, ENST00000376929, | |
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 33 X 21 X 17=11781 |
| # samples | 2 | 38 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(38/11781*10)=-4.95431877505661 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: C9orf96 [Title/Abstract] AND SQSTM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | C9orf96(136253332)-SQSTM1(179263436), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | SQSTM1 | GO:0006914 | autophagy | 20452972 |
| Tgene | SQSTM1 | GO:0007032 | endosome organization | 27368102 |
| Tgene | SQSTM1 | GO:0031397 | negative regulation of protein ubiquitination | 20452972 |
| Tgene | SQSTM1 | GO:0061635 | regulation of protein complex stability | 25127057 |
| Tgene | SQSTM1 | GO:1905719 | protein localization to perinuclear region of cytoplasm | 27368102 |
 Fusion gene breakpoints across C9orf96 (5'-gene) Fusion gene breakpoints across C9orf96 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
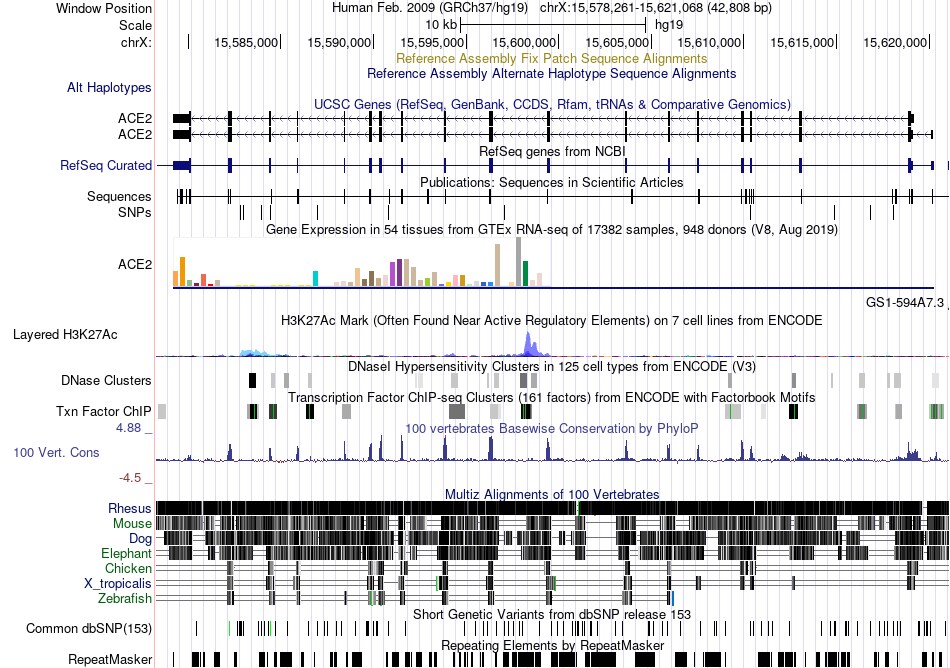 |
 Fusion gene breakpoints across SQSTM1 (3'-gene) Fusion gene breakpoints across SQSTM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
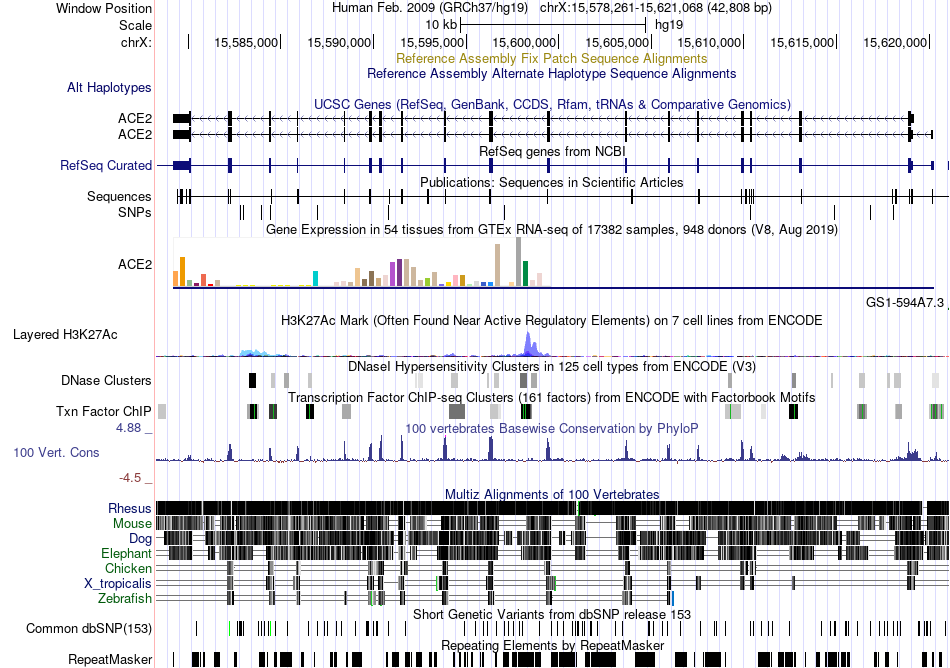 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-02-2486-01A | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
Top |
Fusion Gene ORF analysis for C9orf96-SQSTM1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000371957 | ENST00000360718 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000371957 | ENST00000389805 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000371957 | ENST00000402874 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000371957 | ENST00000506690 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000371957 | ENST00000510187 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000426926 | ENST00000360718 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000426926 | ENST00000389805 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000426926 | ENST00000402874 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000426926 | ENST00000506690 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5CDS-intron | ENST00000426926 | ENST00000510187 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5UTR-3CDS | ENST00000371955 | ENST00000376929 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5UTR-intron | ENST00000371955 | ENST00000360718 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5UTR-intron | ENST00000371955 | ENST00000389805 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5UTR-intron | ENST00000371955 | ENST00000402874 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5UTR-intron | ENST00000371955 | ENST00000506690 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| 5UTR-intron | ENST00000371955 | ENST00000510187 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| In-frame | ENST00000371957 | ENST00000376929 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| In-frame | ENST00000426926 | ENST00000376929 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| intron-3CDS | ENST00000468046 | ENST00000376929 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| intron-intron | ENST00000468046 | ENST00000360718 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| intron-intron | ENST00000468046 | ENST00000389805 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| intron-intron | ENST00000468046 | ENST00000402874 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| intron-intron | ENST00000468046 | ENST00000506690 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
| intron-intron | ENST00000468046 | ENST00000510187 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000426926 | C9orf96 | chr9 | 136253332 | + | ENST00000376929 | SQSTM1 | chr5 | 179263436 | + | 2264 | 624 | 84 | 641 | 185 |
| ENST00000371957 | C9orf96 | chr9 | 136253332 | + | ENST00000376929 | SQSTM1 | chr5 | 179263436 | + | 2143 | 503 | 2006 | 1458 | 182 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000426926 | ENST00000376929 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + | 0.03158172 | 0.9684183 |
| ENST00000371957 | ENST00000376929 | C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263436 | + | 0.03036449 | 0.96963555 |
Top |
Fusion Genomic Features for C9orf96-SQSTM1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263435 | + | 4.98E-09 | 1 |
| C9orf96 | chr9 | 136253332 | + | SQSTM1 | chr5 | 179263435 | + | 4.98E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
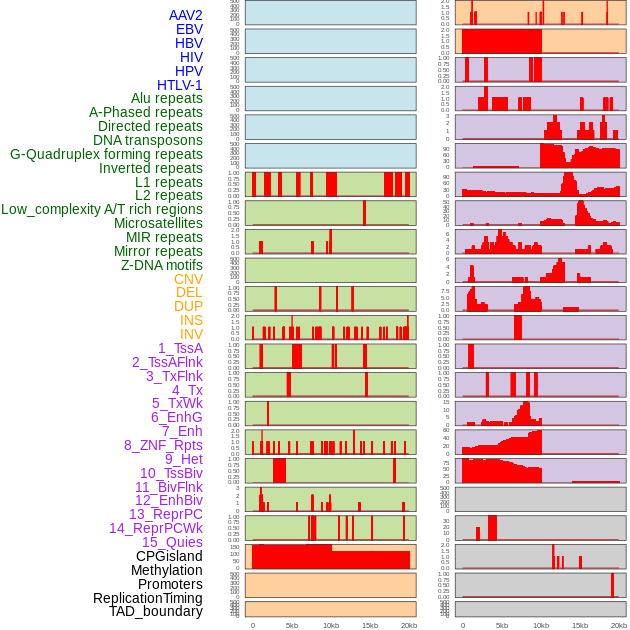 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
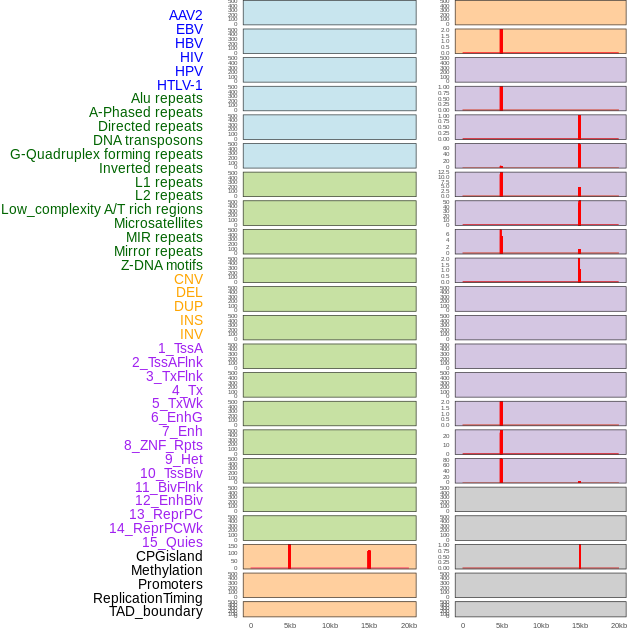 |
Top |
Fusion Protein Features for C9orf96-SQSTM1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:136253332/chr5:179263436) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | SQSTM1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Autophagy receptor required for selective macroautophagy (aggrephagy). Functions as a bridge between polyubiquitinated cargo and autophagosomes. Interacts directly with both the cargo to become degraded and an autophagy modifier of the MAP1 LC3 family (PubMed:16286508, PubMed:20168092, PubMed:24128730, PubMed:28404643, PubMed:22622177). Along with WDFY3, involved in the formation and autophagic degradation of cytoplasmic ubiquitin-containing inclusions (p62 bodies, ALIS/aggresome-like induced structures). Along with WDFY3, required to recruit ubiquitinated proteins to PML bodies in the nucleus (PubMed:24128730, PubMed:20168092). May regulate the activation of NFKB1 by TNF-alpha, nerve growth factor (NGF) and interleukin-1. May play a role in titin/TTN downstream signaling in muscle cells. May regulate signaling cascades through ubiquitination. Adapter that mediates the interaction between TRAF6 and CYLD (By similarity). May be involved in cell differentiation, apoptosis, immune response and regulation of K(+) channels. Involved in endosome organization by retaining vesicles in the perinuclear cloud: following ubiquitination by RNF26, attracts specific vesicle-associated adapters, forming a molecular bridge that restrains cognate vesicles in the perinuclear region and organizes the endosomal pathway for efficient cargo transport (PubMed:27368102). Promotes relocalization of 'Lys-63'-linked ubiquitinated STING1 to autophagosomes (PubMed:29496741). Acts as an activator of the NFE2L2/NRF2 pathway via interaction with KEAP1: interaction inactivates the BCR(KEAP1) complex, promoting nuclear accumulation of NFE2L2/NRF2 and subsequent expression of cytoprotective genes (PubMed:20452972, PubMed:28380357, PubMed:33393215). {ECO:0000250|UniProtKB:O08623, ECO:0000250|UniProtKB:Q64337, ECO:0000269|PubMed:10356400, ECO:0000269|PubMed:10747026, ECO:0000269|PubMed:11244088, ECO:0000269|PubMed:12471037, ECO:0000269|PubMed:15340068, ECO:0000269|PubMed:15802564, ECO:0000269|PubMed:15911346, ECO:0000269|PubMed:15953362, ECO:0000269|PubMed:16079148, ECO:0000269|PubMed:16286508, ECO:0000269|PubMed:19931284, ECO:0000269|PubMed:20168092, ECO:0000269|PubMed:20452972, ECO:0000269|PubMed:22622177, ECO:0000269|PubMed:24128730, ECO:0000269|PubMed:27368102, ECO:0000269|PubMed:28380357, ECO:0000269|PubMed:28404643, ECO:0000269|PubMed:29496741, ECO:0000269|PubMed:33393215}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | C9orf96 | chr9:136253332 | chr5:179263436 | ENST00000371957 | + | 5 | 18 | 34_42 | 132 | 681.0 | Nucleotide binding | ATP |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 389_434 | 304 | 357.0 | Domain | UBA | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 389_434 | 304 | 357.0 | Domain | UBA | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 389_434 | 388 | 441.0 | Domain | UBA | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 389_434 | 304 | 357.0 | Domain | UBA | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 336_341 | 304 | 357.0 | Motif | Note=LIR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 336_341 | 304 | 357.0 | Motif | Note=LIR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 336_341 | 304 | 357.0 | Motif | Note=LIR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 321_342 | 304 | 357.0 | Region | MAP1LC3B-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 321_342 | 304 | 357.0 | Region | MAP1LC3B-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 321_342 | 304 | 357.0 | Region | MAP1LC3B-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | C9orf96 | chr9:136253332 | chr5:179263436 | ENST00000371955 | + | 5 | 16 | 28_297 | 0 | 214.0 | Domain | Protein kinase |
| Hgene | C9orf96 | chr9:136253332 | chr5:179263436 | ENST00000371957 | + | 5 | 18 | 28_297 | 132 | 681.0 | Domain | Protein kinase |
| Hgene | C9orf96 | chr9:136253332 | chr5:179263436 | ENST00000371955 | + | 5 | 16 | 34_42 | 0 | 214.0 | Nucleotide binding | ATP |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 272_294 | 304 | 357.0 | Compositional bias | Note=Ser-rich | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 272_294 | 304 | 357.0 | Compositional bias | Note=Ser-rich | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 272_294 | 388 | 441.0 | Compositional bias | Note=Ser-rich | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 272_294 | 304 | 357.0 | Compositional bias | Note=Ser-rich | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 3_102 | 304 | 357.0 | Domain | PB1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 3_102 | 304 | 357.0 | Domain | PB1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 3_102 | 388 | 441.0 | Domain | PB1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 3_102 | 304 | 357.0 | Domain | PB1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 228_233 | 304 | 357.0 | Motif | Note=TRAF6-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 228_233 | 304 | 357.0 | Motif | Note=TRAF6-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 228_233 | 388 | 441.0 | Motif | Note=TRAF6-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 336_341 | 388 | 441.0 | Motif | Note=LIR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 228_233 | 304 | 357.0 | Motif | Note=TRAF6-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 170_220 | 304 | 357.0 | Region | Note=LIM protein-binding (LB) | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 170_220 | 304 | 357.0 | Region | Note=LIM protein-binding (LB) | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 170_220 | 388 | 441.0 | Region | Note=LIM protein-binding (LB) | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 321_342 | 388 | 441.0 | Region | MAP1LC3B-binding | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 170_220 | 304 | 357.0 | Region | Note=LIM protein-binding (LB) | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 122_167 | 304 | 357.0 | Zinc finger | ZZ-type | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 122_167 | 304 | 357.0 | Zinc finger | ZZ-type | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 122_167 | 388 | 441.0 | Zinc finger | ZZ-type | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 122_167 | 304 | 357.0 | Zinc finger | ZZ-type |
Top |
Fusion Gene Sequence for C9orf96-SQSTM1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >12165_12165_1_C9orf96-SQSTM1_C9orf96_chr9_136253332_ENST00000371957_SQSTM1_chr5_179263436_ENST00000376929_length(transcript)=2143nt_BP=503nt AGTGCCTCGAGGGCGCCGTTCGGGCGGGGAGGATCCCGCGGGTCCCACTGACCCACGCGGGGTGGGGCCAGGGGTGGACGCTCGCCCGTA CGCGGTCGCTACTGATCATGCTTGGGCCAGGGTCCAATCGCAGGCGCCCCACGCAGGGGGAGCGAGGCCCAGGGTCCCCCGGAGAGCCCA TGGAGAAGTACCAGGTTTTGTACCAGCTGAATCCTGGGGCCTTGGGGGTGAACCTGGTGGTGGAGGAAATGGAAACCAAAGTCAAGCATG TGATAAAGCAGGTGGAATGCATGGATGACCATTACGCCAGTCAGGCCCTGGAGGAGCTGATGCCACTGCTGAAGCTGCGGCACGCCCACA TCTCTGTGTACCAGGAGCTGTTCATCACGTGGAATGGGGAGATCTCTTCTCTGTACCTCTGCCTGGTGATGGAGTTCAATGAGCTCAGCT TCCAGGAGGTCATTGAGGATAAGAGGAAGGCAAAGAAAATCATTGACTCTGAGAGGCTGACCCGCGGCTGATTGAGTCCCTCTCCCAGAT GCTGTCCATGGGCTTCTCTGATGAAGGCGGCTGGCTCACCAGGCTCCTGCAGACCAAGAACTATGACATCGGAGCGGCTCTGGACACCAT CCAGTATTCAAAGCATCCCCCGCCGTTGTGACCACTTTTGCCCACCTCTTCTGCGTGCCCCTCTTCTGTCTCATAGTTGTGTTAAGCTTG CGTAGAATTGCAGGTCTCTGTACGGGCCAGTTTCTCTGCCTTCTTCCAGGATCAGGGGTTAGGGTGCAAGAAGCCATTTAGGGCAGCAAA ACAAGTGACATGAAGGGAGGGTCCCTGTGTGTGTGTGTGCTGATGTTTCCTGGGTGCCCTGGCTCCTTGCAGCAGGGCTGGGCCTGCGAG ACCCAAGGCTCACTGCAGCGCGCTCCTGACCCCTCCCTGCAGGGGCTACGTTAGCAGCCCAGCACATAGCTTGCCTAATGGCTTTCACTT TCTCTTTTGTTTTAAATGACTCATAGGTCCCTGACATTTAGTTGATTATTTTCTGCTACAGACCTGGTACACTCTGATTTTAGATAAAGT AAGCCTAGGTGTTGTCAGCAGGCAGGCTGGGGAGGCCAGTGTTGTGGGCTTCCTGCTGGGACTGAGAAGGCTCACGAAGGGCATCCGCAA TGTTGGTTTCACTGAGAGCTGCCTCCTGGTCTCTTCACCACTGTAGTTCTCTCATTTCCAAACCATCAGCTGCTTTTAAAATAAGATCTC TTTGTAGCCATCCTGTTAAATTTGTAAACAATCTAATTAAATGGCATCAGCACTTTAACCAATGACGTTTGCATAGAGAGAAATGATTGA CAGTAAGTTTATTGTTAATGGTTCTTACAGAGTATCTTTAAAAGTGCCTTAGGGGAACCCTGTCCCTCCTAACAAGTGTATCTCGATTAA TAACCTGCCAGTCCCAGATCACACATCATCATCGAAGTCTTCCCCAGTTATAAAGAGGTCACATAGTCGTGTGGGTCGAGGATTCTGTGC CTCCAGGACCAGGGGCCCACCCTCTGCCCAGGGAGTCCTTGCGTCCCATGAGGTCTTCCCGCAAGGCCTCTCAGACCCAGATGTGACGGG GTGTGTGGCCCGAGGAAGCTGGACAGCGGCAGTGGGCCTGCTGAGGCCTTCTCTTGAGGCCTGTGCTCTGGGGGTCCCTTGCTTAGCCTG TGCTGGACCAGCTGGCCTGGGGTCCCTCTGAAGAGACCTTGGCTGCTCACTGTCCACATGTGAACTTTTTCTAGGTGGCAGGACAAATTG CGCCCATTTAGAGGATGTGGCTGTAACCTGCTGGATGGGACTCCATAGCTCCTTCCCAGGACCCCTCAGCTCCCCGGCACTGCAGTCTGC AGAGTTCTCCTGGAGGCAGGGGCTGCTGCCTTGTTTCACCTTCCATGTCAGGCCAGCCTGTCCCTGAAAGAGAAGATGGCCATGCCCTCC ATGTGTAAGAACAATGCCAGGGCCCAGGAGGACCGCCTGCCCTGCCTGGGCCTTGGCTGGGCCTCTGGTTCTGACACTTTCTGCTGGAAG >12165_12165_1_C9orf96-SQSTM1_C9orf96_chr9_136253332_ENST00000371957_SQSTM1_chr5_179263436_ENST00000376929_length(amino acids)=182AA_BP= MGPGIVLTHGGHGHLLFQGQAGLTWKVKQGSSPCLQENSADCSAGELRGPGKELWSPIQQVTATSSKWAQFVLPPRKSSHVDSEQPRSLQ RDPRPAGPAQAKQGTPRAQASREGLSRPTAAVQLPRATHPVTSGSERPCGKTSWDARTPWAEGGPLVLEAQNPRPTRLCDLFITGEDFDD -------------------------------------------------------------- >12165_12165_2_C9orf96-SQSTM1_C9orf96_chr9_136253332_ENST00000426926_SQSTM1_chr5_179263436_ENST00000376929_length(transcript)=2264nt_BP=624nt ACGCTGGAGGCCCCGCCCGCCGCGCGCAGGCCCTGCGGTCCCTCCCGGCCCGGCGGAACGCGTCCCTTTTAAGGGGGCGGGGACCTGGGG GTCTGGGGCCAGCGCGCGGGAGGGACGCCTGAGTGCCTCGAGGGCGCCGTTCGGGCGGGGAGGATCCCGCGGGTCCCACTGACCCACGCG GGGTGGGGCCAGGGGTGGACGCTCGCCCGTACGCGGTCGCTACTGATCATGCTTGGGCCAGGGTCCAATCGCAGGCGCCCCACGCAGGGG GAGCGAGGCCCAGGGTCCCCCGGAGAGCCCATGGAGAAGTACCAGGTTTTGTACCAGCTGAATCCTGGGGCCTTGGGGGTGAACCTGGTG GTGGAGGAAATGGAAACCAAAGTCAAGCATGTGATAAAGCAGGTGGAATGCATGGATGACCATTACGCCAGTCAGGCCCTGGAGGAGCTG ATGCCACTGCTGAAGCTGCGGCACGCCCACATCTCTGTGTACCAGGAGCTGTTCATCACGTGGAATGGGGAGATCTCTTCTCTGTACCTC TGCCTGGTGATGGAGTTCAATGAGCTCAGCTTCCAGGAGGTCATTGAGGATAAGAGGAAGGCAAAGAAAATCATTGACTCTGAGAGGCTG ACCCGCGGCTGATTGAGTCCCTCTCCCAGATGCTGTCCATGGGCTTCTCTGATGAAGGCGGCTGGCTCACCAGGCTCCTGCAGACCAAGA ACTATGACATCGGAGCGGCTCTGGACACCATCCAGTATTCAAAGCATCCCCCGCCGTTGTGACCACTTTTGCCCACCTCTTCTGCGTGCC CCTCTTCTGTCTCATAGTTGTGTTAAGCTTGCGTAGAATTGCAGGTCTCTGTACGGGCCAGTTTCTCTGCCTTCTTCCAGGATCAGGGGT TAGGGTGCAAGAAGCCATTTAGGGCAGCAAAACAAGTGACATGAAGGGAGGGTCCCTGTGTGTGTGTGTGCTGATGTTTCCTGGGTGCCC TGGCTCCTTGCAGCAGGGCTGGGCCTGCGAGACCCAAGGCTCACTGCAGCGCGCTCCTGACCCCTCCCTGCAGGGGCTACGTTAGCAGCC CAGCACATAGCTTGCCTAATGGCTTTCACTTTCTCTTTTGTTTTAAATGACTCATAGGTCCCTGACATTTAGTTGATTATTTTCTGCTAC AGACCTGGTACACTCTGATTTTAGATAAAGTAAGCCTAGGTGTTGTCAGCAGGCAGGCTGGGGAGGCCAGTGTTGTGGGCTTCCTGCTGG GACTGAGAAGGCTCACGAAGGGCATCCGCAATGTTGGTTTCACTGAGAGCTGCCTCCTGGTCTCTTCACCACTGTAGTTCTCTCATTTCC AAACCATCAGCTGCTTTTAAAATAAGATCTCTTTGTAGCCATCCTGTTAAATTTGTAAACAATCTAATTAAATGGCATCAGCACTTTAAC CAATGACGTTTGCATAGAGAGAAATGATTGACAGTAAGTTTATTGTTAATGGTTCTTACAGAGTATCTTTAAAAGTGCCTTAGGGGAACC CTGTCCCTCCTAACAAGTGTATCTCGATTAATAACCTGCCAGTCCCAGATCACACATCATCATCGAAGTCTTCCCCAGTTATAAAGAGGT CACATAGTCGTGTGGGTCGAGGATTCTGTGCCTCCAGGACCAGGGGCCCACCCTCTGCCCAGGGAGTCCTTGCGTCCCATGAGGTCTTCC CGCAAGGCCTCTCAGACCCAGATGTGACGGGGTGTGTGGCCCGAGGAAGCTGGACAGCGGCAGTGGGCCTGCTGAGGCCTTCTCTTGAGG CCTGTGCTCTGGGGGTCCCTTGCTTAGCCTGTGCTGGACCAGCTGGCCTGGGGTCCCTCTGAAGAGACCTTGGCTGCTCACTGTCCACAT GTGAACTTTTTCTAGGTGGCAGGACAAATTGCGCCCATTTAGAGGATGTGGCTGTAACCTGCTGGATGGGACTCCATAGCTCCTTCCCAG GACCCCTCAGCTCCCCGGCACTGCAGTCTGCAGAGTTCTCCTGGAGGCAGGGGCTGCTGCCTTGTTTCACCTTCCATGTCAGGCCAGCCT GTCCCTGAAAGAGAAGATGGCCATGCCCTCCATGTGTAAGAACAATGCCAGGGCCCAGGAGGACCGCCTGCCCTGCCTGGGCCTTGGCTG GGCCTCTGGTTCTGACACTTTCTGCTGGAAGCTGTCAGGCTGGGACAGGCTTTGATTTTGAGGGTTAGCAAGACAAAGCAAATAAATGCC >12165_12165_2_C9orf96-SQSTM1_C9orf96_chr9_136253332_ENST00000426926_SQSTM1_chr5_179263436_ENST00000376929_length(amino acids)=185AA_BP= MGVWGQRAGGTPECLEGAVRAGRIPRVPLTHAGWGQGWTLARTRSLLIMLGPGSNRRRPTQGERGPGSPGEPMEKYQVLYQLNPGALGVN LVVEEMETKVKHVIKQVECMDDHYASQALEELMPLLKLRHAHISVYQELFITWNGEISSLYLCLVMEFNELSFQEVIEDKRKAKKIIDSE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for C9orf96-SQSTM1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 347_352 | 304.3333333333333 | 357.0 | KEAP1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 347_352 | 304.3333333333333 | 357.0 | KEAP1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 347_352 | 304.3333333333333 | 357.0 | KEAP1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 122_224 | 304.3333333333333 | 357.0 | GABRR3 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 122_224 | 304.3333333333333 | 357.0 | GABRR3 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 122_224 | 388.3333333333333 | 441.0 | GABRR3 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 122_224 | 304.3333333333333 | 357.0 | GABRR3 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 347_352 | 388.3333333333333 | 441.0 | KEAP1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 2_50 | 304.3333333333333 | 357.0 | LCK | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 2_50 | 304.3333333333333 | 357.0 | LCK | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 2_50 | 388.3333333333333 | 441.0 | LCK | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 2_50 | 304.3333333333333 | 357.0 | LCK | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 269_440 | 304.3333333333333 | 357.0 | NTRK1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 269_440 | 304.3333333333333 | 357.0 | NTRK1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 269_440 | 388.3333333333333 | 441.0 | NTRK1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 269_440 | 304.3333333333333 | 357.0 | NTRK1 | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 50_80 | 304.3333333333333 | 357.0 | PAWR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 50_80 | 304.3333333333333 | 357.0 | PAWR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 50_80 | 388.3333333333333 | 441.0 | PAWR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 50_80 | 304.3333333333333 | 357.0 | PAWR | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000360718 | 5 | 7 | 43_107 | 304.3333333333333 | 357.0 | PRKCZ and dimerization | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000376929 | 7 | 9 | 43_107 | 304.3333333333333 | 357.0 | PRKCZ and dimerization | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000389805 | 6 | 8 | 43_107 | 388.3333333333333 | 441.0 | PRKCZ and dimerization | |
| Tgene | SQSTM1 | chr9:136253332 | chr5:179263436 | ENST00000402874 | 6 | 8 | 43_107 | 304.3333333333333 | 357.0 | PRKCZ and dimerization |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for C9orf96-SQSTM1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for C9orf96-SQSTM1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
