
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CASK-GLIPR1L1 (FusionGDB2 ID:13195) |
Fusion Gene Summary for CASK-GLIPR1L1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CASK-GLIPR1L1 | Fusion gene ID: 13195 | Hgene | Tgene | Gene symbol | CASK | GLIPR1L1 | Gene ID | 8573 | 256710 |
| Gene name | calcium/calmodulin dependent serine protein kinase | GLIPR1 like 1 | |
| Synonyms | CAGH39|CAMGUK|CMG|FGS4|LIN2|MICPCH|MRXSNA|TNRC8|hCASK | ALKN2972|PRO7434 | |
| Cytomap | Xp11.4 | 12q21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | peripheral plasma membrane protein CASKcalcium/calmodulin-dependent serin protein kinasecalcium/calmodulin-dependent serine protein kinase (MAGUK family)calcium/calmodulin-dependent serine protein kinase membrane-associated guanylate kinaseprotein lin | GLIPR1-like protein 1GLI pathogenesis related 1 like 1 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | O14936 | Q6UWM5 | |
| Ensembl transtripts involved in fusion gene | ENST00000318588, ENST00000361962, ENST00000378154, ENST00000378158, ENST00000378163, ENST00000378166, ENST00000421587, ENST00000442742, ENST00000472704, | ENST00000548623, ENST00000312442, ENST00000378695, | |
| Fusion gene scores | * DoF score | 13 X 12 X 5=780 | 6 X 5 X 4=120 |
| # samples | 14 | 7 | |
| ** MAII score | log2(14/780*10)=-2.47804729680464 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/120*10)=-0.777607578663552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CASK [Title/Abstract] AND GLIPR1L1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CASK(41782183)-GLIPR1L1(75737473), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CASK | GO:0010839 | negative regulation of keratinocyte proliferation | 18664494 |
 Fusion gene breakpoints across CASK (5'-gene) Fusion gene breakpoints across CASK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
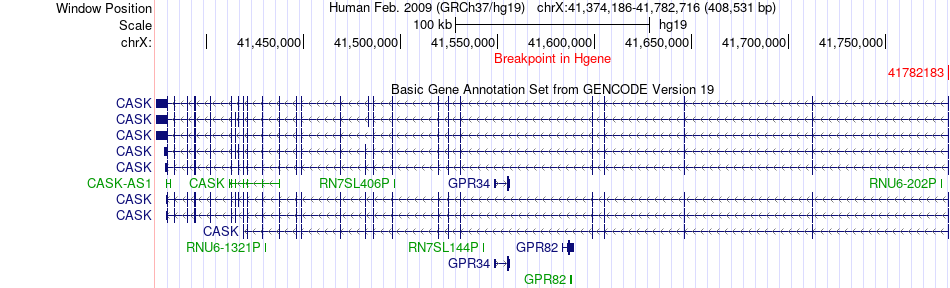 |
 Fusion gene breakpoints across GLIPR1L1 (3'-gene) Fusion gene breakpoints across GLIPR1L1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
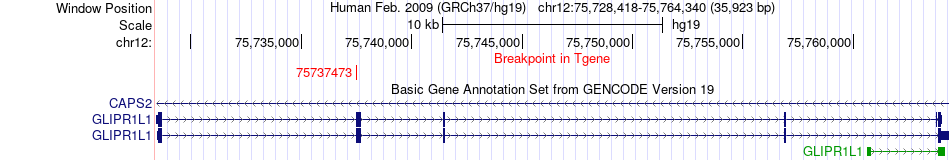 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A14T-01A | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
Top |
Fusion Gene ORF analysis for CASK-GLIPR1L1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000318588 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000361962 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000378154 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000378158 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000378163 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000378166 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000421587 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| 5CDS-intron | ENST00000442742 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000318588 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000318588 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000361962 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000361962 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378154 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378154 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378158 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378158 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378163 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378163 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378166 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000378166 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000421587 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000421587 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000442742 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| In-frame | ENST00000442742 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| intron-3CDS | ENST00000472704 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| intron-3CDS | ENST00000472704 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
| intron-intron | ENST00000472704 | ENST00000548623 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000421587 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 704 | 83 | 107 | 637 | 176 |
| ENST00000421587 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 995 | 83 | 107 | 610 | 167 |
| ENST00000318588 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 726 | 105 | 21 | 659 | 212 |
| ENST00000318588 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 1017 | 105 | 21 | 632 | 203 |
| ENST00000361962 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 726 | 105 | 21 | 659 | 212 |
| ENST00000361962 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 1017 | 105 | 21 | 632 | 203 |
| ENST00000378163 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 1155 | 534 | 297 | 1088 | 263 |
| ENST00000378163 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 1446 | 534 | 297 | 1061 | 254 |
| ENST00000378158 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 1031 | 410 | 173 | 964 | 263 |
| ENST00000378158 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 1322 | 410 | 173 | 937 | 254 |
| ENST00000378166 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 707 | 86 | 2 | 640 | 212 |
| ENST00000378166 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 998 | 86 | 2 | 613 | 203 |
| ENST00000442742 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 699 | 78 | 102 | 632 | 176 |
| ENST00000442742 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 990 | 78 | 102 | 605 | 167 |
| ENST00000378154 | CASK | chrX | 41782183 | - | ENST00000378695 | GLIPR1L1 | chr12 | 75737473 | + | 1155 | 534 | 297 | 1088 | 263 |
| ENST00000378154 | CASK | chrX | 41782183 | - | ENST00000312442 | GLIPR1L1 | chr12 | 75737473 | + | 1446 | 534 | 297 | 1061 | 254 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000421587 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.01178129 | 0.98821867 |
| ENST00000421587 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.01217087 | 0.9878291 |
| ENST00000318588 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.012155275 | 0.98784465 |
| ENST00000318588 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.009748589 | 0.9902514 |
| ENST00000361962 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.012155275 | 0.98784465 |
| ENST00000361962 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.009748589 | 0.9902514 |
| ENST00000378163 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.032874856 | 0.9671252 |
| ENST00000378163 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.022611013 | 0.977389 |
| ENST00000378158 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.032294154 | 0.9677058 |
| ENST00000378158 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.015988277 | 0.98401165 |
| ENST00000378166 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.012768789 | 0.9872312 |
| ENST00000378166 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.012249407 | 0.9877506 |
| ENST00000442742 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.011960974 | 0.98803896 |
| ENST00000442742 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.01156156 | 0.9884384 |
| ENST00000378154 | ENST00000378695 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.032874856 | 0.9671252 |
| ENST00000378154 | ENST00000312442 | CASK | chrX | 41782183 | - | GLIPR1L1 | chr12 | 75737473 | + | 0.022611013 | 0.977389 |
Top |
Fusion Genomic Features for CASK-GLIPR1L1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CASK | chrX | 41782182 | - | GLIPR1L1 | chr12 | 75737472 | + | 1.10E-05 | 0.99998903 |
| CASK | chrX | 41782182 | - | GLIPR1L1 | chr12 | 75737472 | + | 1.10E-05 | 0.99998903 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
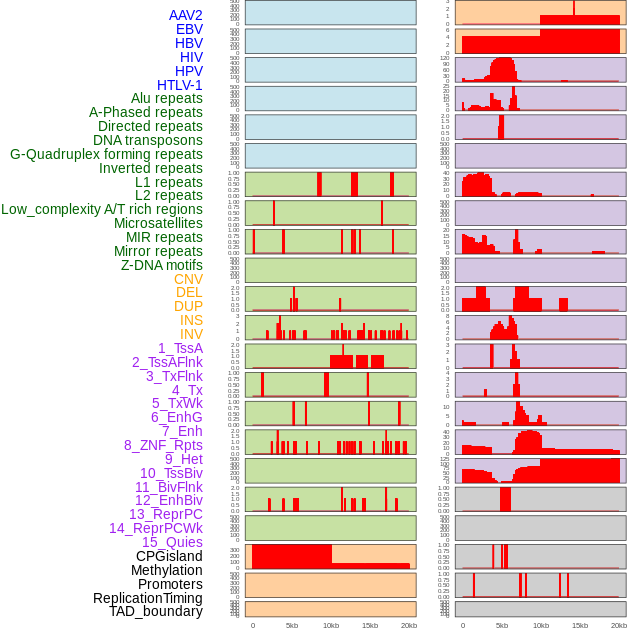 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
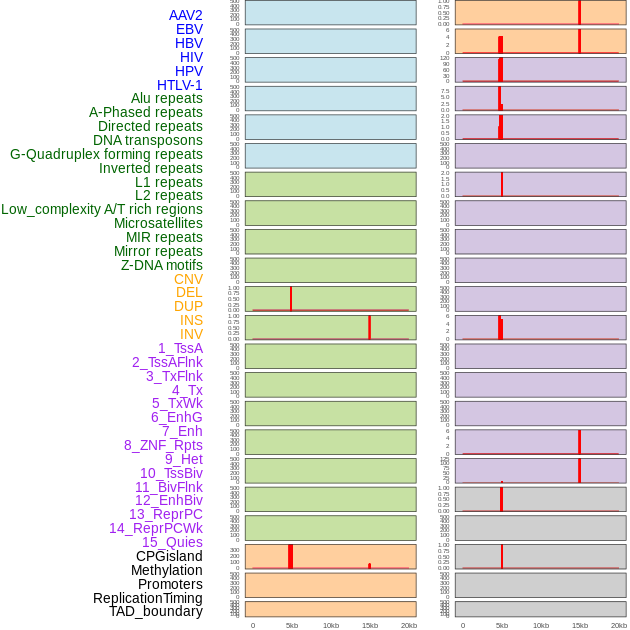 |
Top |
Fusion Protein Features for CASK-GLIPR1L1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:41782183/chr12:75737473) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CASK | GLIPR1L1 |
| FUNCTION: Multidomain scaffolding protein with a role in synaptic transmembrane protein anchoring and ion channel trafficking. Contributes to neural development and regulation of gene expression via interaction with the transcription factor TBR1. Binds to cell-surface proteins, including amyloid precursor protein, neurexins and syndecans. May mediate a link between the extracellular matrix and the actin cytoskeleton via its interaction with syndecan and with the actin/spectrin-binding protein 4.1. Component of the LIN-10-LIN-2-LIN-7 complex, which associates with the motor protein KIF17 to transport vesicles containing N-methyl-D-aspartate (NMDA) receptor subunit NR2B along microtubules (By similarity). {ECO:0000250|UniProtKB:O70589}. | FUNCTION: Plays a role in the binding between sperm and oocytes. Component of epididymosomes, one type of membranous microvesicules which mediate the transfer of lipids and proteins to spermatozoa plasma membrane during epididymal maturation. Also component of the CD9-positive microvesicules found in the cauda region. {ECO:0000250|UniProtKB:Q32LB5, ECO:0000250|UniProtKB:Q9DAG6}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 12_276 | 19 | 910.0 | Domain | Protein kinase |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 343_398 | 19 | 910.0 | Domain | L27 1 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 402_455 | 19 | 910.0 | Domain | L27 2 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 489_564 | 19 | 910.0 | Domain | PDZ |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 612_682 | 19 | 910.0 | Domain | SH3 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 739_911 | 19 | 910.0 | Domain | Guanylate kinase-like |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 12_276 | 19 | 927.0 | Domain | Protein kinase |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 343_398 | 19 | 927.0 | Domain | L27 1 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 402_455 | 19 | 927.0 | Domain | L27 2 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 489_564 | 19 | 927.0 | Domain | PDZ |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 612_682 | 19 | 927.0 | Domain | SH3 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 739_911 | 19 | 927.0 | Domain | Guanylate kinase-like |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 12_276 | 19 | 922.0 | Domain | Protein kinase |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 343_398 | 19 | 922.0 | Domain | L27 1 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 402_455 | 19 | 922.0 | Domain | L27 2 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 489_564 | 19 | 922.0 | Domain | PDZ |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 612_682 | 19 | 922.0 | Domain | SH3 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 739_911 | 19 | 922.0 | Domain | Guanylate kinase-like |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 12_276 | 19 | 898.0 | Domain | Protein kinase |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 343_398 | 19 | 898.0 | Domain | L27 1 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 402_455 | 19 | 898.0 | Domain | L27 2 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 489_564 | 19 | 898.0 | Domain | PDZ |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 612_682 | 19 | 898.0 | Domain | SH3 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 739_911 | 19 | 898.0 | Domain | Guanylate kinase-like |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 12_276 | 19 | 899.0 | Domain | Protein kinase |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 343_398 | 19 | 899.0 | Domain | L27 1 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 402_455 | 19 | 899.0 | Domain | L27 2 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 489_564 | 19 | 899.0 | Domain | PDZ |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 612_682 | 19 | 899.0 | Domain | SH3 |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 739_911 | 19 | 899.0 | Domain | Guanylate kinase-like |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 18_26 | 19 | 910.0 | Nucleotide binding | ATP |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 18_26 | 19 | 927.0 | Nucleotide binding | ATP |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 18_26 | 19 | 922.0 | Nucleotide binding | ATP |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 18_26 | 19 | 898.0 | Nucleotide binding | ATP |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 18_26 | 19 | 899.0 | Nucleotide binding | ATP |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378158 | - | 1 | 26 | 305_315 | 19 | 910.0 | Region | Note=Calmodulin-binding |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378163 | - | 1 | 27 | 305_315 | 19 | 927.0 | Region | Note=Calmodulin-binding |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000378166 | - | 1 | 27 | 305_315 | 19 | 922.0 | Region | Note=Calmodulin-binding |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000421587 | - | 1 | 25 | 305_315 | 19 | 898.0 | Region | Note=Calmodulin-binding |
| Hgene | CASK | chrX:41782183 | chr12:75737473 | ENST00000442742 | - | 1 | 26 | 305_315 | 19 | 899.0 | Region | Note=Calmodulin-binding |
| Tgene | GLIPR1L1 | chrX:41782183 | chr12:75737473 | ENST00000312442 | 0 | 5 | 39_171 | 58 | 234.0 | Domain | SCP | |
| Tgene | GLIPR1L1 | chrX:41782183 | chr12:75737473 | ENST00000378695 | 0 | 6 | 39_171 | 58 | 243.0 | Domain | SCP |
Top |
Fusion Gene Sequence for CASK-GLIPR1L1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >13195_13195_1_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000318588_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=1017nt_BP=105nt CCGTTTTCGAAGCCCTCCACGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTG CGAGGTGATCGGAAAATTTGGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTG GATAAATCATATAAATGCTATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCC ATTACGGCTTGGTATAATGAAACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGG GCCAATTCATTTTATGTCGGTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCT GCAGGAAATTTTGCAAATATGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGC AAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTT TAATGTCATTTATATACAAAAGAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTT TACTGAATCTTCTACACTCTTGCCTGATACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCA GTAGAATAAAGTCTTAAGATTATTTTTTAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGAT TATCATTTTAGTTATAGTACATTTAGAGAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATA >13195_13195_1_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000318588_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=203AA_BP=28 MRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNE TQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNLGGASTAIFVCNYGPAGNFANMPPYVRGESCSLCSKEEKCVKNLCKNPFLKP -------------------------------------------------------------- >13195_13195_2_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000318588_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=726nt_BP=105nt CCGTTTTCGAAGCCCTCCACGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTG CGAGGTGATCGGAAAATTTGGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTG GATAAATCATATAAATGCTATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCC ATTACGGCTTGGTATAATGAAACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGG GCCAATTCATTTTATGTCGGTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCT GCAGGAAATTTTGCAAATATGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGC AGGACTCCACAACTTATTATACCTAACCAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGC TTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAAGAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTA >13195_13195_2_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000318588_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=212AA_BP=28 MRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNE TQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNLGGASTAIFVCNYGPAGNFANMPPYVRGESCSLCSKEEKCVKNLCRTPQLII -------------------------------------------------------------- >13195_13195_3_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000361962_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=1017nt_BP=105nt CCGTTTTCGAAGCCCTCCACGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTG CGAGGTGATCGGAAAATTTGGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTG GATAAATCATATAAATGCTATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCC ATTACGGCTTGGTATAATGAAACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGG GCCAATTCATTTTATGTCGGTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCT GCAGGAAATTTTGCAAATATGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGC AAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTT TAATGTCATTTATATACAAAAGAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTT TACTGAATCTTCTACACTCTTGCCTGATACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCA GTAGAATAAAGTCTTAAGATTATTTTTTAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGAT TATCATTTTAGTTATAGTACATTTAGAGAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATA >13195_13195_3_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000361962_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=203AA_BP=28 MRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNE TQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNLGGASTAIFVCNYGPAGNFANMPPYVRGESCSLCSKEEKCVKNLCKNPFLKP -------------------------------------------------------------- >13195_13195_4_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000361962_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=726nt_BP=105nt CCGTTTTCGAAGCCCTCCACGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTG CGAGGTGATCGGAAAATTTGGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTG GATAAATCATATAAATGCTATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCC ATTACGGCTTGGTATAATGAAACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGG GCCAATTCATTTTATGTCGGTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCT GCAGGAAATTTTGCAAATATGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGC AGGACTCCACAACTTATTATACCTAACCAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGC TTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAAGAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTA >13195_13195_4_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000361962_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=212AA_BP=28 MRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNE TQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNLGGASTAIFVCNYGPAGNFANMPPYVRGESCSLCSKEEKCVKNLCRTPQLII -------------------------------------------------------------- >13195_13195_5_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378154_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=1446nt_BP=534nt GCCGCCGCCACCTCCCTCCGCGGCGATCCTCGCTCCATGGTCCTGCCGCGGCCCCCTCTGTAGCCCGAGCTGCTCCGCGACGCGACGGAG CCGGCCTCGGACAGAGGGAGCGGTCGCGGCCTCGCGGCCCGCGCGGGCTGACGAGCGGAGAGCGCGACGCCGCGCGGTCCCCGAGGATCC GCACGCCGGGCAGCGCGGCCCGAGCAGCGCGAGAAGCAGCGGGCGGGCGGGCGGCGCCGGGCCCGGGCCGCCCTCGCTCCCGCGCCCCGG CCCCGCCGCCCGCGCCGGGGCGGTCGCCTGCAGAGGAAGGCGCCGCAGTCTCTGGGCCTCGCGGCCCCACTCGAGCGTCCTCGGCACCGA GGGCTCGCGGGTCCTGAGGGGAGGAGGCGATCCCGGGGCTGGGCACTGAGCTCTTGGGCGCCCCCACCCCCGTTTTCGAAGCCCTCCACG CTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGG GATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTAT GCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAA ACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGT TGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATG CCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAAAAATCCATTTCTGAAGCCA ACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAA GAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTTTACTGAATCTTCTACACTCTT GCCTGATACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCAGTAGAATAAAGTCTTAAGATT ATTTTTTAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGATTATCATTTTAGTTATAGTACA TTTAGAGAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATACCATCAACATCATTCAGAAAC >13195_13195_5_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378154_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=254AA_BP=79 MQRKAPQSLGLAAPLERPRHRGLAGPEGRRRSRGWALSSWAPPPPFSKPSTLRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAK AWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNL -------------------------------------------------------------- >13195_13195_6_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378154_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=1155nt_BP=534nt GCCGCCGCCACCTCCCTCCGCGGCGATCCTCGCTCCATGGTCCTGCCGCGGCCCCCTCTGTAGCCCGAGCTGCTCCGCGACGCGACGGAG CCGGCCTCGGACAGAGGGAGCGGTCGCGGCCTCGCGGCCCGCGCGGGCTGACGAGCGGAGAGCGCGACGCCGCGCGGTCCCCGAGGATCC GCACGCCGGGCAGCGCGGCCCGAGCAGCGCGAGAAGCAGCGGGCGGGCGGGCGGCGCCGGGCCCGGGCCGCCCTCGCTCCCGCGCCCCGG CCCCGCCGCCCGCGCCGGGGCGGTCGCCTGCAGAGGAAGGCGCCGCAGTCTCTGGGCCTCGCGGCCCCACTCGAGCGTCCTCGGCACCGA GGGCTCGCGGGTCCTGAGGGGAGGAGGCGATCCCGGGGCTGGGCACTGAGCTCTTGGGCGCCCCCACCCCCGTTTTCGAAGCCCTCCACG CTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGG GATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTAT GCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAA ACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGT TGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATG CCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAGGACTCCACAACTTATTATA CCTAACCAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGA >13195_13195_6_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378154_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=263AA_BP=79 MQRKAPQSLGLAAPLERPRHRGLAGPEGRRRSRGWALSSWAPPPPFSKPSTLRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAK AWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNL -------------------------------------------------------------- >13195_13195_7_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378158_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=1322nt_BP=410nt CGGCCCGCGCGGGCTGACGAGCGGAGAGCGCGACGCCGCGCGGTCCCCGAGGATCCGCACGCCGGGCAGCGCGGCCCGAGCAGCGCGAGA AGCAGCGGGCGGGCGGGCGGCGCCGGGCCCGGGCCGCCCTCGCTCCCGCGCCCCGGCCCCGCCGCCCGCGCCGGGGCGGTCGCCTGCAGA GGAAGGCGCCGCAGTCTCTGGGCCTCGCGGCCCCACTCGAGCGTCCTCGGCACCGAGGGCTCGCGGGTCCTGAGGGGAGGAGGCGATCCC GGGGCTGGGCACTGAGCTCTTGGGCGCCCCCACCCCCGTTTTCGAAGCCCTCCACGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGAC GACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGG CAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGT TAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAAACCCAATTTTATGATTTTGATAGTCTATCATGCT CCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAG CTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCT GCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATC CATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAAGAAATTCTCAAATGTTAAAATAAAGGAATAGTTT ATTGCTTAATATAACTTATCATCACTTTGCTTCTTTACTGAATCTTCTACACTCTTGCCTGATACCTAAATTTAATGTTTGTTTTTAACT CAAAAAATGTACTGTAGTATGGAAAATGGATAGCAGTAGAATAAAGTCTTAAGATTATTTTTTAATTACAAATCCATATGTGTATCAAAA GTGTTCCACTCTTTAATTTTCTAATGAGAATGGATTATCATTTTAGTTATAGTACATTTAGAGAACTAAAGACTTTTCACATATCAAATA >13195_13195_7_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378158_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=254AA_BP=79 MQRKAPQSLGLAAPLERPRHRGLAGPEGRRRSRGWALSSWAPPPPFSKPSTLRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAK AWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNL -------------------------------------------------------------- >13195_13195_8_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378158_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=1031nt_BP=410nt CGGCCCGCGCGGGCTGACGAGCGGAGAGCGCGACGCCGCGCGGTCCCCGAGGATCCGCACGCCGGGCAGCGCGGCCCGAGCAGCGCGAGA AGCAGCGGGCGGGCGGGCGGCGCCGGGCCCGGGCCGCCCTCGCTCCCGCGCCCCGGCCCCGCCGCCCGCGCCGGGGCGGTCGCCTGCAGA GGAAGGCGCCGCAGTCTCTGGGCCTCGCGGCCCCACTCGAGCGTCCTCGGCACCGAGGGCTCGCGGGTCCTGAGGGGAGGAGGCGATCCC GGGGCTGGGCACTGAGCTCTTGGGCGCCCCCACCCCCGTTTTCGAAGCCCTCCACGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGAC GACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGG CAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGT TAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAAACCCAATTTTATGATTTTGATAGTCTATCATGCT CCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAG CTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCT GCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAGGACTCCACAACTTATTATACCTAACCAAAATCCATTTCTGAAGCCAACGGGGA GAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAAGAAATTC >13195_13195_8_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378158_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=263AA_BP=79 MQRKAPQSLGLAAPLERPRHRGLAGPEGRRRSRGWALSSWAPPPPFSKPSTLRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAK AWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNL -------------------------------------------------------------- >13195_13195_9_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378163_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=1446nt_BP=534nt GCCGCCGCCACCTCCCTCCGCGGCGATCCTCGCTCCATGGTCCTGCCGCGGCCCCCTCTGTAGCCCGAGCTGCTCCGCGACGCGACGGAG CCGGCCTCGGACAGAGGGAGCGGTCGCGGCCTCGCGGCCCGCGCGGGCTGACGAGCGGAGAGCGCGACGCCGCGCGGTCCCCGAGGATCC GCACGCCGGGCAGCGCGGCCCGAGCAGCGCGAGAAGCAGCGGGCGGGCGGGCGGCGCCGGGCCCGGGCCGCCCTCGCTCCCGCGCCCCGG CCCCGCCGCCCGCGCCGGGGCGGTCGCCTGCAGAGGAAGGCGCCGCAGTCTCTGGGCCTCGCGGCCCCACTCGAGCGTCCTCGGCACCGA GGGCTCGCGGGTCCTGAGGGGAGGAGGCGATCCCGGGGCTGGGCACTGAGCTCTTGGGCGCCCCCACCCCCGTTTTCGAAGCCCTCCACG CTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGG GATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTAT GCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAA ACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGT TGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATG CCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAAAAATCCATTTCTGAAGCCA ACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAA GAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTTTACTGAATCTTCTACACTCTT GCCTGATACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCAGTAGAATAAAGTCTTAAGATT ATTTTTTAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGATTATCATTTTAGTTATAGTACA TTTAGAGAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATACCATCAACATCATTCAGAAAC >13195_13195_9_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378163_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=254AA_BP=79 MQRKAPQSLGLAAPLERPRHRGLAGPEGRRRSRGWALSSWAPPPPFSKPSTLRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAK AWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNL -------------------------------------------------------------- >13195_13195_10_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378163_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=1155nt_BP=534nt GCCGCCGCCACCTCCCTCCGCGGCGATCCTCGCTCCATGGTCCTGCCGCGGCCCCCTCTGTAGCCCGAGCTGCTCCGCGACGCGACGGAG CCGGCCTCGGACAGAGGGAGCGGTCGCGGCCTCGCGGCCCGCGCGGGCTGACGAGCGGAGAGCGCGACGCCGCGCGGTCCCCGAGGATCC GCACGCCGGGCAGCGCGGCCCGAGCAGCGCGAGAAGCAGCGGGCGGGCGGGCGGCGCCGGGCCCGGGCCGCCCTCGCTCCCGCGCCCCGG CCCCGCCGCCCGCGCCGGGGCGGTCGCCTGCAGAGGAAGGCGCCGCAGTCTCTGGGCCTCGCGGCCCCACTCGAGCGTCCTCGGCACCGA GGGCTCGCGGGTCCTGAGGGGAGGAGGCGATCCCGGGGCTGGGCACTGAGCTCTTGGGCGCCCCCACCCCCGTTTTCGAAGCCCTCCACG CTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGG GATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTAT GCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAA ACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGT TGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATG CCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAGGACTCCACAACTTATTATA CCTAACCAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGA >13195_13195_10_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378163_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=263AA_BP=79 MQRKAPQSLGLAAPLERPRHRGLAGPEGRRRSRGWALSSWAPPPPFSKPSTLRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAK AWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNL -------------------------------------------------------------- >13195_13195_11_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378166_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=998nt_BP=86nt CGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTT GGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCT ATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATG AAACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCG GTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATA TGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAAAAATCCATTTCTGAAGC CAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAA AAGAAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTTTACTGAATCTTCTACACTC TTGCCTGATACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCAGTAGAATAAAGTCTTAAGA TTATTTTTTAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGATTATCATTTTAGTTATAGTA CATTTAGAGAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATACCATCAACATCATTCAGAA >13195_13195_11_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378166_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=203AA_BP=28 LRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNE TQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNLGGASTAIFVCNYGPAGNFANMPPYVRGESCSLCSKEEKCVKNLCKNPFLKP -------------------------------------------------------------- >13195_13195_12_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378166_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=707nt_BP=86nt CGCTGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTT GGGATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCT ATGCAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATG AAACCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCG GTTGTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATA TGCCTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAGGACTCCACAACTTATTA TACCTAACCAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGA >13195_13195_12_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000378166_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=212AA_BP=28 LRPLSPPDHGRRRRAVRGCVRAVRGDRKIWDKGLAKMAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNE TQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMCPNLGGASTAIFVCNYGPAGNFANMPPYVRGESCSLCSKEEKCVKNLCRTPQLII -------------------------------------------------------------- >13195_13195_13_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000421587_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=995nt_BP=83nt TGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGGG ATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTATG CAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAAA CCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGTT GTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATGC CTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAAAAATCCATTTCTGAAGCCAA CGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAAG AAATTCTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTTTACTGAATCTTCTACACTCTTG CCTGATACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCAGTAGAATAAAGTCTTAAGATTA TTTTTTAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGATTATCATTTTAGTTATAGTACAT TTAGAGAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATACCATCAACATCATTCAGAAACA >13195_13195_13_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000421587_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=167AA_BP= MAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMC -------------------------------------------------------------- >13195_13195_14_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000421587_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=704nt_BP=83nt TGCGGCCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGGG ATAAAGGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTATG CAGCTTTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAAA CCCAATTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGTT GTGCAGTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATGC CTCCTTACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAGGACTCCACAACTTATTATAC CTAACCAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAA >13195_13195_14_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000421587_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=176AA_BP= MAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMC -------------------------------------------------------------- >13195_13195_15_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000442742_GLIPR1L1_chr12_75737473_ENST00000312442_length(transcript)=990nt_BP=78nt CCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGGGATAAA GGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTATGCAGCT TTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAAACCCAA TTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGTTGTGCA GTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATGCCTCCT TACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAAAAATCCATTTCTGAAGCCAACGGGG AGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTTTAATGTCATTTATATACAAAAGAAATT CTCAAATGTTAAAATAAAGGAATAGTTTATTGCTTAATATAACTTATCATCACTTTGCTTCTTTACTGAATCTTCTACACTCTTGCCTGA TACCTAAATTTAATGTTTGTTTTTAACTCAAAAAATGTACTGTAGTATGGAAAATGGATAGCAGTAGAATAAAGTCTTAAGATTATTTTT TAATTACAAATCCATATGTGTATCAAAAGTGTTCCACTCTTTAATTTTCTAATGAGAATGGATTATCATTTTAGTTATAGTACATTTAGA GAACTAAAGACTTTTCACATATCAAATAGTTTCCTTAGATACCAATAGTTTCCTTTATGAATACCATCAACATCATTCAGAAACAAAATG >13195_13195_15_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000442742_GLIPR1L1_chr12_75737473_ENST00000312442_length(amino acids)=167AA_BP= MAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMC -------------------------------------------------------------- >13195_13195_16_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000442742_GLIPR1L1_chr12_75737473_ENST00000378695_length(transcript)=699nt_BP=78nt CCGCTATCCCCTCCGGACCATGGCCGACGACGACGTGCTGTTCGAGGATGTGTACGAGCTGTGCGAGGTGATCGGAAAATTTGGGATAAA GGTTTAGCAAAGATGGCTAAAGCATGGGCAAACCAGTGCAAATTTGAACATAATGACTGTTTGGATAAATCATATAAATGCTATGCAGCT TTTGAATATGTTGGAGAAAATATCTGGTTAGGTGGAATAAAGTCATTCACACCAAGACATGCCATTACGGCTTGGTATAATGAAACCCAA TTTTATGATTTTGATAGTCTATCATGCTCCAGAGTCTGTGGCCATTATACACAGTTAGTTTGGGCCAATTCATTTTATGTCGGTTGTGCA GTTGCAATGTGTCCTAACCTTGGGGGAGCTTCAACTGCAATATTTGTATGCAACTACGGACCTGCAGGAAATTTTGCAAATATGCCTCCT TACGTAAGAGGAGAATCTTGCTCTCTCTGCTCAAAAGAAGAGAAATGTGTAAAGAACCTCTGCAGGACTCCACAACTTATTATACCTAAC CAAAATCCATTTCTGAAGCCAACGGGGAGAGCACCTCAGCAGACAGCCTTTAATCCATTCAGCTTAGGTTTTCTTCTTCTGAGAATCTTT >13195_13195_16_CASK-GLIPR1L1_CASK_chrX_41782183_ENST00000442742_GLIPR1L1_chr12_75737473_ENST00000378695_length(amino acids)=176AA_BP= MAKAWANQCKFEHNDCLDKSYKCYAAFEYVGENIWLGGIKSFTPRHAITAWYNETQFYDFDSLSCSRVCGHYTQLVWANSFYVGCAVAMC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CASK-GLIPR1L1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CASK-GLIPR1L1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CASK-GLIPR1L1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
