
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACER2-C8orf37 (FusionGDB2 ID:1283) |
Fusion Gene Summary for ACER2-C8orf37 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACER2-C8orf37 | Fusion gene ID: 1283 | Hgene | Tgene | Gene symbol | ACER2 | C8orf37 | Gene ID | 340485 | 157657 |
| Gene name | alkaline ceramidase 2 | chromosome 8 open reading frame 37 | |
| Synonyms | ALKCDase2|ASAH3L | BBS21|CORD16|FAP418|MOT25|RP64|smalltalk | |
| Cytomap | 9p22.1 | 8q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | alkaline ceramidase 2alkCDase 2alkaline CDase 2ceramide hydrolasehaCER2 | protein C8orf37Bardet-Biedl syndrome 21 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q5QJU3 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000340967, ENST00000380376, | ENST00000286688, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 1 X 1 X 1=1 |
| # samples | 3 | 1 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: ACER2 [Title/Abstract] AND C8orf37 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACER2(19435082)-C8orf37(96276002), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACER2 | GO:0001953 | negative regulation of cell-matrix adhesion | 18945876 |
| Hgene | ACER2 | GO:0006974 | cellular response to DNA damage stimulus | 26943039 |
| Hgene | ACER2 | GO:0008284 | positive regulation of cell proliferation | 16940153 |
| Hgene | ACER2 | GO:0032526 | response to retinoic acid | 18945876 |
| Hgene | ACER2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 18945876 |
| Hgene | ACER2 | GO:0046512 | sphingosine biosynthetic process | 16940153|20089856|20628055 |
 Fusion gene breakpoints across ACER2 (5'-gene) Fusion gene breakpoints across ACER2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
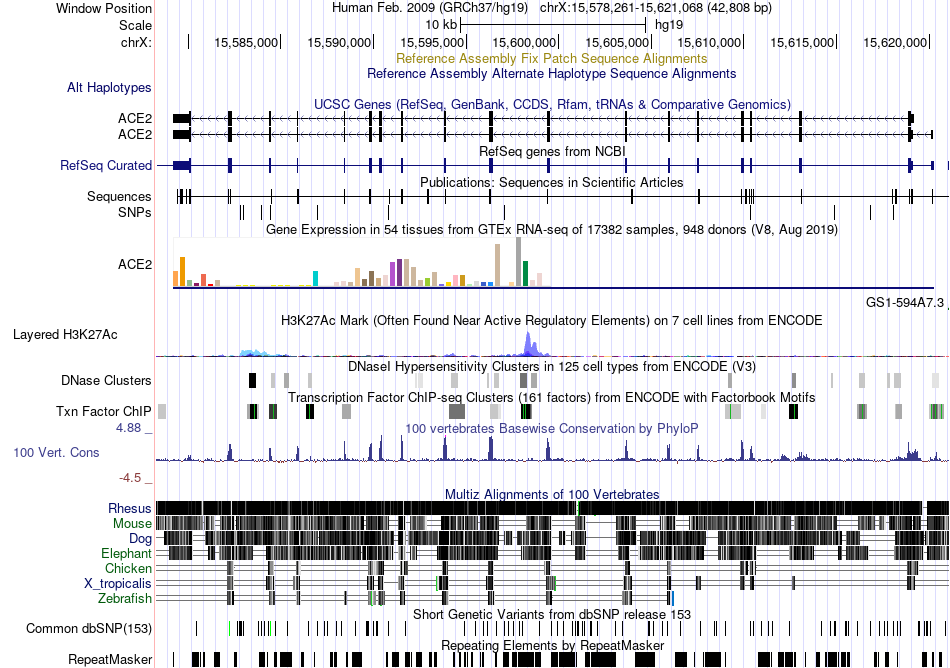 |
 Fusion gene breakpoints across C8orf37 (3'-gene) Fusion gene breakpoints across C8orf37 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
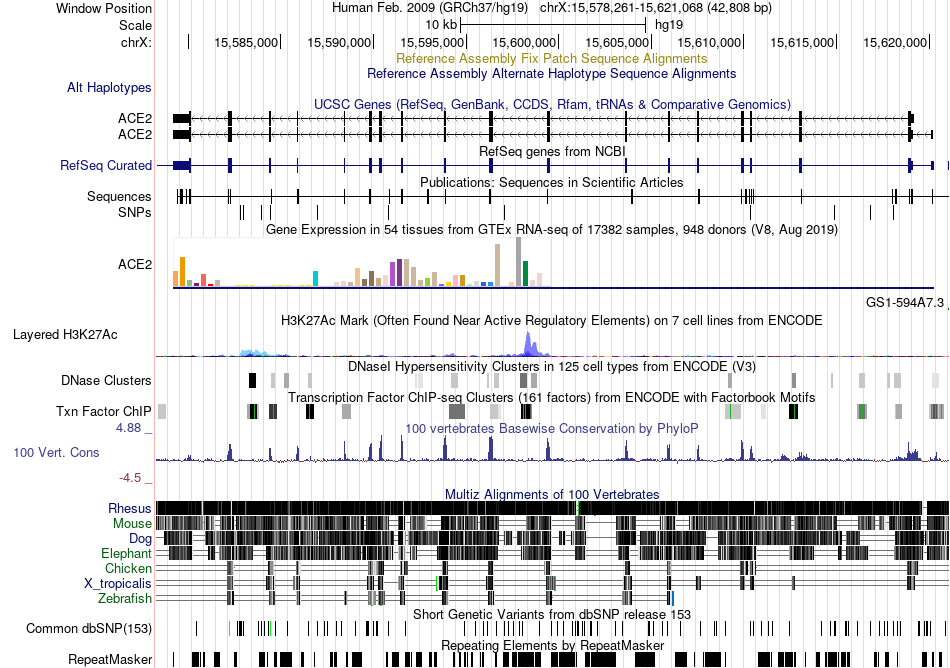 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-A7NF-01A | ACER2 | chr9 | 19435082 | - | C8orf37 | chr8 | 96276002 | - |
| ChimerDB4 | PRAD | TCGA-EJ-A7NF-01A | ACER2 | chr9 | 19435082 | + | C8orf37 | chr8 | 96276002 | - |
Top |
Fusion Gene ORF analysis for ACER2-C8orf37 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000340967 | ENST00000286688 | ACER2 | chr9 | 19435082 | + | C8orf37 | chr8 | 96276002 | - |
| In-frame | ENST00000380376 | ENST00000286688 | ACER2 | chr9 | 19435082 | + | C8orf37 | chr8 | 96276002 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000380376 | ACER2 | chr9 | 19435082 | + | ENST00000286688 | C8orf37 | chr8 | 96276002 | - | 3828 | 661 | 77 | 1129 | 350 |
| ENST00000340967 | ACER2 | chr9 | 19435082 | + | ENST00000286688 | C8orf37 | chr8 | 96276002 | - | 3696 | 529 | 2 | 997 | 331 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000380376 | ENST00000286688 | ACER2 | chr9 | 19435082 | + | C8orf37 | chr8 | 96276002 | - | 0.000950006 | 0.99904996 |
| ENST00000340967 | ENST00000286688 | ACER2 | chr9 | 19435082 | + | C8orf37 | chr8 | 96276002 | - | 0.000850993 | 0.99914896 |
Top |
Fusion Genomic Features for ACER2-C8orf37 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
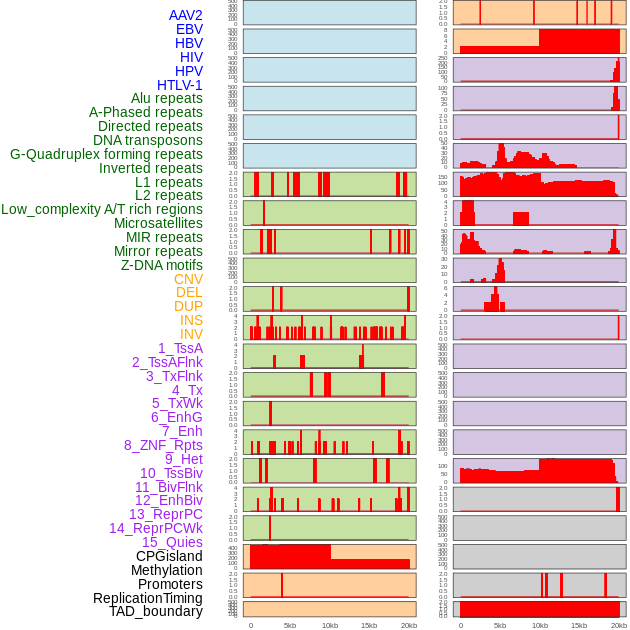 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ACER2-C8orf37 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:19435082/chr8:96276002) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ACER2 | . |
| FUNCTION: Golgi ceramidase that catalyzes the hydrolysis of ceramides into sphingoid bases like sphingosine and free fatty acids at alkaline pH (PubMed:16940153, PubMed:18945876, PubMed:20207939, PubMed:20089856). Ceramides, sphingosine, and its phosphorylated form sphingosine-1-phosphate are bioactive lipids that mediate cellular signaling pathways regulating several biological processes including cell proliferation, apoptosis and differentiation (PubMed:20207939). Has a better catalytic efficiency towards unsaturated long-chain ceramides, including C18:1-, C20:1- and C24:1-ceramides (PubMed:16940153, PubMed:18945876, PubMed:20207939, PubMed:20089856). Saturated long-chain ceramides and unsaturated very long-chain ceramides are also good substrates, whereas saturated very long-chain ceramides and short-chain ceramides are poor substrates (PubMed:20089856). Also hydrolyzes dihydroceramides to produce dihydrosphingosine (PubMed:20207939, PubMed:20628055). It is the ceramidase that controls the levels of circulating sphingosine-1-phosphate and dihydrosphingosine-1-phosphate in plasma through their production by hematopoietic cells (By similarity). Regulates cell proliferation, autophagy and apoptosis by the production of sphingosine and sphingosine-1-phosphate (PubMed:16940153, PubMed:26943039, PubMed:28294157, PubMed:29229990). As part of a p53/TP53-dependent pathway, promotes for instance autophagy and apoptosis in response to DNA damage (PubMed:26943039, PubMed:28294157, PubMed:29229990). Through the production of sphingosine, may also regulate the function of the Golgi complex and regulate the glycosylation of proteins (PubMed:18945876). {ECO:0000250|UniProtKB:Q8VD53, ECO:0000269|PubMed:16940153, ECO:0000269|PubMed:18945876, ECO:0000269|PubMed:20089856, ECO:0000269|PubMed:20207939, ECO:0000269|PubMed:20628055, ECO:0000269|PubMed:26943039, ECO:0000269|PubMed:28294157, ECO:0000269|PubMed:29229990, ECO:0000303|PubMed:20207939}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 1_13 | 167 | 276.0 | Region | Required for proper localization to the Golgi apparatus |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 1_13 | 118 | 141.0 | Region | Required for proper localization to the Golgi apparatus |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 108_124 | 167 | 276.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 143_143 | 167 | 276.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 1_32 | 167 | 276.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 54_62 | 167 | 276.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 84_86 | 167 | 276.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 1_32 | 118 | 141.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 54_62 | 118 | 141.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 84_86 | 118 | 141.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 125_142 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 144_164 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 33_53 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 63_83 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 87_107 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 33_53 | 118 | 141.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 63_83 | 118 | 141.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 87_107 | 118 | 141.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 165_173 | 167 | 276.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 195_211 | 167 | 276.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 233_275 | 167 | 276.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 108_124 | 118 | 141.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 143_143 | 118 | 141.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 165_173 | 118 | 141.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 195_211 | 118 | 141.0 | Topological domain | Lumenal |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 233_275 | 118 | 141.0 | Topological domain | Cytoplasmic |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 174_194 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000340967 | + | 4 | 6 | 212_232 | 167 | 276.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 125_142 | 118 | 141.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 144_164 | 118 | 141.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 174_194 | 118 | 141.0 | Transmembrane | Helical |
| Hgene | ACER2 | chr9:19435082 | chr8:96276002 | ENST00000380376 | + | 4 | 5 | 212_232 | 118 | 141.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ACER2-C8orf37 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1283_1283_1_ACER2-C8orf37_ACER2_chr9_19435082_ENST00000340967_C8orf37_chr8_96276002_ENST00000286688_length(transcript)=3696nt_BP=529nt AGCTGCTCCAATGCCCCGGAGTGGCCATGGGCGCCCCGCACTGGTGGGACCAGCTGCAGGCTGGTAGCTCGGAGGTGGACTGGTGCGAGG ACAACTACACCATCGTGCCTGCTATCGCCGAGTTCTACAACACGATCAGCAATGTCTTATTTTTCATTTTACCGCCCATCTGCATGTGCT TGTTTCGTCAGTATGCAACATGCTTCAACAGTGGCATCTACTTAATCTGGACTCTTTTGGTTGTAGTGGGAATTGGATCCGTCTACTTCC ATGCAACCCTTAGTTTCTTGGGTCAGATGCTTGATGAACTTGCAGTCCTTTGGGTTCTGATGTGTGCTTTGGCCATGTGGTTCCCCAGAA GGTATCTACCAAAGATCTTTCGGAATGACCGGGGTAGGTTCAAGGTGGTGGTCAGTGTCCTGTCTGCGGTTACGACGTGCCTGGCATTTG TCAAGCCTGCCATCAACAACATCTCTCTGATGACCCTGGGAGTTCCTTGCACTGCACTGCTCATCGCAGAGCTAAAGAGATCAACAGAAA CATTTAAAAAAGAAGATGATCTTGACAGTCTTATTAATGAAATACTTGAAGAGCCCAACTTGGACAAAAAACCCTCTAAATTAAAATCTA AATCTTCAGGTAACACATCTGTCAGAGCTTCCATTGAAGGCCTTGGTAAAAGTTGCAGTCCGGTGTACCTTGGTGGAAGCTCTATTCCAT GTGGGATTGGAACAAATATTTCATGGAGAGCATGTGACCATCTGCGTTGTATAGCCTGTGATTTCTTGGTAGTCAGCTATGATGACTATA TGTGGGACAAATCGTGTGATTATCTGTTTTTCAGGAACAACATGCCAGAATTTCACAAATTAAAAGCAAAGTTGATAAAGAAGAAAGGAA CACGGGCATATGCCTGCCAGTGTAGCTGGAGAACTATTGAAGAAGTGACTGACCTTCAGACAGATCATCAGCTTCGCTGGGTTTGTGGTA AACATTAAGAATGCAGACTGCACATTCGGACAGATGCATCCATGAGAGTGGTCTCCATGAATCATCATCATGTTTAGACAATAGTCCCTA GCAATACCATTCCCTGTGGGAAAAGGCAGTGAATTTTATTGATACAACTACATACATTTTTGAAGACCAAATGGACTAAGGAAGATCATG TATTATAACACTTTGGAATTATTCCTGATACATGGATGTTTCACTTAGTAACAGGCATCTACATTCTTTACTAATCTGTCTTCTGTTTAA TAGAGGTTTGATTATGGAGTTCCATACAAATATCTAGTTTTTCATTAGCAACTGCAATATTGATTGCTAATACGTTTAGCAAATCATTCC CCAACTTGTAGCAGTTAAAGTGGATAAAGCCGGCCAGGGCAAAGAGATCATTTAGGGGTGTCTACAACACTTCTTCCAGTCAGATCTTTT GTTGCCAAAGATCATAATTTGGACATGGCAGAAACCAGGTGATCAGTGGCTGACTGAAGGGTGTTCAGAAGTAGTTACAGGTATTTTGCA AAAACACTGCCTCATAGCTGATCTTACCTCATCAACTCAAAGCAATGTTATGCACAATATTAAATACATGTTACCAGTTTTAGCTTATTT TTCCCTCTTTTGTTCATTTAACTATCAAAAGTTATTTTCTATTTTGATTACCCAATAATATATAAAAAGAGTTTGATGCACCCAAAATCA CAAAAATCCTAATACATACAGGTTATTGTTGGATTTTCCAAGTACTTTCTATCCTTTAATGCCTAAAACTCCATCAAGGGAAGTAAACAG CAAGTAATATAATTTTTCAGGCAGCCTGTGGACTTCTCTGGTAGGATCAGGTTGACATATCGTTTATTAGGTGAGTAAGTGAAAAGACAA GTAAAGCATAACTGAACCAACAGGTAACCTGTTGGGCTCCTTCCTGACATCACTGAGATCTGAGGAGCCCTACAATAGCAGCACATTCAG GAAATCAGGTCCCTCAAAATTTCTAGCTCCCAACCCCATTGAGAGATTTTGGGAGCTTTCCAATCTGCCACAAACTCCCTGCTACATATG TACTATATAGTTCAAATGTGTGATACAGAGCACCCTGAAAGGGAATAATTGAAACAAAGTTATAATCTTCTTAGTCTTATGCTAAATTTA TTCCAGCCACCCTACTGAAGGGAGGAAGATCTGATCATTAGAATCCTTTTCCTAGCCATGACACATTTACATACTTGTAAAATTTATTGT AGTAAGTCTAGCATTGCAACAGAATAGATCTTATAATGGCCAATCCTCACACTTTTGTATTAAGGATATTATGACAGACTAAGCATGGTA GCTCACGCCTGTAATCCCAGCACTTTGGGAGGCTGAGGCAGGCAGATCAGCTGAGGTCAGGAGTTCAAGACCAGCCTGGCCAACATGATA AAACACCATCTCTACTAAAAATACAAAAATTAGCTGGGCGCGGTGGTGCATGCCTGTAATCCCAACTACTCGGGAGGCCGAGGCAGGAAA ATTTATTGAACCTGGGAGGCAGAGGTTGTAGTGAGCCAAGGTCGCGCCACTGCGCTCCAGCCTGGACAACACAGTGAGACTCTGTCTCAA AAAAAAAAAAAAGACATTATGACAGATTTTATCTTTTAATAAAAATGTATTATATCCCTAAAGTTATATCTATATGTAATTGAGTCAGAA ACCTAGGGGGATGTCGATTGAAGTCGACTGCTGGAAAAAAATAGCAGAGCAACAAGATCCACACATTTAGAACCTCATAGAATTTGACAC ATTATTCATAATGGTGCAATTCTTTCTTTTTTATTCCAGTAAGCAAATTTACTAAATCACTAATTTGAAAAAAAAAATTTTTTTTTTGAG ATGGAGTGTCACTCTGTTGCCGGGCTGGAGTGCAGTGGCACGATCTCTGCTCACTGCAACCTCCGCCTCGCAGCTTCAAGTGATTGTCCT GCCTCAGCCTCCCCAGTAGCTGGGATTACAGGCACACACCACCACGCCCAGCTAATTTTTGTATTTTTAATAGAGACAGGGTTTCACCAT GTTGGCCAGAATGATGTCGATCTCCTGAACTCGTGATCCGCCCGCCTCAGCCTCCCAAAGTAGCCTACATTGCCTCTACTTTGAAGGGAC AAAGTTGTCCTGGTTTTTTTTTCCCTTTTATTATTTCATTATAACTATCCTAAAACTTACATACATTGCCTATGTTTGGCATTCCATAAC ATAATAATTATAATAATCCCCCAATGTCAGAAATGTATGTGTGAGTTCCCATGTTTATTGGCAATTTATAAGCTTTTTAATATTGTACTA TTTTAAACTAAAATGTAGAAAGAATATTATGTTTATATACAGTCAGATAAATTATTAACTGTTGTAATTTAAGCTGAAAATGAAACTATT TTAGTATCAGTATAGAAGAAAAGTATAGTTTCTAATCATCTATAATTTTAAAATGCAAAGCCTATAAAACTTTCAAGATGTTTTGCCTGT TTGTTTTTAAGATTTATAATAATTTTCTATCTTTAGCAAGTAGCTATTTTTAAGCCAGGTAACTTCTTTATTTGAACTGTATTATACTCT CAGACCCTCATCCTGCCTAGAAATGAATAAGTAATAAATAATTACTCTGTATTATTTACTTCTTTTATTTTATAGTAAAATATTCATTTG >1283_1283_1_ACER2-C8orf37_ACER2_chr9_19435082_ENST00000340967_C8orf37_chr8_96276002_ENST00000286688_length(amino acids)=331AA_BP=173 LLQCPGVAMGAPHWWDQLQAGSSEVDWCEDNYTIVPAIAEFYNTISNVLFFILPPICMCLFRQYATCFNSGIYLIWTLLVVVGIGSVYFH ATLSFLGQMLDELAVLWVLMCALAMWFPRRYLPKIFRNDRGRFKVVVSVLSAVTTCLAFVKPAINNISLMTLGVPCTALLIAELKRSTET FKKEDDLDSLINEILEEPNLDKKPSKLKSKSSGNTSVRASIEGLGKSCSPVYLGGSSIPCGIGTNISWRACDHLRCIACDFLVVSYDDYM -------------------------------------------------------------- >1283_1283_2_ACER2-C8orf37_ACER2_chr9_19435082_ENST00000380376_C8orf37_chr8_96276002_ENST00000286688_length(transcript)=3828nt_BP=661nt GCGGAGCGCGGGAGGCCAGTTGGGAGGCGCACATCCGGCGGTTACCCGGTGCTTCATAAAGCCGCTTTCGCCGCTGGCTGTCGCCGCGTT TTGCCTCCGCAGCAGCTCTGGGCTCTTCTCAGCTGCGCGAGCAGCTGCTCCAATGCCCCGGAGTGGCCATGGGCGCCCCGCACTGGTGGG ACCAGCTGCAGGCTGGTAGCTCGGAGGTGGACTGGTGCGAGGACAACTACACCATCGTGCCTGCTATCGCCGAGTTCTACAACACGATCA GCAATGTCTTATTTTTCATTTTACCGCCCATCTGCATGTGCTTGTTTCGTCAGTATGCAACATGCTTCAACAGTGGCATCTACTTAATCT GGACTCTTTTGGTTGTAGTGGGAATTGGATCCGTCTACTTCCATGCAACCCTTAGTTTCTTGGGTCAGATGCTTGATGAACTTGCAGTCC TTTGGGTTCTGATGTGTGCTTTGGCCATGTGGTTCCCCAGAAGGTATCTACCAAAGATCTTTCGGAATGACCGGGGTAGGTTCAAGGTGG TGGTCAGTGTCCTGTCTGCGGTTACGACGTGCCTGGCATTTGTCAAGCCTGCCATCAACAACATCTCTCTGATGACCCTGGGAGTTCCTT GCACTGCACTGCTCATCGCAGAGCTAAAGAGATCAACAGAAACATTTAAAAAAGAAGATGATCTTGACAGTCTTATTAATGAAATACTTG AAGAGCCCAACTTGGACAAAAAACCCTCTAAATTAAAATCTAAATCTTCAGGTAACACATCTGTCAGAGCTTCCATTGAAGGCCTTGGTA AAAGTTGCAGTCCGGTGTACCTTGGTGGAAGCTCTATTCCATGTGGGATTGGAACAAATATTTCATGGAGAGCATGTGACCATCTGCGTT GTATAGCCTGTGATTTCTTGGTAGTCAGCTATGATGACTATATGTGGGACAAATCGTGTGATTATCTGTTTTTCAGGAACAACATGCCAG AATTTCACAAATTAAAAGCAAAGTTGATAAAGAAGAAAGGAACACGGGCATATGCCTGCCAGTGTAGCTGGAGAACTATTGAAGAAGTGA CTGACCTTCAGACAGATCATCAGCTTCGCTGGGTTTGTGGTAAACATTAAGAATGCAGACTGCACATTCGGACAGATGCATCCATGAGAG TGGTCTCCATGAATCATCATCATGTTTAGACAATAGTCCCTAGCAATACCATTCCCTGTGGGAAAAGGCAGTGAATTTTATTGATACAAC TACATACATTTTTGAAGACCAAATGGACTAAGGAAGATCATGTATTATAACACTTTGGAATTATTCCTGATACATGGATGTTTCACTTAG TAACAGGCATCTACATTCTTTACTAATCTGTCTTCTGTTTAATAGAGGTTTGATTATGGAGTTCCATACAAATATCTAGTTTTTCATTAG CAACTGCAATATTGATTGCTAATACGTTTAGCAAATCATTCCCCAACTTGTAGCAGTTAAAGTGGATAAAGCCGGCCAGGGCAAAGAGAT CATTTAGGGGTGTCTACAACACTTCTTCCAGTCAGATCTTTTGTTGCCAAAGATCATAATTTGGACATGGCAGAAACCAGGTGATCAGTG GCTGACTGAAGGGTGTTCAGAAGTAGTTACAGGTATTTTGCAAAAACACTGCCTCATAGCTGATCTTACCTCATCAACTCAAAGCAATGT TATGCACAATATTAAATACATGTTACCAGTTTTAGCTTATTTTTCCCTCTTTTGTTCATTTAACTATCAAAAGTTATTTTCTATTTTGAT TACCCAATAATATATAAAAAGAGTTTGATGCACCCAAAATCACAAAAATCCTAATACATACAGGTTATTGTTGGATTTTCCAAGTACTTT CTATCCTTTAATGCCTAAAACTCCATCAAGGGAAGTAAACAGCAAGTAATATAATTTTTCAGGCAGCCTGTGGACTTCTCTGGTAGGATC AGGTTGACATATCGTTTATTAGGTGAGTAAGTGAAAAGACAAGTAAAGCATAACTGAACCAACAGGTAACCTGTTGGGCTCCTTCCTGAC ATCACTGAGATCTGAGGAGCCCTACAATAGCAGCACATTCAGGAAATCAGGTCCCTCAAAATTTCTAGCTCCCAACCCCATTGAGAGATT TTGGGAGCTTTCCAATCTGCCACAAACTCCCTGCTACATATGTACTATATAGTTCAAATGTGTGATACAGAGCACCCTGAAAGGGAATAA TTGAAACAAAGTTATAATCTTCTTAGTCTTATGCTAAATTTATTCCAGCCACCCTACTGAAGGGAGGAAGATCTGATCATTAGAATCCTT TTCCTAGCCATGACACATTTACATACTTGTAAAATTTATTGTAGTAAGTCTAGCATTGCAACAGAATAGATCTTATAATGGCCAATCCTC ACACTTTTGTATTAAGGATATTATGACAGACTAAGCATGGTAGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCTGAGGCAGGCAGATC AGCTGAGGTCAGGAGTTCAAGACCAGCCTGGCCAACATGATAAAACACCATCTCTACTAAAAATACAAAAATTAGCTGGGCGCGGTGGTG CATGCCTGTAATCCCAACTACTCGGGAGGCCGAGGCAGGAAAATTTATTGAACCTGGGAGGCAGAGGTTGTAGTGAGCCAAGGTCGCGCC ACTGCGCTCCAGCCTGGACAACACAGTGAGACTCTGTCTCAAAAAAAAAAAAAAGACATTATGACAGATTTTATCTTTTAATAAAAATGT ATTATATCCCTAAAGTTATATCTATATGTAATTGAGTCAGAAACCTAGGGGGATGTCGATTGAAGTCGACTGCTGGAAAAAAATAGCAGA GCAACAAGATCCACACATTTAGAACCTCATAGAATTTGACACATTATTCATAATGGTGCAATTCTTTCTTTTTTATTCCAGTAAGCAAAT TTACTAAATCACTAATTTGAAAAAAAAAATTTTTTTTTTGAGATGGAGTGTCACTCTGTTGCCGGGCTGGAGTGCAGTGGCACGATCTCT GCTCACTGCAACCTCCGCCTCGCAGCTTCAAGTGATTGTCCTGCCTCAGCCTCCCCAGTAGCTGGGATTACAGGCACACACCACCACGCC CAGCTAATTTTTGTATTTTTAATAGAGACAGGGTTTCACCATGTTGGCCAGAATGATGTCGATCTCCTGAACTCGTGATCCGCCCGCCTC AGCCTCCCAAAGTAGCCTACATTGCCTCTACTTTGAAGGGACAAAGTTGTCCTGGTTTTTTTTTCCCTTTTATTATTTCATTATAACTAT CCTAAAACTTACATACATTGCCTATGTTTGGCATTCCATAACATAATAATTATAATAATCCCCCAATGTCAGAAATGTATGTGTGAGTTC CCATGTTTATTGGCAATTTATAAGCTTTTTAATATTGTACTATTTTAAACTAAAATGTAGAAAGAATATTATGTTTATATACAGTCAGAT AAATTATTAACTGTTGTAATTTAAGCTGAAAATGAAACTATTTTAGTATCAGTATAGAAGAAAAGTATAGTTTCTAATCATCTATAATTT TAAAATGCAAAGCCTATAAAACTTTCAAGATGTTTTGCCTGTTTGTTTTTAAGATTTATAATAATTTTCTATCTTTAGCAAGTAGCTATT TTTAAGCCAGGTAACTTCTTTATTTGAACTGTATTATACTCTCAGACCCTCATCCTGCCTAGAAATGAATAAGTAATAAATAATTACTCT >1283_1283_2_ACER2-C8orf37_ACER2_chr9_19435082_ENST00000380376_C8orf37_chr8_96276002_ENST00000286688_length(amino acids)=350AA_BP=192 MSPRFASAAALGSSQLREQLLQCPGVAMGAPHWWDQLQAGSSEVDWCEDNYTIVPAIAEFYNTISNVLFFILPPICMCLFRQYATCFNSG IYLIWTLLVVVGIGSVYFHATLSFLGQMLDELAVLWVLMCALAMWFPRRYLPKIFRNDRGRFKVVVSVLSAVTTCLAFVKPAINNISLMT LGVPCTALLIAELKRSTETFKKEDDLDSLINEILEEPNLDKKPSKLKSKSSGNTSVRASIEGLGKSCSPVYLGGSSIPCGIGTNISWRAC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACER2-C8orf37 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACER2-C8orf37 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ACER2-C8orf37 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
