
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CASZ1-EXOSC10 (FusionGDB2 ID:13282) |
Fusion Gene Summary for CASZ1-EXOSC10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CASZ1-EXOSC10 | Fusion gene ID: 13282 | Hgene | Tgene | Gene symbol | CASZ1 | EXOSC10 | Gene ID | 54897 | 5394 |
| Gene name | castor zinc finger 1 | exosome component 10 | |
| Synonyms | CAS11|CST|SRG|ZNF693|dJ734G22.1 | PM-Scl|PM/Scl-100|PMSCL|PMSCL2|RRP6|Rrp6p|p2|p3|p4 | |
| Cytomap | 1p36.22 | 1p36.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein castor homolog 1castor homolog 1, zinc fingercastor-related proteinsurvival-relatedzinc finger protein 693 | exosome component 10P100 polymyositis-scleroderma overlap syndrome-associated autoantigenautoantigen PM-SCLpolymyositis/scleroderma autoantigen 100 kDapolymyositis/scleroderma autoantigen 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86V15 | Q01780 | |
| Ensembl transtripts involved in fusion gene | ENST00000344008, ENST00000377022, ENST00000478728, | ENST00000544779, ENST00000485606, ENST00000304457, ENST00000376936, | |
| Fusion gene scores | * DoF score | 23 X 19 X 12=5244 | 10 X 11 X 7=770 |
| # samples | 30 | 16 | |
| ** MAII score | log2(30/5244*10)=-4.12763327972587 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/770*10)=-2.2667865406949 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CASZ1 [Title/Abstract] AND EXOSC10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CASZ1(10856621)-EXOSC10(11131030), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CASZ1-EXOSC10 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CASZ1-EXOSC10 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. CASZ1-EXOSC10 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CASZ1 | GO:0045893 | positive regulation of transcription, DNA-templated | 27693370 |
 Fusion gene breakpoints across CASZ1 (5'-gene) Fusion gene breakpoints across CASZ1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
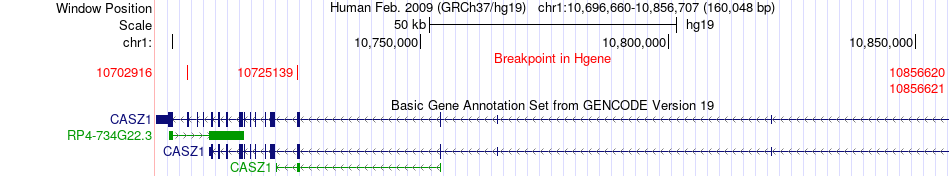 |
 Fusion gene breakpoints across EXOSC10 (3'-gene) Fusion gene breakpoints across EXOSC10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
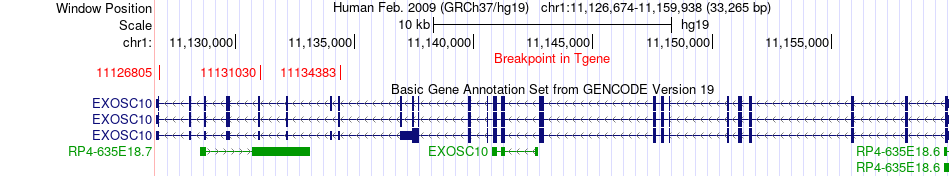 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-1846 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| ChimerDB4 | UCEC | TCGA-AX-A3G6-01A | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| ChimerDB4 | UCEC | TCGA-AX-A3G6 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| ChimerDB4 | UCEC | TCGA-D1-A16I-01A | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
Top |
Fusion Gene ORF analysis for CASZ1-EXOSC10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000344008 | ENST00000544779 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5CDS-3UTR | ENST00000377022 | ENST00000544779 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5CDS-3UTR | ENST00000377022 | ENST00000544779 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| 5CDS-intron | ENST00000344008 | ENST00000485606 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5CDS-intron | ENST00000377022 | ENST00000485606 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5CDS-intron | ENST00000377022 | ENST00000485606 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| 5UTR-3CDS | ENST00000344008 | ENST00000304457 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000344008 | ENST00000304457 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000344008 | ENST00000376936 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000344008 | ENST00000376936 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000377022 | ENST00000304457 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000377022 | ENST00000304457 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000377022 | ENST00000376936 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000377022 | ENST00000376936 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3CDS | ENST00000478728 | ENST00000304457 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5UTR-3CDS | ENST00000478728 | ENST00000376936 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5UTR-3UTR | ENST00000344008 | ENST00000544779 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3UTR | ENST00000344008 | ENST00000544779 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3UTR | ENST00000377022 | ENST00000544779 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3UTR | ENST00000377022 | ENST00000544779 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-3UTR | ENST00000478728 | ENST00000544779 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| 5UTR-intron | ENST00000344008 | ENST00000485606 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-intron | ENST00000344008 | ENST00000485606 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-intron | ENST00000377022 | ENST00000485606 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-intron | ENST00000377022 | ENST00000485606 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| 5UTR-intron | ENST00000478728 | ENST00000485606 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| Frame-shift | ENST00000344008 | ENST00000304457 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| Frame-shift | ENST00000344008 | ENST00000376936 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| Frame-shift | ENST00000377022 | ENST00000304457 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| Frame-shift | ENST00000377022 | ENST00000376936 | CASZ1 | chr1 | 10725139 | - | EXOSC10 | chr1 | 11134383 | - |
| In-frame | ENST00000377022 | ENST00000304457 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| In-frame | ENST00000377022 | ENST00000376936 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-3CDS | ENST00000344008 | ENST00000304457 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-3CDS | ENST00000344008 | ENST00000376936 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-3CDS | ENST00000478728 | ENST00000304457 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-3CDS | ENST00000478728 | ENST00000304457 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-3CDS | ENST00000478728 | ENST00000304457 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-3CDS | ENST00000478728 | ENST00000376936 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-3CDS | ENST00000478728 | ENST00000376936 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-3CDS | ENST00000478728 | ENST00000376936 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-3UTR | ENST00000344008 | ENST00000544779 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-3UTR | ENST00000478728 | ENST00000544779 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-3UTR | ENST00000478728 | ENST00000544779 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-3UTR | ENST00000478728 | ENST00000544779 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-intron | ENST00000344008 | ENST00000485606 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
| intron-intron | ENST00000478728 | ENST00000485606 | CASZ1 | chr1 | 10856621 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-intron | ENST00000478728 | ENST00000485606 | CASZ1 | chr1 | 10856620 | - | EXOSC10 | chr1 | 11131030 | - |
| intron-intron | ENST00000478728 | ENST00000485606 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000377022 | CASZ1 | chr1 | 10702916 | - | ENST00000304457 | EXOSC10 | chr1 | 11126805 | - | 4611 | 4480 | 285 | 4610 | 1442 |
| ENST00000377022 | CASZ1 | chr1 | 10702916 | - | ENST00000376936 | EXOSC10 | chr1 | 11126805 | - | 4611 | 4480 | 285 | 4610 | 1442 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000377022 | ENST00000304457 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - | 0.15922229 | 0.8407777 |
| ENST00000377022 | ENST00000376936 | CASZ1 | chr1 | 10702916 | - | EXOSC10 | chr1 | 11126805 | - | 0.15922229 | 0.8407777 |
Top |
Fusion Genomic Features for CASZ1-EXOSC10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
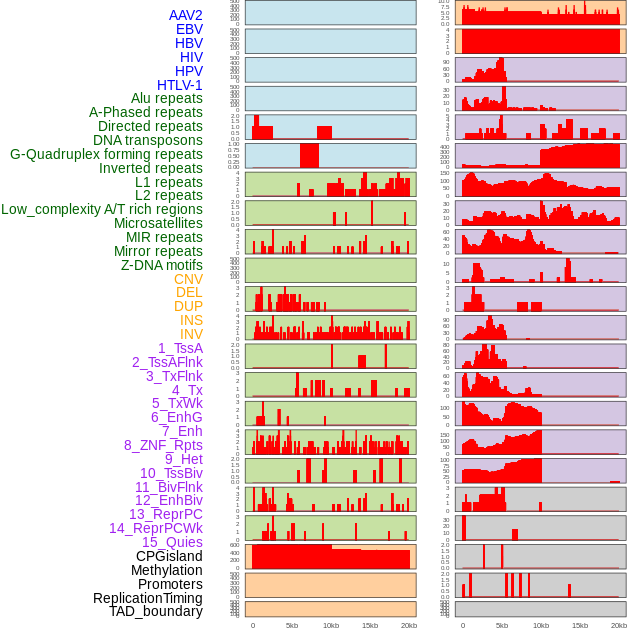 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CASZ1-EXOSC10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:10856621/chr1:11131030) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CASZ1 | EXOSC10 |
| FUNCTION: Transcriptional activator (PubMed:23639441, PubMed:27693370). Involved in vascular assembly and morphogenesis through direct transcriptional regulation of EGFL7 (PubMed:23639441). {ECO:0000269|PubMed:23639441, ECO:0000269|PubMed:27693370}. | FUNCTION: Putative catalytic component of the RNA exosome complex which has 3'->5' exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. In the nucleus, the RNA exosome complex is involved in proper maturation of stable RNA species such as rRNA, snRNA and snoRNA, in the elimination of RNA processing by-products and non-coding 'pervasive' transcripts, such as antisense RNA species and promoter-upstream transcripts (PROMPTs), and of mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm. The RNA exosome may be involved in Ig class switch recombination (CSR) and/or Ig variable region somatic hypermutation (SHM) by targeting AICDA deamination activity to transcribed dsDNA substrates. In the cytoplasm, the RNA exosome complex is involved in general mRNA turnover and specifically degrades inherently unstable mRNAs containing AU-rich elements (AREs) within their 3' untranslated regions, and in RNA surveillance pathways, preventing translation of aberrant mRNAs. It seems to be involved in degradation of histone mRNA. EXOSC10 has 3'-5' exonuclease activity (By similarity). EXOSC10 is required for nucleolar localization of C1D and probably mediates the association of MTREX, C1D and MPP6 wth the RNA exosome involved in the maturation of 5.8S rRNA. {ECO:0000250, ECO:0000269|PubMed:14527413, ECO:0000269|PubMed:16455498, ECO:0000269|PubMed:17412707, ECO:0000269|PubMed:17545563, ECO:0000269|PubMed:18172165, ECO:0000269|PubMed:19056938, ECO:0000269|PubMed:20368444, ECO:0000269|PubMed:20699273}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1080_1143 | 1387 | 1760.0 | Compositional bias | Note=Pro-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 384_417 | 1387 | 1760.0 | Compositional bias | Note=Pro-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1031_1055 | 1387 | 1760.0 | Zinc finger | C2H2-type 4 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1300_1324 | 1387 | 1760.0 | Zinc finger | C2H2-type 5 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 551_575 | 1387 | 1760.0 | Zinc finger | C2H2-type 1 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 610_634 | 1387 | 1760.0 | Zinc finger | C2H2-type 2 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 668_692 | 1387 | 1760.0 | Zinc finger | C2H2-type 3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1080_1143 | 0 | 1167.0 | Compositional bias | Note=Pro-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1635_1670 | 0 | 1167.0 | Compositional bias | Note=Ala-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1672_1729 | 0 | 1167.0 | Compositional bias | Note=Glu-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1691_1718 | 0 | 1167.0 | Compositional bias | Note=Asp-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 384_417 | 0 | 1167.0 | Compositional bias | Note=Pro-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1635_1670 | 1387 | 1760.0 | Compositional bias | Note=Ala-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1672_1729 | 1387 | 1760.0 | Compositional bias | Note=Glu-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1691_1718 | 1387 | 1760.0 | Compositional bias | Note=Asp-rich |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1031_1055 | 0 | 1167.0 | Zinc finger | C2H2-type 4 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1300_1324 | 0 | 1167.0 | Zinc finger | C2H2-type 5 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1457_1481 | 0 | 1167.0 | Zinc finger | C2H2-type 6 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1515_1537 | 0 | 1167.0 | Zinc finger | C2H2-type 7 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 1571_1595 | 0 | 1167.0 | Zinc finger | C2H2-type 8 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 551_575 | 0 | 1167.0 | Zinc finger | C2H2-type 1 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 610_634 | 0 | 1167.0 | Zinc finger | C2H2-type 2 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000344008 | - | 1 | 16 | 668_692 | 0 | 1167.0 | Zinc finger | C2H2-type 3 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1457_1481 | 1387 | 1760.0 | Zinc finger | C2H2-type 6 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1515_1537 | 1387 | 1760.0 | Zinc finger | C2H2-type 7 |
| Hgene | CASZ1 | chr1:10702916 | chr1:11126805 | ENST00000377022 | - | 20 | 21 | 1571_1595 | 1387 | 1760.0 | Zinc finger | C2H2-type 8 |
| Tgene | EXOSC10 | chr1:10702916 | chr1:11126805 | ENST00000304457 | 22 | 24 | 503_583 | 850 | 861.0 | Domain | HRDC | |
| Tgene | EXOSC10 | chr1:10702916 | chr1:11126805 | ENST00000376936 | 23 | 25 | 503_583 | 875 | 886.0 | Domain | HRDC |
Top |
Fusion Gene Sequence for CASZ1-EXOSC10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >13282_13282_1_CASZ1-EXOSC10_CASZ1_chr1_10702916_ENST00000377022_EXOSC10_chr1_11126805_ENST00000304457_length(transcript)=4611nt_BP=4480nt ATGCGCGCCCGCGGCGCTCCATCCCAAACACACACACATTTTTCCCCCGGACTTGGAAAGTCACCCAAAGCCGCCCCAAGTGCAGGCAAA CAGACCTTGCTGACTGAAGGCAAAGACTCCTCCATCCCACCTGCCCTCCCAGCTGCCACGGCCATCGACCCCTCCTTTACAAGGGGCTCC TGCAACTCCCCAGCAGCACGTGTTGGAATGGACTTCGGACCCTGGCCTCTTGGGATGCAAAGGATGAAGTGACACCCCCAGCTACATCCG AGGAGGTTCTAGGACCTGCTACGAGAGTTTGGGACCAAGGAGAAGAGAATGGATCTTGGAACAGCTGAGGGCACCCGGTGCACGGACCCG CCTGCAGGCAAGCCCGCCATGGCGCCCAAACGCAAGGGTGGCCTGAAGCTGAACGCCATCTGCGCCAAGCTGAGCCGCCAGGTGGTGGTG GAGAAGCGAGCTGACGCCGGCTCCCACACGGAGGGCAGCCCATCGCAGCCCCGGGACCAAGAGCGCAGTGGCCCTGAGTCTGGGGCAGCC CGGGCCCCCCGCAGCGAGGAAGACAAGAGACGGGCAGTGATCGAGAAGTGGGTGAACGGGGAGTACAGCGAGGAGCCGGCACCCACACCC GTGTTGGGGCGGATTGCCCGCGAGGGCCTGGAGCTGCCTCCCGAGGGTGTCTACATGGTGCAGCCCCAGGGGTGCAGCGATGAGGAAGAC CACGCGGAGGAGCCCTCCAAGGACGGCGGTGCCCTGGAGGAGAAGGATTCGGACGGGGCAGCCTCCAAGGAGGACAGCGGCCCCAGCACC AGGCAGGCTTCAGGAGAGGCCTCCTCGCTGCGGGACTACGCGGCCTCCACCATGACCGAGTTCCTCGGCATGTTTGGCTATGATGACCAG AACACGCGGGACGAGCTGGCCAGGAAGATCAGCTTTGAGAAGCTGCACGCGGGCTCCACCCCGGAGGCAGCCACCTCCTCCATGCTGCCC ACCTCCGAGGATACCCTCAGCAAGCGGGCGCGGTTCTCTAAGTATGAGGAGTACATCCGCAAGCTCAAGGCTGGCGAGCAGCTCTCCTGG CCGGCCCCCAGCACCAAGACCGAGGAGCGGGTGGGCAAGGAGGTGGTGGGCACCCTGCCCGGCCTGCGGCTGCCCAGCAGCACGGCCCAC CTGGAGACCAAGGCCACCATCCTGCCCCTGCCGTCGCACAGCAGTGTCCAGATGCAGAACCTGGTAGCCCGGGCCTCCAAGTACGACTTC TTCATCCAAAAACTGAAGACCGGCGAGAATCTGCGGCCCCAGAACGGGAGCACCTACAAGAAGCCATCCAAGTACGACCTGGAGAATGTC AAGTACCTGCACCTCTTCAAACCCGGGGAGGGCAGCCCCGACATGGGCGGGGCCATCGCCTTCAAGACAGGCAAGGTGGGGCGCCCTTCC AAGTACGACGTCCGGGGCATCCAGAAGCCAGGCCCCGCCAAGGTTCCGCCCACCCCCAGCCTGGCTCCCGCACCCCTCGCCAGCGTGCCC AGTGCCCCCAGCGCCCCCGGGCCAGGGCCAGAGCCTCCTGCCTCCCTGTCCTTCAACACTCCCGAGTACCTGAAGTCAACCTTCTCCAAA ACAGACTCCATCACCACGGGGACCGTCTCCACTGTCAAGAACGGACTGCCCACAGATAAACCAGCCGTCACTGAAGATGTAAACATTTAC CAGAAATATATTGCCAGGTTCTCGGGCAGCCAGCACTGTGGCCACATCCACTGTGCCTACCAGTACCGCGAGCACTACCACTGCCTTGAC CCTGAGTGTAACTACCAGAGGTTCACGAGTAAGCAGGACGTGATCCGCCACTACAACATGCACAAGAAGCGCGACAACTCCCTGCAGCAC GGCTTCATGCGTTTCAGCCCGCTGGACGACTGCAGCGTCTACTACCACGGCTGCCACCTCAATGGGAAGAGCACCCACTATCACTGCATG CAGGTGGGCTGTAACAAGGTGTACACGAGCACGTCTGACGTGATGACCCACGAGAACTTCCACAAGAAGAATACCCAGCTCATTAACGAC GGCTTCCAGCGCTTCCGAGCCACCGAAGACTGTGGCACAGCCGACTGCCAGTTCTACGGACAGAAGACCACGCACTTCCACTGCAGGCGC CCCGGCTGCACATTCACTTTCAAGAACAAGTGTGACATCGAGAAGCACAAGAGCTACCACATCAAGGACGATGCCTACGCCAAGGACGGC TTCAAGAAGTTCTACAAGTACGAGGAGTGCAAGTACGAGGGCTGCGTGTACAGCAAGGCTACCAACCACTTCCACTGCATCCGCGCCGGC TGCGGCTTCACCTTCACCTCCACCAGCCAGATGACCTCTCACAAGCGCAAGCATGAGCGCCGGCACATCCGCTCCTCGGGCGCGCTGGGG CTGCCGCCCTCGCTGCTGGGCGCCAAGGACACGGAGCACGAGGAGTCCAGCAACGACGACCTTGTTGACTTCTCCGCCCTGAGCAGCAAG AACTCCAGCCTGAGCGCCTCCCCTACCAGCCAGCAGTCCTCTGCGTCCCTGGCTGCCGCCACTGCCGCCACCGAGGCTGGGCCCAGTGCC ACCAAACCTCCCAACAGCAAGATCTCGGGGCTGCTGCCCCAGGGCCTGCCTGGCTCAATCCCCCTGGCCCTGGCCCTCTCCAACTCGGGC CTGCCCACCCCCACGCCCTACTTCCCCATACTGGCTGGCCGTGGGAGCACCTCCCTGCCTGTGGGCACCCCCAGCCTCCTGGGTGCCGTG TCGTCTGGGTCAGCAGCCTCAGCCACCCCTGACACACCCACGCTGGTCGCCTCGGGAGCTGGAGACTCAGCCCCCGTGGCTGCCGCCTCT GTCCCGGCACCACCCGCCTCCATCATGGAGAGGATCTCTGCAAGCAAGGGCCTCATCTCGCCCATGATGGCCAGGCTGGCTGCAGCTGCC CTCAAGCCCTCTGCCACCTTTGACCCAGGAAGCGGGCAGCAGGTCACCCCAGCCAGGTTCCCCCCGGCCCAAGTGAAGCCGGAACCCGGT GAGAGCACCGGCGCCCCAGGCCCCCACGAAGCCTCCCAGGACCGCAGTCTAGACCTGACTGTGAAGGAGCCCAGCAACGAATCAAATGGC CACGCAGTCCCGGCAAATTCATCTCTTTTATCCTCGCTTATGAATAAGATGTCTCAGGGCAACCCTGGCCTGGGCAGCCTGCTGAACATC AAGGCGGAAGCGGAGGGGAGCCCCGCTGCGGAGCCCTCGCCCTTCCTAGGCAAGGCCGTGAAGGCGCTGGTTCAGGAGAAGTTGGCAGAG CCCTGGAAGGTGTACCTGCGCAGGTTTGGTACAAAGGACTTCTGTGACGGCCAGTGTGACTTCCTCCACAAGGCCCACTTCCACTGCGTG GTGGAGGAATGCGGCGCGCTCTTCAGCACCTTGGACGGGGCCATCAAGCACGCAAACTTCCACTTCCGGACAGAGGGAGGAGCAGCAAAA GGAAACACAGAGGCTGCCTTTCCGGCCTCGGCCGCCGAGACCAAACCTCCCATGGCCCCCTCGTCCCCTCCGGTCCCTCCTGTCACCACG GCCACGGTGTCCTCTCTGGAGGGGCCCGCTCCCAGCCCGGCCTCCGTGCCCTCCACCCCCACCCTGCTCGCCTGGAAGCAGCTGGCTTCC ACCATACCCCAGATGCCTCAGATCCCAGCGTCAGTGCCTCACCTGCCCGCCTCGCCCTTGGCAACGACTTCTCTAGAGAACGCCAAGCCC CAGGTCAAACCCGGATTCCTCCAGTTCCAGGAGAACGATCCTTGCCTCGCCACGGACTGCAAGTACGCCAACAAGTTCCACTTCCACTGT CTCTTTGGGAACTGCAAGTACGTCTGCAAAACGTCTGGCAAGGCCGAATCCCACTGCCTGGACCACATCAACCCCAACAACAACCTGGTG AACGTGCGAGACCAGTTTGCATACTACTCTCTGCAGTGTCTCTGTCCCAACCAGCACTGCGAGTTCCGAATGCGTGGGCACTACCACTGC CTCCGCACCGGCTGCTATTTTGTGACCAACATCACCACCAAGCTCCCCTGGCACATCAAGAAGCATGAGAAGGCGGAGCGGCGGGCAGCC AATGGCTTCAAATACTTCACCAAGCGCGAGGAGTGTGGCAGGCTAGGTTGCAAGTACAACCAGGTGAACAGCCACTTCCACTGCATCCGG GAGGGCTGCCAGTTCTCCTTCCTCCTCAAGCACCAGATGACCTCCCACGCGCGGAAGCACATGCGGAGGATGCTGGGGAAGAACTTCGAC CGCGTGCCCCCCTCCCAGGGCCCCCCAGGCCTGATGGATGCTGAGACAGATGAGTGCATGGACTACACTGGCTGCAGCCCAGGCGCCATG TCCTCTGAGTCATCCACCATGGACCGGAGCTGCTCCAGCACCCCCGTGGGTAACGAGAGCACCGCGGCAGAGGCTTCAGGTACAACTGGC CACAGAGATAGTCCTGGAAGACACGTGGCGCCTGTGGACCGGAAGCACCAAATGCTGGTGCTGCTTTTGTACATACATATTTTTAAACCA >13282_13282_1_CASZ1-EXOSC10_CASZ1_chr1_10702916_ENST00000377022_EXOSC10_chr1_11126805_ENST00000304457_length(amino acids)=1442AA_BP=1397 MLREFGTKEKRMDLGTAEGTRCTDPPAGKPAMAPKRKGGLKLNAICAKLSRQVVVEKRADAGSHTEGSPSQPRDQERSGPESGAARAPRS EEDKRRAVIEKWVNGEYSEEPAPTPVLGRIAREGLELPPEGVYMVQPQGCSDEEDHAEEPSKDGGALEEKDSDGAASKEDSGPSTRQASG EASSLRDYAASTMTEFLGMFGYDDQNTRDELARKISFEKLHAGSTPEAATSSMLPTSEDTLSKRARFSKYEEYIRKLKAGEQLSWPAPST KTEERVGKEVVGTLPGLRLPSSTAHLETKATILPLPSHSSVQMQNLVARASKYDFFIQKLKTGENLRPQNGSTYKKPSKYDLENVKYLHL FKPGEGSPDMGGAIAFKTGKVGRPSKYDVRGIQKPGPAKVPPTPSLAPAPLASVPSAPSAPGPGPEPPASLSFNTPEYLKSTFSKTDSIT TGTVSTVKNGLPTDKPAVTEDVNIYQKYIARFSGSQHCGHIHCAYQYREHYHCLDPECNYQRFTSKQDVIRHYNMHKKRDNSLQHGFMRF SPLDDCSVYYHGCHLNGKSTHYHCMQVGCNKVYTSTSDVMTHENFHKKNTQLINDGFQRFRATEDCGTADCQFYGQKTTHFHCRRPGCTF TFKNKCDIEKHKSYHIKDDAYAKDGFKKFYKYEECKYEGCVYSKATNHFHCIRAGCGFTFTSTSQMTSHKRKHERRHIRSSGALGLPPSL LGAKDTEHEESSNDDLVDFSALSSKNSSLSASPTSQQSSASLAAATAATEAGPSATKPPNSKISGLLPQGLPGSIPLALALSNSGLPTPT PYFPILAGRGSTSLPVGTPSLLGAVSSGSAASATPDTPTLVASGAGDSAPVAAASVPAPPASIMERISASKGLISPMMARLAAAALKPSA TFDPGSGQQVTPARFPPAQVKPEPGESTGAPGPHEASQDRSLDLTVKEPSNESNGHAVPANSSLLSSLMNKMSQGNPGLGSLLNIKAEAE GSPAAEPSPFLGKAVKALVQEKLAEPWKVYLRRFGTKDFCDGQCDFLHKAHFHCVVEECGALFSTLDGAIKHANFHFRTEGGAAKGNTEA AFPASAAETKPPMAPSSPPVPPVTTATVSSLEGPAPSPASVPSTPTLLAWKQLASTIPQMPQIPASVPHLPASPLATTSLENAKPQVKPG FLQFQENDPCLATDCKYANKFHFHCLFGNCKYVCKTSGKAESHCLDHINPNNNLVNVRDQFAYYSLQCLCPNQHCEFRMRGHYHCLRTGC YFVTNITTKLPWHIKKHEKAERRAANGFKYFTKREECGRLGCKYNQVNSHFHCIREGCQFSFLLKHQMTSHARKHMRRMLGKNFDRVPPS QGPPGLMDAETDECMDYTGCSPGAMSSESSTMDRSCSSTPVGNESTAAEASGTTGHRDSPGRHVAPVDRKHQMLVLLLYIHIFKPLKFFL -------------------------------------------------------------- >13282_13282_2_CASZ1-EXOSC10_CASZ1_chr1_10702916_ENST00000377022_EXOSC10_chr1_11126805_ENST00000376936_length(transcript)=4611nt_BP=4480nt ATGCGCGCCCGCGGCGCTCCATCCCAAACACACACACATTTTTCCCCCGGACTTGGAAAGTCACCCAAAGCCGCCCCAAGTGCAGGCAAA CAGACCTTGCTGACTGAAGGCAAAGACTCCTCCATCCCACCTGCCCTCCCAGCTGCCACGGCCATCGACCCCTCCTTTACAAGGGGCTCC TGCAACTCCCCAGCAGCACGTGTTGGAATGGACTTCGGACCCTGGCCTCTTGGGATGCAAAGGATGAAGTGACACCCCCAGCTACATCCG AGGAGGTTCTAGGACCTGCTACGAGAGTTTGGGACCAAGGAGAAGAGAATGGATCTTGGAACAGCTGAGGGCACCCGGTGCACGGACCCG CCTGCAGGCAAGCCCGCCATGGCGCCCAAACGCAAGGGTGGCCTGAAGCTGAACGCCATCTGCGCCAAGCTGAGCCGCCAGGTGGTGGTG GAGAAGCGAGCTGACGCCGGCTCCCACACGGAGGGCAGCCCATCGCAGCCCCGGGACCAAGAGCGCAGTGGCCCTGAGTCTGGGGCAGCC CGGGCCCCCCGCAGCGAGGAAGACAAGAGACGGGCAGTGATCGAGAAGTGGGTGAACGGGGAGTACAGCGAGGAGCCGGCACCCACACCC GTGTTGGGGCGGATTGCCCGCGAGGGCCTGGAGCTGCCTCCCGAGGGTGTCTACATGGTGCAGCCCCAGGGGTGCAGCGATGAGGAAGAC CACGCGGAGGAGCCCTCCAAGGACGGCGGTGCCCTGGAGGAGAAGGATTCGGACGGGGCAGCCTCCAAGGAGGACAGCGGCCCCAGCACC AGGCAGGCTTCAGGAGAGGCCTCCTCGCTGCGGGACTACGCGGCCTCCACCATGACCGAGTTCCTCGGCATGTTTGGCTATGATGACCAG AACACGCGGGACGAGCTGGCCAGGAAGATCAGCTTTGAGAAGCTGCACGCGGGCTCCACCCCGGAGGCAGCCACCTCCTCCATGCTGCCC ACCTCCGAGGATACCCTCAGCAAGCGGGCGCGGTTCTCTAAGTATGAGGAGTACATCCGCAAGCTCAAGGCTGGCGAGCAGCTCTCCTGG CCGGCCCCCAGCACCAAGACCGAGGAGCGGGTGGGCAAGGAGGTGGTGGGCACCCTGCCCGGCCTGCGGCTGCCCAGCAGCACGGCCCAC CTGGAGACCAAGGCCACCATCCTGCCCCTGCCGTCGCACAGCAGTGTCCAGATGCAGAACCTGGTAGCCCGGGCCTCCAAGTACGACTTC TTCATCCAAAAACTGAAGACCGGCGAGAATCTGCGGCCCCAGAACGGGAGCACCTACAAGAAGCCATCCAAGTACGACCTGGAGAATGTC AAGTACCTGCACCTCTTCAAACCCGGGGAGGGCAGCCCCGACATGGGCGGGGCCATCGCCTTCAAGACAGGCAAGGTGGGGCGCCCTTCC AAGTACGACGTCCGGGGCATCCAGAAGCCAGGCCCCGCCAAGGTTCCGCCCACCCCCAGCCTGGCTCCCGCACCCCTCGCCAGCGTGCCC AGTGCCCCCAGCGCCCCCGGGCCAGGGCCAGAGCCTCCTGCCTCCCTGTCCTTCAACACTCCCGAGTACCTGAAGTCAACCTTCTCCAAA ACAGACTCCATCACCACGGGGACCGTCTCCACTGTCAAGAACGGACTGCCCACAGATAAACCAGCCGTCACTGAAGATGTAAACATTTAC CAGAAATATATTGCCAGGTTCTCGGGCAGCCAGCACTGTGGCCACATCCACTGTGCCTACCAGTACCGCGAGCACTACCACTGCCTTGAC CCTGAGTGTAACTACCAGAGGTTCACGAGTAAGCAGGACGTGATCCGCCACTACAACATGCACAAGAAGCGCGACAACTCCCTGCAGCAC GGCTTCATGCGTTTCAGCCCGCTGGACGACTGCAGCGTCTACTACCACGGCTGCCACCTCAATGGGAAGAGCACCCACTATCACTGCATG CAGGTGGGCTGTAACAAGGTGTACACGAGCACGTCTGACGTGATGACCCACGAGAACTTCCACAAGAAGAATACCCAGCTCATTAACGAC GGCTTCCAGCGCTTCCGAGCCACCGAAGACTGTGGCACAGCCGACTGCCAGTTCTACGGACAGAAGACCACGCACTTCCACTGCAGGCGC CCCGGCTGCACATTCACTTTCAAGAACAAGTGTGACATCGAGAAGCACAAGAGCTACCACATCAAGGACGATGCCTACGCCAAGGACGGC TTCAAGAAGTTCTACAAGTACGAGGAGTGCAAGTACGAGGGCTGCGTGTACAGCAAGGCTACCAACCACTTCCACTGCATCCGCGCCGGC TGCGGCTTCACCTTCACCTCCACCAGCCAGATGACCTCTCACAAGCGCAAGCATGAGCGCCGGCACATCCGCTCCTCGGGCGCGCTGGGG CTGCCGCCCTCGCTGCTGGGCGCCAAGGACACGGAGCACGAGGAGTCCAGCAACGACGACCTTGTTGACTTCTCCGCCCTGAGCAGCAAG AACTCCAGCCTGAGCGCCTCCCCTACCAGCCAGCAGTCCTCTGCGTCCCTGGCTGCCGCCACTGCCGCCACCGAGGCTGGGCCCAGTGCC ACCAAACCTCCCAACAGCAAGATCTCGGGGCTGCTGCCCCAGGGCCTGCCTGGCTCAATCCCCCTGGCCCTGGCCCTCTCCAACTCGGGC CTGCCCACCCCCACGCCCTACTTCCCCATACTGGCTGGCCGTGGGAGCACCTCCCTGCCTGTGGGCACCCCCAGCCTCCTGGGTGCCGTG TCGTCTGGGTCAGCAGCCTCAGCCACCCCTGACACACCCACGCTGGTCGCCTCGGGAGCTGGAGACTCAGCCCCCGTGGCTGCCGCCTCT GTCCCGGCACCACCCGCCTCCATCATGGAGAGGATCTCTGCAAGCAAGGGCCTCATCTCGCCCATGATGGCCAGGCTGGCTGCAGCTGCC CTCAAGCCCTCTGCCACCTTTGACCCAGGAAGCGGGCAGCAGGTCACCCCAGCCAGGTTCCCCCCGGCCCAAGTGAAGCCGGAACCCGGT GAGAGCACCGGCGCCCCAGGCCCCCACGAAGCCTCCCAGGACCGCAGTCTAGACCTGACTGTGAAGGAGCCCAGCAACGAATCAAATGGC CACGCAGTCCCGGCAAATTCATCTCTTTTATCCTCGCTTATGAATAAGATGTCTCAGGGCAACCCTGGCCTGGGCAGCCTGCTGAACATC AAGGCGGAAGCGGAGGGGAGCCCCGCTGCGGAGCCCTCGCCCTTCCTAGGCAAGGCCGTGAAGGCGCTGGTTCAGGAGAAGTTGGCAGAG CCCTGGAAGGTGTACCTGCGCAGGTTTGGTACAAAGGACTTCTGTGACGGCCAGTGTGACTTCCTCCACAAGGCCCACTTCCACTGCGTG GTGGAGGAATGCGGCGCGCTCTTCAGCACCTTGGACGGGGCCATCAAGCACGCAAACTTCCACTTCCGGACAGAGGGAGGAGCAGCAAAA GGAAACACAGAGGCTGCCTTTCCGGCCTCGGCCGCCGAGACCAAACCTCCCATGGCCCCCTCGTCCCCTCCGGTCCCTCCTGTCACCACG GCCACGGTGTCCTCTCTGGAGGGGCCCGCTCCCAGCCCGGCCTCCGTGCCCTCCACCCCCACCCTGCTCGCCTGGAAGCAGCTGGCTTCC ACCATACCCCAGATGCCTCAGATCCCAGCGTCAGTGCCTCACCTGCCCGCCTCGCCCTTGGCAACGACTTCTCTAGAGAACGCCAAGCCC CAGGTCAAACCCGGATTCCTCCAGTTCCAGGAGAACGATCCTTGCCTCGCCACGGACTGCAAGTACGCCAACAAGTTCCACTTCCACTGT CTCTTTGGGAACTGCAAGTACGTCTGCAAAACGTCTGGCAAGGCCGAATCCCACTGCCTGGACCACATCAACCCCAACAACAACCTGGTG AACGTGCGAGACCAGTTTGCATACTACTCTCTGCAGTGTCTCTGTCCCAACCAGCACTGCGAGTTCCGAATGCGTGGGCACTACCACTGC CTCCGCACCGGCTGCTATTTTGTGACCAACATCACCACCAAGCTCCCCTGGCACATCAAGAAGCATGAGAAGGCGGAGCGGCGGGCAGCC AATGGCTTCAAATACTTCACCAAGCGCGAGGAGTGTGGCAGGCTAGGTTGCAAGTACAACCAGGTGAACAGCCACTTCCACTGCATCCGG GAGGGCTGCCAGTTCTCCTTCCTCCTCAAGCACCAGATGACCTCCCACGCGCGGAAGCACATGCGGAGGATGCTGGGGAAGAACTTCGAC CGCGTGCCCCCCTCCCAGGGCCCCCCAGGCCTGATGGATGCTGAGACAGATGAGTGCATGGACTACACTGGCTGCAGCCCAGGCGCCATG TCCTCTGAGTCATCCACCATGGACCGGAGCTGCTCCAGCACCCCCGTGGGTAACGAGAGCACCGCGGCAGAGGCTTCAGGTACAACTGGC CACAGAGATAGTCCTGGAAGACACGTGGCGCCTGTGGACCGGAAGCACCAAATGCTGGTGCTGCTTTTGTACATACATATTTTTAAACCA >13282_13282_2_CASZ1-EXOSC10_CASZ1_chr1_10702916_ENST00000377022_EXOSC10_chr1_11126805_ENST00000376936_length(amino acids)=1442AA_BP=1397 MLREFGTKEKRMDLGTAEGTRCTDPPAGKPAMAPKRKGGLKLNAICAKLSRQVVVEKRADAGSHTEGSPSQPRDQERSGPESGAARAPRS EEDKRRAVIEKWVNGEYSEEPAPTPVLGRIAREGLELPPEGVYMVQPQGCSDEEDHAEEPSKDGGALEEKDSDGAASKEDSGPSTRQASG EASSLRDYAASTMTEFLGMFGYDDQNTRDELARKISFEKLHAGSTPEAATSSMLPTSEDTLSKRARFSKYEEYIRKLKAGEQLSWPAPST KTEERVGKEVVGTLPGLRLPSSTAHLETKATILPLPSHSSVQMQNLVARASKYDFFIQKLKTGENLRPQNGSTYKKPSKYDLENVKYLHL FKPGEGSPDMGGAIAFKTGKVGRPSKYDVRGIQKPGPAKVPPTPSLAPAPLASVPSAPSAPGPGPEPPASLSFNTPEYLKSTFSKTDSIT TGTVSTVKNGLPTDKPAVTEDVNIYQKYIARFSGSQHCGHIHCAYQYREHYHCLDPECNYQRFTSKQDVIRHYNMHKKRDNSLQHGFMRF SPLDDCSVYYHGCHLNGKSTHYHCMQVGCNKVYTSTSDVMTHENFHKKNTQLINDGFQRFRATEDCGTADCQFYGQKTTHFHCRRPGCTF TFKNKCDIEKHKSYHIKDDAYAKDGFKKFYKYEECKYEGCVYSKATNHFHCIRAGCGFTFTSTSQMTSHKRKHERRHIRSSGALGLPPSL LGAKDTEHEESSNDDLVDFSALSSKNSSLSASPTSQQSSASLAAATAATEAGPSATKPPNSKISGLLPQGLPGSIPLALALSNSGLPTPT PYFPILAGRGSTSLPVGTPSLLGAVSSGSAASATPDTPTLVASGAGDSAPVAAASVPAPPASIMERISASKGLISPMMARLAAAALKPSA TFDPGSGQQVTPARFPPAQVKPEPGESTGAPGPHEASQDRSLDLTVKEPSNESNGHAVPANSSLLSSLMNKMSQGNPGLGSLLNIKAEAE GSPAAEPSPFLGKAVKALVQEKLAEPWKVYLRRFGTKDFCDGQCDFLHKAHFHCVVEECGALFSTLDGAIKHANFHFRTEGGAAKGNTEA AFPASAAETKPPMAPSSPPVPPVTTATVSSLEGPAPSPASVPSTPTLLAWKQLASTIPQMPQIPASVPHLPASPLATTSLENAKPQVKPG FLQFQENDPCLATDCKYANKFHFHCLFGNCKYVCKTSGKAESHCLDHINPNNNLVNVRDQFAYYSLQCLCPNQHCEFRMRGHYHCLRTGC YFVTNITTKLPWHIKKHEKAERRAANGFKYFTKREECGRLGCKYNQVNSHFHCIREGCQFSFLLKHQMTSHARKHMRRMLGKNFDRVPPS QGPPGLMDAETDECMDYTGCSPGAMSSESSTMDRSCSSTPVGNESTAAEASGTTGHRDSPGRHVAPVDRKHQMLVLLLYIHIFKPLKFFL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CASZ1-EXOSC10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CASZ1-EXOSC10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CASZ1-EXOSC10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
