
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CCAR1-CAAP1 (FusionGDB2 ID:13515) |
Fusion Gene Summary for CCAR1-CAAP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CCAR1-CAAP1 | Fusion gene ID: 13515 | Hgene | Tgene | Gene symbol | CCAR1 | CAAP1 | Gene ID | 55749 | 79886 |
| Gene name | cell division cycle and apoptosis regulator 1 | caspase activity and apoptosis inhibitor 1 | |
| Synonyms | - | C9orf82|CAAP | |
| Cytomap | 10q21.3 | 9p21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cell division cycle and apoptosis regulator protein 1cell cycle and apoptosis regulatory protein 1death inducer with SAP domain | caspase activity and apoptosis inhibitor 1conserved anti-apoptotic protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8IX12 | Q9H8G2 | |
| Ensembl transtripts involved in fusion gene | ENST00000265872, ENST00000535016, ENST00000543719, ENST00000483264, | ENST00000495958, ENST00000535437, ENST00000520187, ENST00000333916, | |
| Fusion gene scores | * DoF score | 13 X 11 X 8=1144 | 3 X 2 X 3=18 |
| # samples | 17 | 4 | |
| ** MAII score | log2(17/1144*10)=-2.75048040064069 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/18*10)=1.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: CCAR1 [Title/Abstract] AND CAAP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
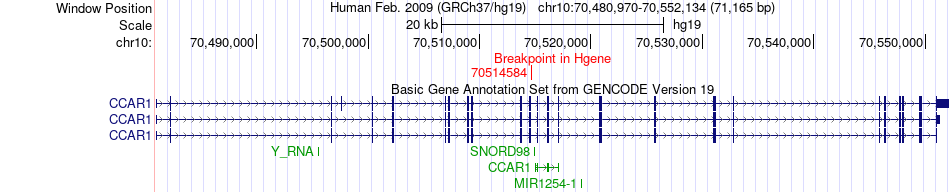
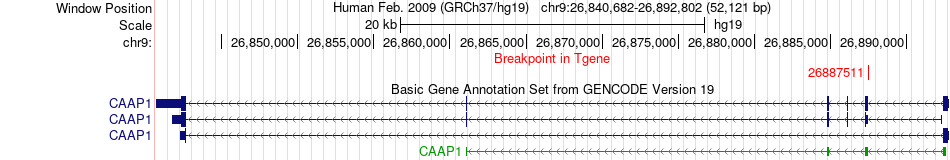
| Most frequent breakpoint | CCAR1(70514584)-CAAP1(26887511), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CCAR1 | GO:0043065 | positive regulation of apoptotic process | 12816952 |
 Fusion gene breakpoints across CCAR1 (5'-gene) Fusion gene breakpoints across CCAR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CAAP1 (3'-gene) Fusion gene breakpoints across CAAP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCS | TCGA-N8-A4PL | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
Top |
Fusion Gene ORF analysis for CCAR1-CAAP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000265872 | ENST00000495958 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-5UTR | ENST00000265872 | ENST00000535437 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-5UTR | ENST00000535016 | ENST00000495958 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-5UTR | ENST00000535016 | ENST00000535437 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-5UTR | ENST00000543719 | ENST00000495958 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-5UTR | ENST00000543719 | ENST00000535437 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-intron | ENST00000265872 | ENST00000520187 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-intron | ENST00000535016 | ENST00000520187 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| 5CDS-intron | ENST00000543719 | ENST00000520187 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| In-frame | ENST00000265872 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| In-frame | ENST00000535016 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| In-frame | ENST00000543719 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| intron-3CDS | ENST00000483264 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| intron-5UTR | ENST00000483264 | ENST00000495958 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| intron-5UTR | ENST00000483264 | ENST00000535437 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
| intron-intron | ENST00000483264 | ENST00000520187 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535016 | CCAR1 | chr10 | 70514584 | + | ENST00000333916 | CAAP1 | chr9 | 26887511 | - | 3931 | 1532 | 119 | 2314 | 731 |
| ENST00000265872 | CCAR1 | chr10 | 70514584 | + | ENST00000333916 | CAAP1 | chr9 | 26887511 | - | 3976 | 1577 | 119 | 2359 | 746 |
| ENST00000543719 | CCAR1 | chr10 | 70514584 | + | ENST00000333916 | CAAP1 | chr9 | 26887511 | - | 3925 | 1526 | 113 | 2308 | 731 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535016 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - | 0.000848914 | 0.9991511 |
| ENST00000265872 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - | 0.001050107 | 0.99894994 |
| ENST00000543719 | ENST00000333916 | CCAR1 | chr10 | 70514584 | + | CAAP1 | chr9 | 26887511 | - | 0.000830184 | 0.9991698 |
Top |
Fusion Genomic Features for CCAR1-CAAP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
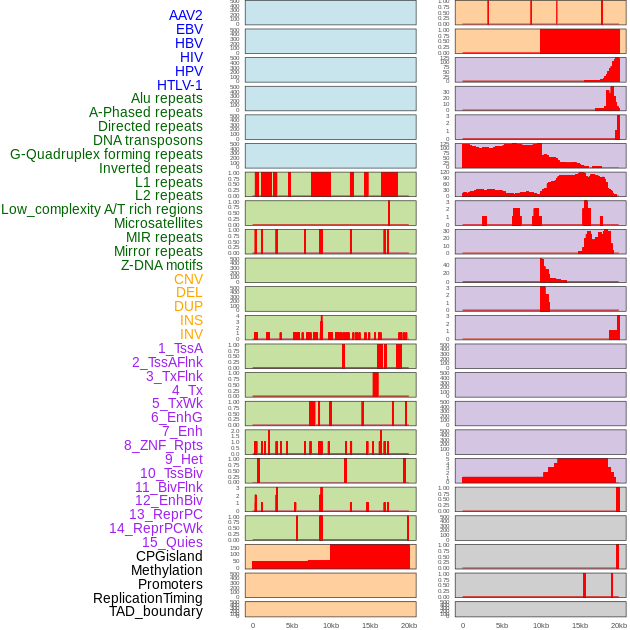
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CCAR1-CAAP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:70514584/chr9:26887511) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CCAR1 | CAAP1 |
| FUNCTION: Associates with components of the Mediator and p160 coactivator complexes that play a role as intermediaries transducing regulatory signals from upstream transcriptional activator proteins to basal transcription machinery at the core promoter. Recruited to endogenous nuclear receptor target genes in response to the appropriate hormone. Also functions as a p53 coactivator. May thus play an important role in transcriptional regulation (By similarity). May be involved in apoptosis signaling in the presence of the reinoid CD437. Apoptosis induction involves sequestration of 14-3-3 protein(s) and mediated altered expression of multiple cell cycle regulatory genes including MYC, CCNB1 and CDKN1A. Plays a role in cell cycle progression and/or cell proliferation (PubMed:12816952). In association with CALCOCO1 enhances GATA1- and MED1-mediated transcriptional activation from the gamma-globin promoter during erythroid differentiation of K562 erythroleukemia cells (PubMed:24245781). Can act as a both a coactivator and corepressor of AR-mediated transcription. Contributes to chromatin looping and AR transcription complex assembly by stabilizing AR-GATA2 association on chromatin and facilitating MED1 and RNA polymerase II recruitment to AR-binding sites. May play an important role in the growth and tumorigenesis of prostate cancer cells (PubMed:23887938). {ECO:0000250|UniProtKB:Q8CH18, ECO:0000269|PubMed:12816952, ECO:0000269|PubMed:23887938, ECO:0000269|PubMed:24245781}. | FUNCTION: Anti-apoptotic protein that modulates a caspase-10 dependent mitochondrial caspase-3/9 feedback amplification loop. {ECO:0000269|PubMed:21980415}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 293_361 | 486 | 1151.0 | Compositional bias | Note=Arg-rich |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 73_79 | 486 | 1151.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 293_361 | 471 | 1136.0 | Compositional bias | Note=Arg-rich |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 73_79 | 471 | 1136.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 293_361 | 471 | 1136.0 | Compositional bias | Note=Arg-rich |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 73_79 | 471 | 1136.0 | Compositional bias | Note=Poly-Ala |
| Tgene | CAAP1 | chr10:70514584 | chr9:26887511 | ENST00000333916 | 0 | 6 | 281_311 | 101 | 362.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CAAP1 | chr10:70514584 | chr9:26887511 | ENST00000535437 | 0 | 6 | 281_311 | 0 | 217.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CAAP1 | chr10:70514584 | chr9:26887511 | ENST00000535437 | 0 | 6 | 19_25 | 0 | 217.0 | Compositional bias | Note=Poly-Ala |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 1033_1114 | 486 | 1151.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 594_618 | 486 | 1151.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 1033_1114 | 471 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 594_618 | 471 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 1033_1114 | 471 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 594_618 | 471 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 673_889 | 486 | 1151.0 | Compositional bias | Note=Glu-rich |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 673_889 | 471 | 1136.0 | Compositional bias | Note=Glu-rich |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 673_889 | 471 | 1136.0 | Compositional bias | Note=Glu-rich |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 636_670 | 486 | 1151.0 | Domain | SAP |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 636_670 | 471 | 1136.0 | Domain | SAP |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 636_670 | 471 | 1136.0 | Domain | SAP |
| Tgene | CAAP1 | chr10:70514584 | chr9:26887511 | ENST00000333916 | 0 | 6 | 19_25 | 101 | 362.0 | Compositional bias | Note=Poly-Ala |
Top |
Fusion Gene Sequence for CCAR1-CAAP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >13515_13515_1_CCAR1-CAAP1_CCAR1_chr10_70514584_ENST00000265872_CAAP1_chr9_26887511_ENST00000333916_length(transcript)=3976nt_BP=1577nt GAAGTTGGCGCATGCGCCTAAAGCTGACGGGTTTGAAATGGCTTCGATGTTAGCCGGGACCCGACTCAGATCGATGCTATAGAAGACAAA CAAGGGAAGGTTTTTTTTCCTTTTGCATCATGGCTCAATTTGGAGGACAGAAGAATCCGCCATGGGCTACTCAGTTTACAGCCACTGCAG TATCACAGCCAGCTGCACTGGGTGTTCAACAGCCATCACTCCTTGGAGCATCTCCTACCATTTATACACAGCAAACTGCATTGGCAGCAG CAGGCCTTACCACACAAACTCCAGCAAACTATCAGTTAACACAAACTGCTGCATTGCAGCAACAAGCCGCAGCTGCAGCAGCTGCATTAC AACAGCAATATTCACAACCTCAGCAGGCCCTGTATAGTGTGCAACAACAGTTACAGCAACCCCAGCAAACCCTCTTAACACAGCCAGCTG TTGCACTGCCTACAAGCCTTAGCCTGTCTACTCCTCAGCCAACAGCACAAATAACTGTATCATATCCAACACCAAGGTCCAGTCAACAGC AAACCCAGCCTCAGAAGCAGCGTGTTTTCACAGGGGTGGTTACAAAACTACATGATACGTTTGGATTTGTGGATGAAGATGTATTCTTTC AGCTTAGTGCTGTCAAAGGGAAAACCCCCCAAGTAGGTGACAGAGTATTGGTTGAAGCTACTTATAATCCTAATATGCCTTTTAAATGGA ATGCACAGAGAATTCAAACACTACCAAATCAGAATCAGTCGCAAACCCAGCCATTACTGAAGACTCCTCCTGCTGTACTTCAGCCAATTG CACCACAGACAACATTTGGTGTTCAGACTCAGCCCCAGCCCCAGTCACTGCTGCAGGCACAGATTTCAGCAGCTTCTATTACACCACTAT TGCAGACTCAACCACAGCCCTTATTACAGCAGCCTCAGCAAAAAGCTGGTTTATTGCAGCCTCCTGTTCGTATAGTTTCACAGCCACAAC CGGCACGACGATTAGATCCCCCATCCCGATTTTCAGGAAGAAATGACAGAGGGGATCAAGTGCCTAACAGAAAAGATGATCGAAGTCGTG AGAGAGAGAGAGAAAGACGTAGATCGAGAGAAAGATCACCTCAGAGGAAACGTTCCCGGGAAAGATCTCCACGAAGAGAGCGAGAGCGAT CACCTCGGAGAGTTCGACGTGTTGTTCCACGTTACACAGTTCAGTTTTCAAAGTTTTCTTTAGATTGTCCCAGTTGTGACATGATGGAAC TAAGGCGCCGTTATCAAAATTTGTATATACCTAGTGACTTTTTTGATGCTCAATTTACATGGGTGGATGCTTTCCCTTTGTCAAGACCAT TTCAGCTGGGAAATTACTGCAATTTTTATGTAATGCACAGAGAAGTAGAGTCCTTAGAAAAAAATATGGCCATTCTTGATCCACCAGATG CTGACCACTTATACAGTGCAAAGGTAATGCTGATGGCTAGCCCTAGTATGGAAGATTTATATCATAAGTCATGTGCTCTTGCTGAGGACC CACAAGAACTTCGAGATGGATTCCAACATCCTGCTAGACTTGTTAAGGAAACTAAATATATTTTGCCAACTTTGGAAAAAGAATTATTCT TGGCAGAGCACAGTGACCTTGAAGAAGGTGGACTGGACCTGACTGTGTCATTGAAACCAGTTAGTTTCTATATATCAGACAAAAAAGAAA TGCTTCAGCAGTGCTTCTGTATTATAGGAGAGAAAAAGTTACAGAAGATGCTTCCTGATGTGTTAAAGAACTGTTCAATAGAAGAAATTA AAAAACTATGCCAGGAACAGTTAGAGCTCCTGTCTGAAAAAAAAATTTTGAAGATTCTTGAGGGTGACAATGGAATGGACTCTGATATGG AAGAGGAAGCAGATGATGGTTCTAAGATGGGATCTGATTTAGTCAGTCAGCAAGACATCTGTATAGATTCTGCTTCATCCGTGAGAGAGA ATAAGCAACCTGAAGGTTTGGAATTAAAACAAGGAAAAGGGGAAGATAGTGATGTACTCAGTATAAATGCAGATGCTTATGACAGCGACA TAGAAGGCCCATGCAACGAAGAAGCAGCTGCTCCCGAGGCACCAGAAAATACAGTCCAAAGTGAAGCTGGTCAGATAGATGACCTGGAGA AAGACATTGAGAAAAGTGTGAATGAGATTCTAGGACTGGCAGAGTCTAGCCCAAACGAACCCAAAGCAGCCACCCTGGCTGTTCCTCCAC CAGAAGATGTTCAACCTTCTGCACAGCAACTGGAGCTGCTAGAACTTGAGATGAGGGCAAGAGCGATTAAAGCCCTAATGAAAGCTGGTG ATATAAAAAAGCCAGCCTAGGTATTTAACTTGATTTTGAATTTTAGGTATGTTTGAACAAAGCCACATCATTTAATTTTGTATCTAAAAT TTATTTGGGGTCTTATATGTTATTTCTCATGTAACCCTTATTAGGACTCATTTTAGCCCTAAATTACCTGTGGCTGTTTCTTTTTATTTT TTTGACTACTTTTATATTATAAATGTGTGTTACTGTCTTATGAATTCATGGCAATATAGTTGGATAGCCTGGATACTTTGTTAGATGAGT ATTTAGCTGTGTCTGCAAATCTTAAAAGCCATTAGCAAAGAGTCGTGGTATTTTTTTCTTTATTTTTAAATGTTTGGGCACCAAACCTAA AAGCAAAAGATTGACGAAGCATGTTTCTCTTAAGGCTACTTGTATTTTACAATACAATATTAAATTATTTAATTTGAGAAATTTAGTTTT GCTTATATGCACTTTTTAAATATATACTATTTTGAAGATTCCTTATGTAAATGCAAATTTCCTAGTTAAAACCGAATAACAGAGATCTGA AATGACTGAGAAAAACTTTTTTATTAAAGGAAGGAATTAATTTAAGGCAATTTTTAACTATGTAGAACTAATTGCCCATGTTTAATTATA GCAGACACGCCATTCTAACAGGTATTTGATACCATTGGATGCATTATTCTAGGTTTTTTCTTTAATAAAAATGGAACAAGTTTTCATTTA CATTCCAAGCTGTCAGGAAATGAAGAATATTTTATTATCTAGGATTTTATCTGATGTAGTTGCTTAAAGATCTGATGTGCTATAATTCCA TGAATCAGAAATAATAAAATGCTATCATTCTGGATCTGAAGACTTTTGATACTTTTTCAAAAGCAAAATTAATTTCAGGAACCTTTGATA AGTTGTTGTTATAATTAATCTAATTTTGTATAGTTTTTGTAAATAAATTACCATCCTTCCACAATTAGGGATGCTTTTATCCCCCCATCA CTAATTGCAGTTGTTTGATACCAAAATAAATTTACGTAGAGATCCTTAACTTAAAATAAATTAATTTTTTCAAAAAACATAAATCTGGAA CTGTTGTTTCTATATTTGATAACAAATACAGTATATTTTATTTATAAGCCATGGTCTACTGATACTGTATGAGGACTTTCCTTATATATA AAAGTTGCAGGGATTGTGTTTTATTAGCTGCTTTAATTATGTTAATTTTAGAGAGTTTTTAAATGGAAATAGAGGACATTTATGAAACGC TGGAATTGCAGTTACAAATTCTTTTTGTTGTTGTTGTTCCTGAACATGCCTTGGAATAATTCTACCATTTTTTCCCCCTCCATAAATCTT TCTAATAAAGCATAGAAAAAGCCTATATGATTTTAAATGCTTCTCTTAAGCTGGTAAACAGATTTGAGTTATGAGTTCATTGTTATTGCT TCAAGATGAAAAGACAGTGATATAATTTTTCTATTTCAACTTAAAAGTAATAGTTAATATGCTAAAGTAGTACAGAATAAACTTTATTGC TGCTTACTAACTACAAAATACTGTAGATGGCATCTGTATGATTAAACATATAAAGTAAAACAGGTCTGAGGGCTTTGTAGATGATTAAAG >13515_13515_1_CCAR1-CAAP1_CCAR1_chr10_70514584_ENST00000265872_CAAP1_chr9_26887511_ENST00000333916_length(amino acids)=746AA_BP=486 MAQFGGQKNPPWATQFTATAVSQPAALGVQQPSLLGASPTIYTQQTALAAAGLTTQTPANYQLTQTAALQQQAAAAAAALQQQYSQPQQA LYSVQQQLQQPQQTLLTQPAVALPTSLSLSTPQPTAQITVSYPTPRSSQQQTQPQKQRVFTGVVTKLHDTFGFVDEDVFFQLSAVKGKTP QVGDRVLVEATYNPNMPFKWNAQRIQTLPNQNQSQTQPLLKTPPAVLQPIAPQTTFGVQTQPQPQSLLQAQISAASITPLLQTQPQPLLQ QPQQKAGLLQPPVRIVSQPQPARRLDPPSRFSGRNDRGDQVPNRKDDRSRERERERRRSRERSPQRKRSRERSPRRERERSPRRVRRVVP RYTVQFSKFSLDCPSCDMMELRRRYQNLYIPSDFFDAQFTWVDAFPLSRPFQLGNYCNFYVMHREVESLEKNMAILDPPDADHLYSAKVM LMASPSMEDLYHKSCALAEDPQELRDGFQHPARLVKETKYILPTLEKELFLAEHSDLEEGGLDLTVSLKPVSFYISDKKEMLQQCFCIIG EKKLQKMLPDVLKNCSIEEIKKLCQEQLELLSEKKILKILEGDNGMDSDMEEEADDGSKMGSDLVSQQDICIDSASSVRENKQPEGLELK QGKGEDSDVLSINADAYDSDIEGPCNEEAAAPEAPENTVQSEAGQIDDLEKDIEKSVNEILGLAESSPNEPKAATLAVPPPEDVQPSAQQ -------------------------------------------------------------- >13515_13515_2_CCAR1-CAAP1_CCAR1_chr10_70514584_ENST00000535016_CAAP1_chr9_26887511_ENST00000333916_length(transcript)=3931nt_BP=1532nt GAAGTTGGCGCATGCGCCTAAAGCTGACGGGTTTGAAATGGCTTCGATGTTAGCCGGGACCCGACTCAGATCGATGCTATAGAAGACAAA CAAGGGAAGGTTTTTTTTCCTTTTGCATCATGGCTCAATTTGGAGGACAGAAGAATCCGCCATGGGCTACTCAGTTTACAGCCACTGCAG TATCACAGCCAGCTGCACTGGGTGTTCAACAGCCATCACTCCTTGGAGCATCTCCTACCATTTATACACAGCAAACTGCATTGGCAGCAG CAGGCCTTACCACACAAACTCCAGCAAACTATCAGTTAACACAAACTGCTGCATTGCAGCAACAAGCCGCAGCTGCAGCAGCTGCATTAC AACAGTTACAGCAACCCCAGCAAACCCTCTTAACACAGCCAGCTGTTGCACTGCCTACAAGCCTTAGCCTGTCTACTCCTCAGCCAACAG CACAAATAACTGTATCATATCCAACACCAAGGTCCAGTCAACAGCAAACCCAGCCTCAGAAGCAGCGTGTTTTCACAGGGGTGGTTACAA AACTACATGATACGTTTGGATTTGTGGATGAAGATGTATTCTTTCAGCTTAGTGCTGTCAAAGGGAAAACCCCCCAAGTAGGTGACAGAG TATTGGTTGAAGCTACTTATAATCCTAATATGCCTTTTAAATGGAATGCACAGAGAATTCAAACACTACCAAATCAGAATCAGTCGCAAA CCCAGCCATTACTGAAGACTCCTCCTGCTGTACTTCAGCCAATTGCACCACAGACAACATTTGGTGTTCAGACTCAGCCCCAGCCCCAGT CACTGCTGCAGGCACAGATTTCAGCAGCTTCTATTACACCACTATTGCAGACTCAACCACAGCCCTTATTACAGCAGCCTCAGCAAAAAG CTGGTTTATTGCAGCCTCCTGTTCGTATAGTTTCACAGCCACAACCGGCACGACGATTAGATCCCCCATCCCGATTTTCAGGAAGAAATG ACAGAGGGGATCAAGTGCCTAACAGAAAAGATGATCGAAGTCGTGAGAGAGAGAGAGAAAGACGTAGATCGAGAGAAAGATCACCTCAGA GGAAACGTTCCCGGGAAAGATCTCCACGAAGAGAGCGAGAGCGATCACCTCGGAGAGTTCGACGTGTTGTTCCACGTTACACAGTTCAGT TTTCAAAGTTTTCTTTAGATTGTCCCAGTTGTGACATGATGGAACTAAGGCGCCGTTATCAAAATTTGTATATACCTAGTGACTTTTTTG ATGCTCAATTTACATGGGTGGATGCTTTCCCTTTGTCAAGACCATTTCAGCTGGGAAATTACTGCAATTTTTATGTAATGCACAGAGAAG TAGAGTCCTTAGAAAAAAATATGGCCATTCTTGATCCACCAGATGCTGACCACTTATACAGTGCAAAGGTAATGCTGATGGCTAGCCCTA GTATGGAAGATTTATATCATAAGTCATGTGCTCTTGCTGAGGACCCACAAGAACTTCGAGATGGATTCCAACATCCTGCTAGACTTGTTA AGGAAACTAAATATATTTTGCCAACTTTGGAAAAAGAATTATTCTTGGCAGAGCACAGTGACCTTGAAGAAGGTGGACTGGACCTGACTG TGTCATTGAAACCAGTTAGTTTCTATATATCAGACAAAAAAGAAATGCTTCAGCAGTGCTTCTGTATTATAGGAGAGAAAAAGTTACAGA AGATGCTTCCTGATGTGTTAAAGAACTGTTCAATAGAAGAAATTAAAAAACTATGCCAGGAACAGTTAGAGCTCCTGTCTGAAAAAAAAA TTTTGAAGATTCTTGAGGGTGACAATGGAATGGACTCTGATATGGAAGAGGAAGCAGATGATGGTTCTAAGATGGGATCTGATTTAGTCA GTCAGCAAGACATCTGTATAGATTCTGCTTCATCCGTGAGAGAGAATAAGCAACCTGAAGGTTTGGAATTAAAACAAGGAAAAGGGGAAG ATAGTGATGTACTCAGTATAAATGCAGATGCTTATGACAGCGACATAGAAGGCCCATGCAACGAAGAAGCAGCTGCTCCCGAGGCACCAG AAAATACAGTCCAAAGTGAAGCTGGTCAGATAGATGACCTGGAGAAAGACATTGAGAAAAGTGTGAATGAGATTCTAGGACTGGCAGAGT CTAGCCCAAACGAACCCAAAGCAGCCACCCTGGCTGTTCCTCCACCAGAAGATGTTCAACCTTCTGCACAGCAACTGGAGCTGCTAGAAC TTGAGATGAGGGCAAGAGCGATTAAAGCCCTAATGAAAGCTGGTGATATAAAAAAGCCAGCCTAGGTATTTAACTTGATTTTGAATTTTA GGTATGTTTGAACAAAGCCACATCATTTAATTTTGTATCTAAAATTTATTTGGGGTCTTATATGTTATTTCTCATGTAACCCTTATTAGG ACTCATTTTAGCCCTAAATTACCTGTGGCTGTTTCTTTTTATTTTTTTGACTACTTTTATATTATAAATGTGTGTTACTGTCTTATGAAT TCATGGCAATATAGTTGGATAGCCTGGATACTTTGTTAGATGAGTATTTAGCTGTGTCTGCAAATCTTAAAAGCCATTAGCAAAGAGTCG TGGTATTTTTTTCTTTATTTTTAAATGTTTGGGCACCAAACCTAAAAGCAAAAGATTGACGAAGCATGTTTCTCTTAAGGCTACTTGTAT TTTACAATACAATATTAAATTATTTAATTTGAGAAATTTAGTTTTGCTTATATGCACTTTTTAAATATATACTATTTTGAAGATTCCTTA TGTAAATGCAAATTTCCTAGTTAAAACCGAATAACAGAGATCTGAAATGACTGAGAAAAACTTTTTTATTAAAGGAAGGAATTAATTTAA GGCAATTTTTAACTATGTAGAACTAATTGCCCATGTTTAATTATAGCAGACACGCCATTCTAACAGGTATTTGATACCATTGGATGCATT ATTCTAGGTTTTTTCTTTAATAAAAATGGAACAAGTTTTCATTTACATTCCAAGCTGTCAGGAAATGAAGAATATTTTATTATCTAGGAT TTTATCTGATGTAGTTGCTTAAAGATCTGATGTGCTATAATTCCATGAATCAGAAATAATAAAATGCTATCATTCTGGATCTGAAGACTT TTGATACTTTTTCAAAAGCAAAATTAATTTCAGGAACCTTTGATAAGTTGTTGTTATAATTAATCTAATTTTGTATAGTTTTTGTAAATA AATTACCATCCTTCCACAATTAGGGATGCTTTTATCCCCCCATCACTAATTGCAGTTGTTTGATACCAAAATAAATTTACGTAGAGATCC TTAACTTAAAATAAATTAATTTTTTCAAAAAACATAAATCTGGAACTGTTGTTTCTATATTTGATAACAAATACAGTATATTTTATTTAT AAGCCATGGTCTACTGATACTGTATGAGGACTTTCCTTATATATAAAAGTTGCAGGGATTGTGTTTTATTAGCTGCTTTAATTATGTTAA TTTTAGAGAGTTTTTAAATGGAAATAGAGGACATTTATGAAACGCTGGAATTGCAGTTACAAATTCTTTTTGTTGTTGTTGTTCCTGAAC ATGCCTTGGAATAATTCTACCATTTTTTCCCCCTCCATAAATCTTTCTAATAAAGCATAGAAAAAGCCTATATGATTTTAAATGCTTCTC TTAAGCTGGTAAACAGATTTGAGTTATGAGTTCATTGTTATTGCTTCAAGATGAAAAGACAGTGATATAATTTTTCTATTTCAACTTAAA AGTAATAGTTAATATGCTAAAGTAGTACAGAATAAACTTTATTGCTGCTTACTAACTACAAAATACTGTAGATGGCATCTGTATGATTAA >13515_13515_2_CCAR1-CAAP1_CCAR1_chr10_70514584_ENST00000535016_CAAP1_chr9_26887511_ENST00000333916_length(amino acids)=731AA_BP=471 MAQFGGQKNPPWATQFTATAVSQPAALGVQQPSLLGASPTIYTQQTALAAAGLTTQTPANYQLTQTAALQQQAAAAAAALQQLQQPQQTL LTQPAVALPTSLSLSTPQPTAQITVSYPTPRSSQQQTQPQKQRVFTGVVTKLHDTFGFVDEDVFFQLSAVKGKTPQVGDRVLVEATYNPN MPFKWNAQRIQTLPNQNQSQTQPLLKTPPAVLQPIAPQTTFGVQTQPQPQSLLQAQISAASITPLLQTQPQPLLQQPQQKAGLLQPPVRI VSQPQPARRLDPPSRFSGRNDRGDQVPNRKDDRSRERERERRRSRERSPQRKRSRERSPRRERERSPRRVRRVVPRYTVQFSKFSLDCPS CDMMELRRRYQNLYIPSDFFDAQFTWVDAFPLSRPFQLGNYCNFYVMHREVESLEKNMAILDPPDADHLYSAKVMLMASPSMEDLYHKSC ALAEDPQELRDGFQHPARLVKETKYILPTLEKELFLAEHSDLEEGGLDLTVSLKPVSFYISDKKEMLQQCFCIIGEKKLQKMLPDVLKNC SIEEIKKLCQEQLELLSEKKILKILEGDNGMDSDMEEEADDGSKMGSDLVSQQDICIDSASSVRENKQPEGLELKQGKGEDSDVLSINAD AYDSDIEGPCNEEAAAPEAPENTVQSEAGQIDDLEKDIEKSVNEILGLAESSPNEPKAATLAVPPPEDVQPSAQQLELLELEMRARAIKA -------------------------------------------------------------- >13515_13515_3_CCAR1-CAAP1_CCAR1_chr10_70514584_ENST00000543719_CAAP1_chr9_26887511_ENST00000333916_length(transcript)=3925nt_BP=1526nt TGCGCCTAAAGCTGACGGGTTTGAAATGGCTTCGATGTTAGCCGGGACCCGACTCAGGTGAAGATCGATGCTATAGAAGACAAACAAGGG AAGGTTTTTTTTCCTTTTGCATCATGGCTCAATTTGGAGGACAGAAGAATCCGCCATGGGCTACTCAGTTTACAGCCACTGCAGTATCAC AGCCAGCTGCACTGGGTGTTCAACAGCCATCACTCCTTGGAGCATCTCCTACCATTTATACACAGCAAACTGCATTGGCAGCAGCAGGCC TTACCACACAAACTCCAGCAAACTATCAGTTAACACAAACTGCTGCATTGCAGCAACAAGCCGCAGCTGCAGCAGCTGCATTACAACAGT TACAGCAACCCCAGCAAACCCTCTTAACACAGCCAGCTGTTGCACTGCCTACAAGCCTTAGCCTGTCTACTCCTCAGCCAACAGCACAAA TAACTGTATCATATCCAACACCAAGGTCCAGTCAACAGCAAACCCAGCCTCAGAAGCAGCGTGTTTTCACAGGGGTGGTTACAAAACTAC ATGATACGTTTGGATTTGTGGATGAAGATGTATTCTTTCAGCTTAGTGCTGTCAAAGGGAAAACCCCCCAAGTAGGTGACAGAGTATTGG TTGAAGCTACTTATAATCCTAATATGCCTTTTAAATGGAATGCACAGAGAATTCAAACACTACCAAATCAGAATCAGTCGCAAACCCAGC CATTACTGAAGACTCCTCCTGCTGTACTTCAGCCAATTGCACCACAGACAACATTTGGTGTTCAGACTCAGCCCCAGCCCCAGTCACTGC TGCAGGCACAGATTTCAGCAGCTTCTATTACACCACTATTGCAGACTCAACCACAGCCCTTATTACAGCAGCCTCAGCAAAAAGCTGGTT TATTGCAGCCTCCTGTTCGTATAGTTTCACAGCCACAACCGGCACGACGATTAGATCCCCCATCCCGATTTTCAGGAAGAAATGACAGAG GGGATCAAGTGCCTAACAGAAAAGATGATCGAAGTCGTGAGAGAGAGAGAGAAAGACGTAGATCGAGAGAAAGATCACCTCAGAGGAAAC GTTCCCGGGAAAGATCTCCACGAAGAGAGCGAGAGCGATCACCTCGGAGAGTTCGACGTGTTGTTCCACGTTACACAGTTCAGTTTTCAA AGTTTTCTTTAGATTGTCCCAGTTGTGACATGATGGAACTAAGGCGCCGTTATCAAAATTTGTATATACCTAGTGACTTTTTTGATGCTC AATTTACATGGGTGGATGCTTTCCCTTTGTCAAGACCATTTCAGCTGGGAAATTACTGCAATTTTTATGTAATGCACAGAGAAGTAGAGT CCTTAGAAAAAAATATGGCCATTCTTGATCCACCAGATGCTGACCACTTATACAGTGCAAAGGTAATGCTGATGGCTAGCCCTAGTATGG AAGATTTATATCATAAGTCATGTGCTCTTGCTGAGGACCCACAAGAACTTCGAGATGGATTCCAACATCCTGCTAGACTTGTTAAGGAAA CTAAATATATTTTGCCAACTTTGGAAAAAGAATTATTCTTGGCAGAGCACAGTGACCTTGAAGAAGGTGGACTGGACCTGACTGTGTCAT TGAAACCAGTTAGTTTCTATATATCAGACAAAAAAGAAATGCTTCAGCAGTGCTTCTGTATTATAGGAGAGAAAAAGTTACAGAAGATGC TTCCTGATGTGTTAAAGAACTGTTCAATAGAAGAAATTAAAAAACTATGCCAGGAACAGTTAGAGCTCCTGTCTGAAAAAAAAATTTTGA AGATTCTTGAGGGTGACAATGGAATGGACTCTGATATGGAAGAGGAAGCAGATGATGGTTCTAAGATGGGATCTGATTTAGTCAGTCAGC AAGACATCTGTATAGATTCTGCTTCATCCGTGAGAGAGAATAAGCAACCTGAAGGTTTGGAATTAAAACAAGGAAAAGGGGAAGATAGTG ATGTACTCAGTATAAATGCAGATGCTTATGACAGCGACATAGAAGGCCCATGCAACGAAGAAGCAGCTGCTCCCGAGGCACCAGAAAATA CAGTCCAAAGTGAAGCTGGTCAGATAGATGACCTGGAGAAAGACATTGAGAAAAGTGTGAATGAGATTCTAGGACTGGCAGAGTCTAGCC CAAACGAACCCAAAGCAGCCACCCTGGCTGTTCCTCCACCAGAAGATGTTCAACCTTCTGCACAGCAACTGGAGCTGCTAGAACTTGAGA TGAGGGCAAGAGCGATTAAAGCCCTAATGAAAGCTGGTGATATAAAAAAGCCAGCCTAGGTATTTAACTTGATTTTGAATTTTAGGTATG TTTGAACAAAGCCACATCATTTAATTTTGTATCTAAAATTTATTTGGGGTCTTATATGTTATTTCTCATGTAACCCTTATTAGGACTCAT TTTAGCCCTAAATTACCTGTGGCTGTTTCTTTTTATTTTTTTGACTACTTTTATATTATAAATGTGTGTTACTGTCTTATGAATTCATGG CAATATAGTTGGATAGCCTGGATACTTTGTTAGATGAGTATTTAGCTGTGTCTGCAAATCTTAAAAGCCATTAGCAAAGAGTCGTGGTAT TTTTTTCTTTATTTTTAAATGTTTGGGCACCAAACCTAAAAGCAAAAGATTGACGAAGCATGTTTCTCTTAAGGCTACTTGTATTTTACA ATACAATATTAAATTATTTAATTTGAGAAATTTAGTTTTGCTTATATGCACTTTTTAAATATATACTATTTTGAAGATTCCTTATGTAAA TGCAAATTTCCTAGTTAAAACCGAATAACAGAGATCTGAAATGACTGAGAAAAACTTTTTTATTAAAGGAAGGAATTAATTTAAGGCAAT TTTTAACTATGTAGAACTAATTGCCCATGTTTAATTATAGCAGACACGCCATTCTAACAGGTATTTGATACCATTGGATGCATTATTCTA GGTTTTTTCTTTAATAAAAATGGAACAAGTTTTCATTTACATTCCAAGCTGTCAGGAAATGAAGAATATTTTATTATCTAGGATTTTATC TGATGTAGTTGCTTAAAGATCTGATGTGCTATAATTCCATGAATCAGAAATAATAAAATGCTATCATTCTGGATCTGAAGACTTTTGATA CTTTTTCAAAAGCAAAATTAATTTCAGGAACCTTTGATAAGTTGTTGTTATAATTAATCTAATTTTGTATAGTTTTTGTAAATAAATTAC CATCCTTCCACAATTAGGGATGCTTTTATCCCCCCATCACTAATTGCAGTTGTTTGATACCAAAATAAATTTACGTAGAGATCCTTAACT TAAAATAAATTAATTTTTTCAAAAAACATAAATCTGGAACTGTTGTTTCTATATTTGATAACAAATACAGTATATTTTATTTATAAGCCA TGGTCTACTGATACTGTATGAGGACTTTCCTTATATATAAAAGTTGCAGGGATTGTGTTTTATTAGCTGCTTTAATTATGTTAATTTTAG AGAGTTTTTAAATGGAAATAGAGGACATTTATGAAACGCTGGAATTGCAGTTACAAATTCTTTTTGTTGTTGTTGTTCCTGAACATGCCT TGGAATAATTCTACCATTTTTTCCCCCTCCATAAATCTTTCTAATAAAGCATAGAAAAAGCCTATATGATTTTAAATGCTTCTCTTAAGC TGGTAAACAGATTTGAGTTATGAGTTCATTGTTATTGCTTCAAGATGAAAAGACAGTGATATAATTTTTCTATTTCAACTTAAAAGTAAT AGTTAATATGCTAAAGTAGTACAGAATAAACTTTATTGCTGCTTACTAACTACAAAATACTGTAGATGGCATCTGTATGATTAAACATAT >13515_13515_3_CCAR1-CAAP1_CCAR1_chr10_70514584_ENST00000543719_CAAP1_chr9_26887511_ENST00000333916_length(amino acids)=731AA_BP=471 MAQFGGQKNPPWATQFTATAVSQPAALGVQQPSLLGASPTIYTQQTALAAAGLTTQTPANYQLTQTAALQQQAAAAAAALQQLQQPQQTL LTQPAVALPTSLSLSTPQPTAQITVSYPTPRSSQQQTQPQKQRVFTGVVTKLHDTFGFVDEDVFFQLSAVKGKTPQVGDRVLVEATYNPN MPFKWNAQRIQTLPNQNQSQTQPLLKTPPAVLQPIAPQTTFGVQTQPQPQSLLQAQISAASITPLLQTQPQPLLQQPQQKAGLLQPPVRI VSQPQPARRLDPPSRFSGRNDRGDQVPNRKDDRSRERERERRRSRERSPQRKRSRERSPRRERERSPRRVRRVVPRYTVQFSKFSLDCPS CDMMELRRRYQNLYIPSDFFDAQFTWVDAFPLSRPFQLGNYCNFYVMHREVESLEKNMAILDPPDADHLYSAKVMLMASPSMEDLYHKSC ALAEDPQELRDGFQHPARLVKETKYILPTLEKELFLAEHSDLEEGGLDLTVSLKPVSFYISDKKEMLQQCFCIIGEKKLQKMLPDVLKNC SIEEIKKLCQEQLELLSEKKILKILEGDNGMDSDMEEEADDGSKMGSDLVSQQDICIDSASSVRENKQPEGLELKQGKGEDSDVLSINAD AYDSDIEGPCNEEAAAPEAPENTVQSEAGQIDDLEKDIEKSVNEILGLAESSPNEPKAATLAVPPPEDVQPSAQQLELLELEMRARAIKA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CCAR1-CAAP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 1_249 | 486.0 | 1151.0 | AR |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 1_249 | 471.0 | 1136.0 | AR |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 1_249 | 471.0 | 1136.0 | AR |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 643_1150 | 486.0 | 1151.0 | GATA1 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 643_1150 | 471.0 | 1136.0 | GATA1 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 643_1150 | 471.0 | 1136.0 | GATA1 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000265872 | + | 12 | 25 | 203_660 | 486.0 | 1151.0 | GATA2 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000535016 | + | 11 | 24 | 203_660 | 471.0 | 1136.0 | GATA2 |
| Hgene | CCAR1 | chr10:70514584 | chr9:26887511 | ENST00000543719 | + | 11 | 24 | 203_660 | 471.0 | 1136.0 | GATA2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CCAR1-CAAP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CCAR1-CAAP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
