
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CAP1-NARS2 (FusionGDB2 ID:12818) |
Fusion Gene Summary for CAP1-NARS2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CAP1-NARS2 | Fusion gene ID: 12818 | Hgene | Tgene | Gene symbol | CAP1 | NARS2 | Gene ID | 10487 | 79731 |
| Gene name | cyclase associated actin cytoskeleton regulatory protein 1 | asparaginyl-tRNA synthetase 2, mitochondrial | |
| Synonyms | CAP|CAP1-PEN | DFNB94|SLM5|asnRS | |
| Cytomap | 1p34.2 | 11q14.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | adenylyl cyclase-associated protein 1CAP, adenylate cyclase-associated protein 1adenylate cyclase associated protein 1testis secretory sperm-binding protein Li 218p | probable asparagine--tRNA ligase, mitochondrialasparagine tRNA ligase 2, mitochondrial (putative)asparaginyl-tRNA synthetase 2, mitochondrial (putative)deafness, autosomal recessive 94probable asparaginyl-tRNA synthetase, mitochondrial | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q01518 | Q96I59 | |
| Ensembl transtripts involved in fusion gene | ENST00000340450, ENST00000372792, ENST00000372797, ENST00000372798, ENST00000372802, ENST00000372805, ENST00000479759, | ENST00000281038, ENST00000528850, | |
| Fusion gene scores | * DoF score | 16 X 13 X 7=1456 | 29 X 15 X 11=4785 |
| # samples | 19 | 37 | |
| ** MAII score | log2(19/1456*10)=-2.93793903186775 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(37/4785*10)=-3.69292174885708 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CAP1 [Title/Abstract] AND NARS2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CAP1(40525842)-NARS2(78154804), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CAP1 (5'-gene) Fusion gene breakpoints across CAP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
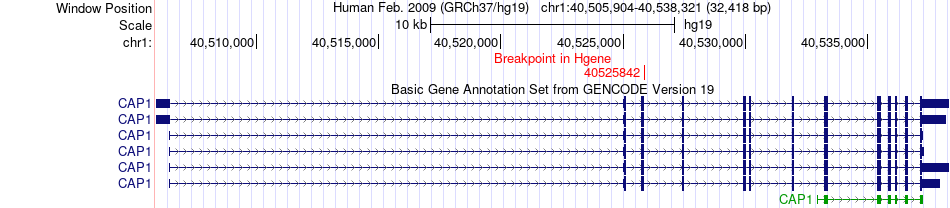 |
 Fusion gene breakpoints across NARS2 (3'-gene) Fusion gene breakpoints across NARS2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
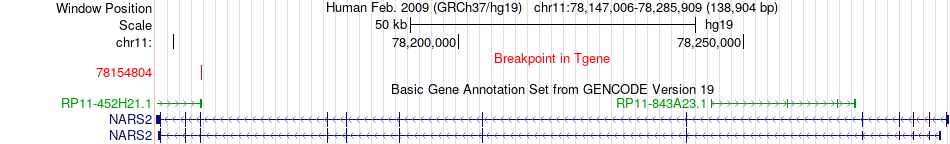 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-29-1778-01A | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| ChimerDB4 | OV | TCGA-29-1778 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
Top |
Fusion Gene ORF analysis for CAP1-NARS2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000340450 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000340450 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372792 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372792 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372797 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372797 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372798 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372798 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372802 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372802 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372805 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| In-frame | ENST00000372805 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| intron-3CDS | ENST00000479759 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
| intron-3CDS | ENST00000479759 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000372797 | CAP1 | chr1 | 40525842 | + | ENST00000281038 | NARS2 | chr11 | 78154804 | - | 1756 | 777 | 534 | 1046 | 170 |
| ENST00000372797 | CAP1 | chr1 | 40525842 | + | ENST00000528850 | NARS2 | chr11 | 78154804 | - | 1494 | 777 | 534 | 1046 | 170 |
| ENST00000372802 | CAP1 | chr1 | 40525842 | + | ENST00000281038 | NARS2 | chr11 | 78154804 | - | 1749 | 770 | 527 | 1039 | 170 |
| ENST00000372802 | CAP1 | chr1 | 40525842 | + | ENST00000528850 | NARS2 | chr11 | 78154804 | - | 1487 | 770 | 527 | 1039 | 170 |
| ENST00000372792 | CAP1 | chr1 | 40525842 | + | ENST00000281038 | NARS2 | chr11 | 78154804 | - | 1261 | 282 | 48 | 551 | 167 |
| ENST00000372792 | CAP1 | chr1 | 40525842 | + | ENST00000528850 | NARS2 | chr11 | 78154804 | - | 999 | 282 | 48 | 551 | 167 |
| ENST00000372798 | CAP1 | chr1 | 40525842 | + | ENST00000281038 | NARS2 | chr11 | 78154804 | - | 1255 | 276 | 45 | 545 | 166 |
| ENST00000372798 | CAP1 | chr1 | 40525842 | + | ENST00000528850 | NARS2 | chr11 | 78154804 | - | 993 | 276 | 45 | 545 | 166 |
| ENST00000340450 | CAP1 | chr1 | 40525842 | + | ENST00000281038 | NARS2 | chr11 | 78154804 | - | 1251 | 272 | 44 | 541 | 165 |
| ENST00000340450 | CAP1 | chr1 | 40525842 | + | ENST00000528850 | NARS2 | chr11 | 78154804 | - | 989 | 272 | 44 | 541 | 165 |
| ENST00000372805 | CAP1 | chr1 | 40525842 | + | ENST00000281038 | NARS2 | chr11 | 78154804 | - | 1246 | 267 | 36 | 536 | 166 |
| ENST00000372805 | CAP1 | chr1 | 40525842 | + | ENST00000528850 | NARS2 | chr11 | 78154804 | - | 984 | 267 | 36 | 536 | 166 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000372797 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.004145061 | 0.9958549 |
| ENST00000372797 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.004834065 | 0.99516594 |
| ENST00000372802 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.004712747 | 0.9952872 |
| ENST00000372802 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.006481415 | 0.99351853 |
| ENST00000372792 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.003903204 | 0.9960968 |
| ENST00000372792 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.004965011 | 0.99503505 |
| ENST00000372798 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.003722312 | 0.99627763 |
| ENST00000372798 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.005639523 | 0.9943605 |
| ENST00000340450 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.003599255 | 0.9964007 |
| ENST00000340450 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.005284438 | 0.9947155 |
| ENST00000372805 | ENST00000281038 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.004300237 | 0.99569976 |
| ENST00000372805 | ENST00000528850 | CAP1 | chr1 | 40525842 | + | NARS2 | chr11 | 78154804 | - | 0.005504046 | 0.9944959 |
Top |
Fusion Genomic Features for CAP1-NARS2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
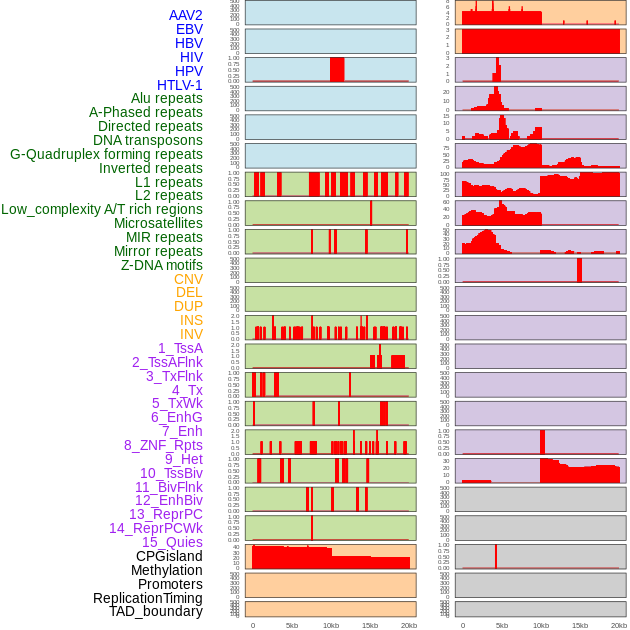 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CAP1-NARS2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:40525842/chr11:78154804) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CAP1 | NARS2 |
| FUNCTION: Directly regulates filament dynamics and has been implicated in a number of complex developmental and morphological processes, including mRNA localization and the establishment of cell polarity. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000340450 | + | 3 | 13 | 218_256 | 71 | 475.0 | Compositional bias | Note=Ala/Pro/Ser-rich |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000340450 | + | 3 | 13 | 230_241 | 71 | 475.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372792 | + | 3 | 13 | 218_256 | 72 | 476.0 | Compositional bias | Note=Ala/Pro/Ser-rich |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372792 | + | 3 | 13 | 230_241 | 72 | 476.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372797 | + | 3 | 13 | 218_256 | 72 | 476.0 | Compositional bias | Note=Ala/Pro/Ser-rich |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372797 | + | 3 | 13 | 230_241 | 72 | 476.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372798 | + | 3 | 13 | 218_256 | 71 | 475.0 | Compositional bias | Note=Ala/Pro/Ser-rich |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372798 | + | 3 | 13 | 230_241 | 71 | 475.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372802 | + | 3 | 13 | 218_256 | 71 | 475.0 | Compositional bias | Note=Ala/Pro/Ser-rich |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372802 | + | 3 | 13 | 230_241 | 71 | 475.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372805 | + | 3 | 13 | 218_256 | 72 | 476.0 | Compositional bias | Note=Ala/Pro/Ser-rich |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372805 | + | 3 | 13 | 230_241 | 72 | 476.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000340450 | + | 3 | 13 | 319_453 | 71 | 475.0 | Domain | C-CAP/cofactor C-like |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372792 | + | 3 | 13 | 319_453 | 72 | 476.0 | Domain | C-CAP/cofactor C-like |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372797 | + | 3 | 13 | 319_453 | 72 | 476.0 | Domain | C-CAP/cofactor C-like |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372798 | + | 3 | 13 | 319_453 | 71 | 475.0 | Domain | C-CAP/cofactor C-like |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372802 | + | 3 | 13 | 319_453 | 71 | 475.0 | Domain | C-CAP/cofactor C-like |
| Hgene | CAP1 | chr1:40525842 | chr11:78154804 | ENST00000372805 | + | 3 | 13 | 319_453 | 72 | 476.0 | Domain | C-CAP/cofactor C-like |
Top |
Fusion Gene Sequence for CAP1-NARS2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >12818_12818_1_CAP1-NARS2_CAP1_chr1_40525842_ENST00000340450_NARS2_chr11_78154804_ENST00000281038_length(transcript)=1251nt_BP=272nt GAAGCGGAGAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGAT TGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGGAGCAGCTC CATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAAC ATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGC GCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGG GATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTAT AGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAG AAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTAT TTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGA ATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAAT TTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTTATCTCATTTT CAGTACTTCATTAAACATTGCAGAAAAAAATATTCCTTAAGGTCTTAATTGATTTAAAGAAGTAGCTATTCTGAATTGAAATCTCCTTTC ATTGAACTGGATGAAAAAATCATGTTTAATAACTGTTGCTTTTCAATTTTCAAAGCTGTTGAGATATTACATTAAGTATTTCAACTCTTT >12818_12818_1_CAP1-NARS2_CAP1_chr1_40525842_ENST00000340450_NARS2_chr11_78154804_ENST00000281038_length(amino acids)=165AA_BP=76 MRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVGEL -------------------------------------------------------------- >12818_12818_2_CAP1-NARS2_CAP1_chr1_40525842_ENST00000340450_NARS2_chr11_78154804_ENST00000528850_length(transcript)=989nt_BP=272nt GAAGCGGAGAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGAT TGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGGAGCAGCTC CATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAAC ATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGC GCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGG GATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTAT AGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAG AAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTAT TTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGA ATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAAT >12818_12818_2_CAP1-NARS2_CAP1_chr1_40525842_ENST00000340450_NARS2_chr11_78154804_ENST00000528850_length(amino acids)=165AA_BP=76 MRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVGEL -------------------------------------------------------------- >12818_12818_3_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372792_NARS2_chr11_78154804_ENST00000281038_length(transcript)=1261nt_BP=282nt GCCGGAAGCGGAGAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGCAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTA GAAAGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGCA GGAGCAGCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGAC GTGCAGAAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTC TTAGAGGAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGT TTTGGGATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCA TGCCTTTTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTT TGGATTTTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAG TTGAGGGTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATG AAGAAATTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTC CCTGTGTAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTT ATCTCATTTTCAGTACTTCATTAAACATTGCAGAAAAAAATATTCCTTAAGGTCTTAATTGATTTAAAGAAGTAGCTATTCTGAATTGAA ATCTCCTTTCATTGAACTGGATGAAAAAATCATGTTTAATAACTGTTGCTTTTCAATTTTCAAAGCTGTTGAGATATTACATTAAGTATT TCAACTCTTTAATCACTGTTGTTATAATTTGTTTATATTTGATGTTTATAATTTGTCTAATAAAATAGATTTTTTTAATAGACACCTATG >12818_12818_3_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372792_NARS2_chr11_78154804_ENST00000281038_length(amino acids)=167AA_BP=78 MSRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKAGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVG -------------------------------------------------------------- >12818_12818_4_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372792_NARS2_chr11_78154804_ENST00000528850_length(transcript)=999nt_BP=282nt GCCGGAAGCGGAGAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGCAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTA GAAAGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGCA GGAGCAGCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGAC GTGCAGAAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTC TTAGAGGAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGT TTTGGGATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCA TGCCTTTTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTT TGGATTTTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAG TTGAGGGTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATG AAGAAATTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTC CCTGTGTAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTT >12818_12818_4_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372792_NARS2_chr11_78154804_ENST00000528850_length(amino acids)=167AA_BP=78 MSRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKAGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVG -------------------------------------------------------------- >12818_12818_5_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372797_NARS2_chr11_78154804_ENST00000281038_length(transcript)=1756nt_BP=777nt GCGGAGCGTGGCCTCGATCACCACCGAGAGCTGACGGCCGCCCGGATTCCCGCCCTCAGAGAATCCTGGCCCCCAGTCGTTCCAAGACCC TTTACGGACTGCAGCTTAAAGGAACCGGCCTCTGCCATTGAGACCAAACTGACAGAGGAACTGGTCACTCTTCTGTTCTTTCCAGAGAGG AGCTTAGCCAGGGACTTCTGCCCTTCCTGGGCCCGGGCCTCGGGGGCGATTCCGGGTAGGTAAAGCGGAACGAGGGCGTGGCCTCTCTTG TGAGCCCGCCCTCCCCGTGTCGCTCCCGCCTTGAGGGCGGGGCTCTTTCCCAAGTGCCCTCTGTGAGAGCCGGGCCGCTCTCTCCGGGGC GTGGCGAAGAGGGGCGGAGTCACGAGCGGGGCGGTGAGACTTCCTGCCCAGTCGCGGGCCAGCCTAGCGCTTCAGCCGGCGGCTCATTTC CGGTGGGGGCGCCGCGCCCAGTGAGGGCCCGGAAGTGGGTCGCGCGGAGATTGCTGGGCGGTTCTTGCCGGAAGCGGAGAGCGGCTGATC GCAGTCCGGAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCAT ACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGCAGGAGCAGCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGT CCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTT GGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAA TGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTT GACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCAT GGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATA AGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTAC TTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAG AATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAA TGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTTATCTCATTTTCAGTACTTCATTAAACATTGCAGAAAAAAATATTC CTTAAGGTCTTAATTGATTTAAAGAAGTAGCTATTCTGAATTGAAATCTCCTTTCATTGAACTGGATGAAAAAATCATGTTTAATAACTG TTGCTTTTCAATTTTCAAAGCTGTTGAGATATTACATTAAGTATTTCAACTCTTTAATCACTGTTGTTATAATTTGTTTATATTTGATGT >12818_12818_5_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372797_NARS2_chr11_78154804_ENST00000281038_length(amino acids)=170AA_BP=81 MIAVRRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKAGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVP -------------------------------------------------------------- >12818_12818_6_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372797_NARS2_chr11_78154804_ENST00000528850_length(transcript)=1494nt_BP=777nt GCGGAGCGTGGCCTCGATCACCACCGAGAGCTGACGGCCGCCCGGATTCCCGCCCTCAGAGAATCCTGGCCCCCAGTCGTTCCAAGACCC TTTACGGACTGCAGCTTAAAGGAACCGGCCTCTGCCATTGAGACCAAACTGACAGAGGAACTGGTCACTCTTCTGTTCTTTCCAGAGAGG AGCTTAGCCAGGGACTTCTGCCCTTCCTGGGCCCGGGCCTCGGGGGCGATTCCGGGTAGGTAAAGCGGAACGAGGGCGTGGCCTCTCTTG TGAGCCCGCCCTCCCCGTGTCGCTCCCGCCTTGAGGGCGGGGCTCTTTCCCAAGTGCCCTCTGTGAGAGCCGGGCCGCTCTCTCCGGGGC GTGGCGAAGAGGGGCGGAGTCACGAGCGGGGCGGTGAGACTTCCTGCCCAGTCGCGGGCCAGCCTAGCGCTTCAGCCGGCGGCTCATTTC CGGTGGGGGCGCCGCGCCCAGTGAGGGCCCGGAAGTGGGTCGCGCGGAGATTGCTGGGCGGTTCTTGCCGGAAGCGGAGAGCGGCTGATC GCAGTCCGGAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCAT ACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGCAGGAGCAGCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGT CCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTT GGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAA TGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTT GACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCAT GGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATA AGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTAC TTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAG AATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAA >12818_12818_6_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372797_NARS2_chr11_78154804_ENST00000528850_length(amino acids)=170AA_BP=81 MIAVRRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKAGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVP -------------------------------------------------------------- >12818_12818_7_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372798_NARS2_chr11_78154804_ENST00000281038_length(transcript)=1255nt_BP=276nt GGAAGCGGAGAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGCAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAA AGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGGAGCA GCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAG AAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAG GAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGG ATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTT TTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATT TTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGG GTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAA TTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTG TAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTTATCTCA TTTTCAGTACTTCATTAAACATTGCAGAAAAAAATATTCCTTAAGGTCTTAATTGATTTAAAGAAGTAGCTATTCTGAATTGAAATCTCC TTTCATTGAACTGGATGAAAAAATCATGTTTAATAACTGTTGCTTTTCAATTTTCAAAGCTGTTGAGATATTACATTAAGTATTTCAACT >12818_12818_7_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372798_NARS2_chr11_78154804_ENST00000281038_length(amino acids)=166AA_BP=77 MSRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVGE -------------------------------------------------------------- >12818_12818_8_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372798_NARS2_chr11_78154804_ENST00000528850_length(transcript)=993nt_BP=276nt GGAAGCGGAGAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGCAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAA AGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGGAGCA GCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAG AAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAG GAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGG ATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTT TTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATT TTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGG GTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAA TTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTG TAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTTATCTCA >12818_12818_8_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372798_NARS2_chr11_78154804_ENST00000528850_length(amino acids)=166AA_BP=77 MSRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVGE -------------------------------------------------------------- >12818_12818_9_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372802_NARS2_chr11_78154804_ENST00000281038_length(transcript)=1749nt_BP=770nt GTGGCCTCGATCACCACCGAGAGCTGACGGCCGCCCGGATTCCCGCCCTCAGAGAATCCTGGCCCCCAGTCGTTCCAAGACCCTTTACGG ACTGCAGCTTAAAGGAACCGGCCTCTGCCATTGAGACCAAACTGACAGAGGAACTGGTCACTCTTCTGTTCTTTCCAGAGAGGAGCTTAG CCAGGGACTTCTGCCCTTCCTGGGCCCGGGCCTCGGGGGCGATTCCGGGTAGGTAAAGCGGAACGAGGGCGTGGCCTCTCTTGTGAGCCC GCCCTCCCCGTGTCGCTCCCGCCTTGAGGGCGGGGCTCTTTCCCAAGTGCCCTCTGTGAGAGCCGGGCCGCTCTCTCCGGGGCGTGGCGA AGAGGGGCGGAGTCACGAGCGGGGCGGTGAGACTTCCTGCCCAGTCGCGGGCCAGCCTAGCGCTTCAGCCGGCGGCTCATTTCCGGTGGG GGCGCCGCGCCCAGTGAGGGCCCGGAAGTGGGTCGCGCGGAGATTGCTGGGCGGTTCTTGCCGGAAGCGGAGAGCGGCTGATCGCAGTCC GGAGCAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCT CTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGGAGCAGCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGG CAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAAC TCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATC TGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATA TCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAG ACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATC ATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGC AAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGA TAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCA TCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTTATCTCATTTTCAGTACTTCATTAAACATTGCAGAAAAAAATATTCCTTAAGG TCTTAATTGATTTAAAGAAGTAGCTATTCTGAATTGAAATCTCCTTTCATTGAACTGGATGAAAAAATCATGTTTAATAACTGTTGCTTT TCAATTTTCAAAGCTGTTGAGATATTACATTAAGTATTTCAACTCTTTAATCACTGTTGTTATAATTTGTTTATATTTGATGTTTATAAT >12818_12818_9_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372802_NARS2_chr11_78154804_ENST00000281038_length(amino acids)=170AA_BP=81 MIAVRSRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVP -------------------------------------------------------------- >12818_12818_10_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372802_NARS2_chr11_78154804_ENST00000528850_length(transcript)=1487nt_BP=770nt GTGGCCTCGATCACCACCGAGAGCTGACGGCCGCCCGGATTCCCGCCCTCAGAGAATCCTGGCCCCCAGTCGTTCCAAGACCCTTTACGG ACTGCAGCTTAAAGGAACCGGCCTCTGCCATTGAGACCAAACTGACAGAGGAACTGGTCACTCTTCTGTTCTTTCCAGAGAGGAGCTTAG CCAGGGACTTCTGCCCTTCCTGGGCCCGGGCCTCGGGGGCGATTCCGGGTAGGTAAAGCGGAACGAGGGCGTGGCCTCTCTTGTGAGCCC GCCCTCCCCGTGTCGCTCCCGCCTTGAGGGCGGGGCTCTTTCCCAAGTGCCCTCTGTGAGAGCCGGGCCGCTCTCTCCGGGGCGTGGCGA AGAGGGGCGGAGTCACGAGCGGGGCGGTGAGACTTCCTGCCCAGTCGCGGGCCAGCCTAGCGCTTCAGCCGGCGGCTCATTTCCGGTGGG GGCGCCGCGCCCAGTGAGGGCCCGGAAGTGGGTCGCGCGGAGATTGCTGGGCGGTTCTTGCCGGAAGCGGAGAGCGGCTGATCGCAGTCC GGAGCAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGATTGGAGAGGGCAGTGGGCCGCCTGGAGGCAGTATCTCATACCT CTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGGAGCAGCTCCATATGTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGG CAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAACATGTTGCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAAC TCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGCGCTTAGCCAGATCGGGACTTACAGAAGTCTACCAATGGTATC TGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGGGATTTGAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATA TCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTATAGCTGGAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAG ACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAGAAATGCAGATTTCAATATGTAATTGTTGTGCCATAAGATATC ATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTATTTCACGTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGC AAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGAATTAGTCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGA TAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAATTTGTGAGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCA >12818_12818_10_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372802_NARS2_chr11_78154804_ENST00000528850_length(amino acids)=170AA_BP=81 MIAVRSRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVP -------------------------------------------------------------- >12818_12818_11_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372805_NARS2_chr11_78154804_ENST00000281038_length(transcript)=1246nt_BP=267nt GAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGATTGGAGAGG GCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGCAGGAGCAGCTCCATAT GTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAACATGTT GCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGCGCTTA GCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGGGATTT GAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTATAGCTG GAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAGAAATG CAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTATTTCAC GTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGAATTAG TCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAATTTGTG AGAACCATCTAGTAGCTCGAAATAAAATAATGTTGCATCTTTTCTCCCTTGCCATATACTTTGTGATAAATCCTTATCTCATTTTCAGTA CTTCATTAAACATTGCAGAAAAAAATATTCCTTAAGGTCTTAATTGATTTAAAGAAGTAGCTATTCTGAATTGAAATCTCCTTTCATTGA ACTGGATGAAAAAATCATGTTTAATAACTGTTGCTTTTCAATTTTCAAAGCTGTTGAGATATTACATTAAGTATTTCAACTCTTTAATCA >12818_12818_11_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372805_NARS2_chr11_78154804_ENST00000281038_length(amino acids)=166AA_BP=77 MRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKAGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVGE -------------------------------------------------------------- >12818_12818_12_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372805_NARS2_chr11_78154804_ENST00000528850_length(transcript)=984nt_BP=267nt GAGCGGCTGATCGCAGTCCGGAGGTGAGGCGGAACTCTGAGGTGGTCCATTATGGCTGACATGCAAAATCTGGTAGAAAGATTGGAGAGG GCAGTGGGCCGCCTGGAGGCAGTATCTCATACCTCTGACATGCACCGTGGGTATGCAGACAGTCCTTCAAAAGCAGGAGCAGCTCCATAT GTGCAGGCATTTGACTCGCTGCTTGCTGGTCCTGTGGCAGAGTACTTGAAGATCAGTAAAGAGATTGGGGGAGACGTGCAGAAACATGTT GCTGCTGTTGATCTTCTGGTTCCTGGAGTTGGGGAACTCTTTGGAGGAGGCCTCAGAGAAGAACGATACCATTTCTTAGAGGAGCGCTTA GCCAGATCGGGACTTACAGAAGTCTACCAATGGTATCTGGACCTTCGTCGATTTGGATCTGTGCCACATGGAGGTTTTGGGATGGGATTT GAACGCTACCTGCAGTGCATCTTGGGTGTTGACAATATCAAAGATGTTATCCCTTTCCCAAGGTTTCCTCATTCATGCCTTTTATAGCTG GAAGATTGGTTAAGGAAAAGCACCCCCCATGGCAGAGACACTGCACATGATTGTGCATACAGCAGAATGCATGTTTGGATTTTAGAAATG CAGATTTCAATATGTAATTGTTGTGCCATAAGATATCATAGAAAAAATATAAGTGGTTGTGATTTTCTTAGAAAGTTGAGGGTATTTCAC GTAAGGATGAGCTCCCGCAAGAAGAGGTACTTATAGCAAGGGGACTCTCAAATCCATTACCTCAATTAAGAAATGAAGAAATTGAATTAG TCTCAAAGTTTCTTTTAAACTCTAAAACAGAATGAGATAATGTATTTTACGTTGTCTATAATCATTAAATCACTCCCTGTGTAATTTGTG >12818_12818_12_CAP1-NARS2_CAP1_chr1_40525842_ENST00000372805_NARS2_chr11_78154804_ENST00000528850_length(amino acids)=166AA_BP=77 MRWSIMADMQNLVERLERAVGRLEAVSHTSDMHRGYADSPSKAGAAPYVQAFDSLLAGPVAEYLKISKEIGGDVQKHVAAVDLLVPGVGE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CAP1-NARS2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CAP1-NARS2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CAP1-NARS2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
