
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CCAR1-DDX21 (FusionGDB2 ID:13517) |
Fusion Gene Summary for CCAR1-DDX21 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CCAR1-DDX21 | Fusion gene ID: 13517 | Hgene | Tgene | Gene symbol | CCAR1 | DDX21 | Gene ID | 55749 | 54606 |
| Gene name | cell division cycle and apoptosis regulator 1 | DEAD-box helicase 56 | |
| Synonyms | - | DDX21|DDX26|NOH61 | |
| Cytomap | 10q21.3 | 7p13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cell division cycle and apoptosis regulator protein 1cell cycle and apoptosis regulatory protein 1death inducer with SAP domain | probable ATP-dependent RNA helicase DDX5661-kd nucleolar helicaseATP-dependent 61 kDa nucleolar RNA helicaseDEAD (Asp-Glu-Ala-Asp) box helicase 56DEAD (Asp-Glu-Ala-Asp) box polypeptide 56DEAD box protein 21DEAD box protein 56DEAD-box RNA helicasen | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8IX12 | Q9NR30 | |
| Ensembl transtripts involved in fusion gene | ENST00000265872, ENST00000535016, ENST00000543719, ENST00000483264, | ENST00000354185, | |
| Fusion gene scores | * DoF score | 13 X 11 X 8=1144 | 36 X 11 X 14=5544 |
| # samples | 17 | 37 | |
| ** MAII score | log2(17/1144*10)=-2.75048040064069 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(37/5544*10)=-3.9053300816209 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CCAR1 [Title/Abstract] AND DDX21 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CCAR1(70482334)-DDX21(70723046), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CCAR1 | GO:0043065 | positive regulation of apoptotic process | 12816952 |
 Fusion gene breakpoints across CCAR1 (5'-gene) Fusion gene breakpoints across CCAR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
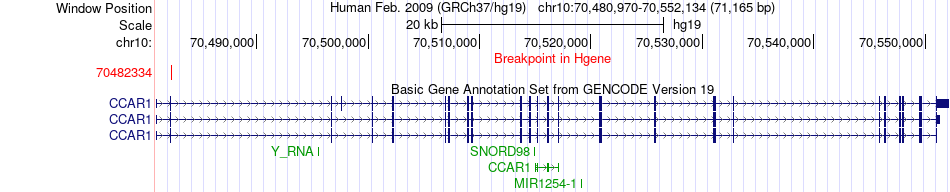 |
 Fusion gene breakpoints across DDX21 (3'-gene) Fusion gene breakpoints across DDX21 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
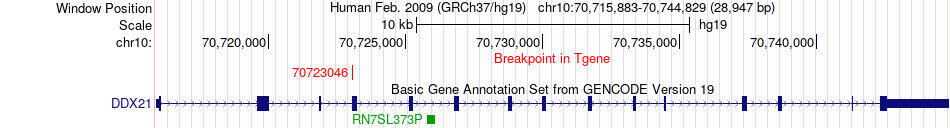 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-VQ-A8PE | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + |
Top |
Fusion Gene ORF analysis for CCAR1-DDX21 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000265872 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + |
| In-frame | ENST00000535016 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + |
| In-frame | ENST00000543719 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + |
| intron-3CDS | ENST00000483264 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535016 | CCAR1 | chr10 | 70482334 | + | ENST00000354185 | DDX21 | chr10 | 70723046 | + | 4198 | 192 | 119 | 1936 | 605 |
| ENST00000265872 | CCAR1 | chr10 | 70482334 | + | ENST00000354185 | DDX21 | chr10 | 70723046 | + | 4198 | 192 | 119 | 1936 | 605 |
| ENST00000543719 | CCAR1 | chr10 | 70482334 | + | ENST00000354185 | DDX21 | chr10 | 70723046 | + | 4192 | 186 | 113 | 1930 | 605 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535016 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + | 0.000734407 | 0.9992656 |
| ENST00000265872 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + | 0.000734407 | 0.9992656 |
| ENST00000543719 | ENST00000354185 | CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + | 0.000725499 | 0.99927455 |
Top |
Fusion Genomic Features for CCAR1-DDX21 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + | 0.000552974 | 0.99944705 |
| CCAR1 | chr10 | 70482334 | + | DDX21 | chr10 | 70723046 | + | 0.000552974 | 0.99944705 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
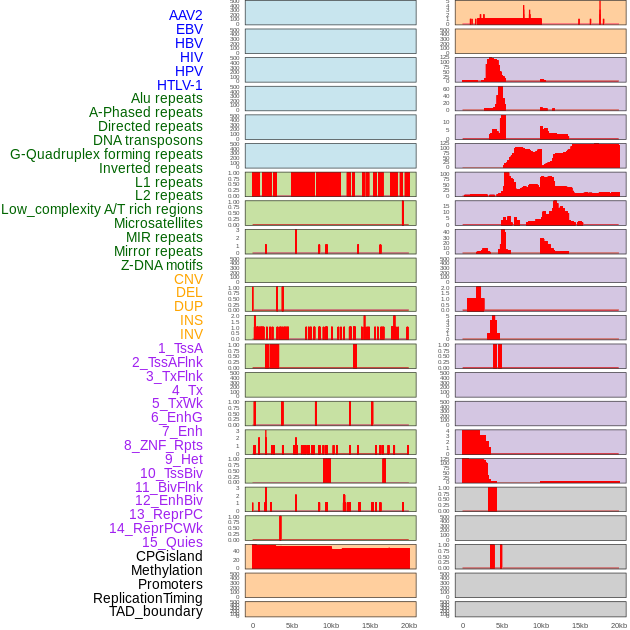 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
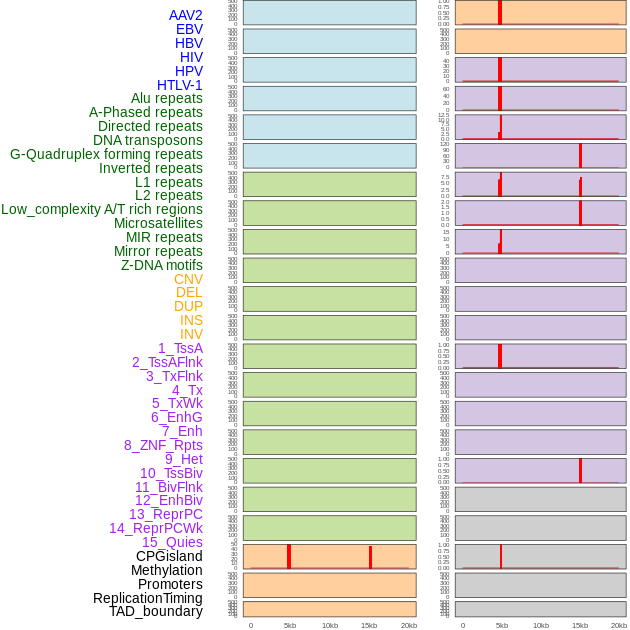 |
Top |
Fusion Protein Features for CCAR1-DDX21 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:70482334/chr10:70723046) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CCAR1 | DDX21 |
| FUNCTION: Associates with components of the Mediator and p160 coactivator complexes that play a role as intermediaries transducing regulatory signals from upstream transcriptional activator proteins to basal transcription machinery at the core promoter. Recruited to endogenous nuclear receptor target genes in response to the appropriate hormone. Also functions as a p53 coactivator. May thus play an important role in transcriptional regulation (By similarity). May be involved in apoptosis signaling in the presence of the reinoid CD437. Apoptosis induction involves sequestration of 14-3-3 protein(s) and mediated altered expression of multiple cell cycle regulatory genes including MYC, CCNB1 and CDKN1A. Plays a role in cell cycle progression and/or cell proliferation (PubMed:12816952). In association with CALCOCO1 enhances GATA1- and MED1-mediated transcriptional activation from the gamma-globin promoter during erythroid differentiation of K562 erythroleukemia cells (PubMed:24245781). Can act as a both a coactivator and corepressor of AR-mediated transcription. Contributes to chromatin looping and AR transcription complex assembly by stabilizing AR-GATA2 association on chromatin and facilitating MED1 and RNA polymerase II recruitment to AR-binding sites. May play an important role in the growth and tumorigenesis of prostate cancer cells (PubMed:23887938). {ECO:0000250|UniProtKB:Q8CH18, ECO:0000269|PubMed:12816952, ECO:0000269|PubMed:23887938, ECO:0000269|PubMed:24245781}. | FUNCTION: RNA helicase that acts as a sensor of the transcriptional status of both RNA polymerase (Pol) I and II: promotes ribosomal RNA (rRNA) processing and transcription from polymerase II (Pol II) (PubMed:25470060, PubMed:28790157). Binds various RNAs, such as rRNAs, snoRNAs, 7SK and, at lower extent, mRNAs (PubMed:25470060). In the nucleolus, localizes to rDNA locus, where it directly binds rRNAs and snoRNAs, and promotes rRNA transcription, processing and modification. Required for rRNA 2'-O-methylation, possibly by promoting the recruitment of late-acting snoRNAs SNORD56 and SNORD58 with pre-ribosomal complexes (PubMed:25470060, PubMed:25477391). In the nucleoplasm, binds 7SK RNA and is recruited to the promoters of Pol II-transcribed genes: acts by facilitating the release of P-TEFb from inhibitory 7SK snRNP in a manner that is dependent on its helicase activity, thereby promoting transcription of its target genes (PubMed:25470060). Functions as cofactor for JUN-activated transcription: required for phosphorylation of JUN at 'Ser-77' (PubMed:11823437, PubMed:25260534). Can unwind double-stranded RNA (helicase) and can fold or introduce a secondary structure to a single-stranded RNA (foldase) (PubMed:9461305). Together with SIRT7, required to prevent R-loop-associated DNA damage and transcription-associated genomic instability: deacetylation by SIRT7 activates the helicase activity, thereby overcoming R-loop-mediated stalling of RNA polymerases (PubMed:28790157). Involved in rRNA processing (PubMed:14559904, PubMed:18180292). May bind to specific miRNA hairpins (PubMed:28431233). Component of a multi-helicase-TICAM1 complex that acts as a cytoplasmic sensor of viral double-stranded RNA (dsRNA) and plays a role in the activation of a cascade of antiviral responses including the induction of proinflammatory cytokines via the adapter molecule TICAM1 (By similarity). {ECO:0000250|UniProtKB:Q9JIK5, ECO:0000269|PubMed:11823437, ECO:0000269|PubMed:14559904, ECO:0000269|PubMed:18180292, ECO:0000269|PubMed:25260534, ECO:0000269|PubMed:25470060, ECO:0000269|PubMed:25477391, ECO:0000269|PubMed:28431233, ECO:0000269|PubMed:28790157, ECO:0000269|PubMed:9461305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 217_396 | 202 | 784.0 | Domain | Helicase ATP-binding | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 429_573 | 202 | 784.0 | Domain | Helicase C-terminal | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 339_342 | 202 | 784.0 | Motif | DEAD box | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 230_237 | 202 | 784.0 | Nucleotide binding | ATP | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 724_748 | 202 | 784.0 | Region | Note=3 X 5 AA repeats | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 724_728 | 202 | 784.0 | Repeat | Note=1 | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 734_738 | 202 | 784.0 | Repeat | Note=2 | |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 744_748 | 202 | 784.0 | Repeat | Note=3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 1033_1114 | 24 | 1151.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 594_618 | 24 | 1151.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 1033_1114 | 24 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 594_618 | 24 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 1033_1114 | 24 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 594_618 | 24 | 1136.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 293_361 | 24 | 1151.0 | Compositional bias | Note=Arg-rich |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 673_889 | 24 | 1151.0 | Compositional bias | Note=Glu-rich |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 73_79 | 24 | 1151.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 293_361 | 24 | 1136.0 | Compositional bias | Note=Arg-rich |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 673_889 | 24 | 1136.0 | Compositional bias | Note=Glu-rich |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 73_79 | 24 | 1136.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 293_361 | 24 | 1136.0 | Compositional bias | Note=Arg-rich |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 673_889 | 24 | 1136.0 | Compositional bias | Note=Glu-rich |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 73_79 | 24 | 1136.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 636_670 | 24 | 1151.0 | Domain | SAP |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 636_670 | 24 | 1136.0 | Domain | SAP |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 636_670 | 24 | 1136.0 | Domain | SAP |
| Tgene | DDX21 | chr10:70482334 | chr10:70723046 | ENST00000354185 | 2 | 15 | 186_214 | 202 | 784.0 | Motif | Q motif |
Top |
Fusion Gene Sequence for CCAR1-DDX21 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >13517_13517_1_CCAR1-DDX21_CCAR1_chr10_70482334_ENST00000265872_DDX21_chr10_70723046_ENST00000354185_length(transcript)=4198nt_BP=192nt GAAGTTGGCGCATGCGCCTAAAGCTGACGGGTTTGAAATGGCTTCGATGTTAGCCGGGACCCGACTCAGATCGATGCTATAGAAGACAAA CAAGGGAAGGTTTTTTTTCCTTTTGCATCATGGCTCAATTTGGAGGACAGAAGAATCCGCCATGGGCTACTCAGTTTACAGCCACTGCAG TATCACAGCCAGGCCGAGGAGTGACCTTCCTATTTCCTATACAAGCAAAGACATTCCATCATGTTTACAGCGGGAAGGACTTAATTGCAC AGGCACGGACAGGAACTGGGAAGACATTCTCCTTTGCCATCCCTTTGATTGAGAAACTTCATGGGGAACTGCAAGACAGGAAGAGAGGCC GTGCCCCTCAGGTACTGGTTCTTGCACCTACAAGAGAGTTGGCAAATCAAGTAAGCAAAGACTTCAGTGACATCACAAAAAAGCTGTCAG TGGCTTGTTTTTATGGTGGAACTCCCTATGGAGGTCAATTTGAACGCATGAGGAATGGGATTGATATCCTGGTTGGAACACCAGGTCGTA TCAAAGACCACATACAGAATGGCAAACTAGATCTCACCAAACTTAAGCATGTTGTCCTGGATGAAGTGGACCAGATGTTGGATATGGGAT TTGCTGATCAAGTGGAAGAGATTTTAAGTGTGGCATACAAGAAAGATTCTGAAGACAATCCCCAAACATTGCTTTTTTCTGCAACTTGCC CTCATTGGGTATTTAATGTTGCCAAGAAATACATGAAATCTACATATGAACAGGTGGACCTGATTGGTAAAAAGACTCAGAAAACGGCAA TAACTGTGGAGCATCTGGCTATTAAGTGCCACTGGACTCAGAGGGCAGCAGTTATTGGGGATGTCATCCGAGTATATAGTGGTCATCAAG GACGCACTATCATCTTTTGTGAAACCAAGAAAGAAGCCCAGGAGCTGTCCCAGAATTCAGCTATAAAGCAGGATGCTCAGTCCTTGCATG GAGACATTCCACAGAAGCAAAGGGAAATCACCCTGAAAGGTTTTAGAAATGGTAGTTTTGGAGTTTTGGTGGCAACCAATGTTGCTGCAC GTGGGTTAGACATCCCTGAGGTTGATTTGGTTATACAAAGCTCTCCACCAAAGGATGTAGAGTCCTACATTCATCGATCCGGGCGGACAG GCAGAGCTGGAAGGACGGGGGTGTGCATCTGCTTTTATCAGCACAAGGAAGAATATCAGTTAGTACAAGTGGAGCAAAAAGCGGGAATTA AGTTCAAACGAATAGGTGTTCCTTCTGCAACAGAAATAATAAAAGCTTCCAGCAAAGATGCCATCAGGCTTTTGGATTCCGTGCCTCCCA CTGCCATTAGTCACTTCAAACAATCAGCTGAGAAGCTGATAGAGGAGAAGGGAGCTGTGGAAGCTCTGGCAGCAGCACTGGCCCATATTT CAGGTGCCACGTCCGTAGACCAGCGCTCCTTGATCAACTCAAATGTGGGTTTTGTGACCATGATCTTGCAGTGCTCAATTGAAATGCCAA ATATTAGTTATGCTTGGAAAGAACTTAAAGAGCAGCTGGGCGAGGAGATTGATTCCAAAGTGAAGGGAATGGTTTTTCTCAAAGGAAAGC TGGGTGTTTGCTTTGATGTACCTACCGCATCAGTAACAGAAATACAGGAGAAATGGCATGATTCACGACGCTGGCAGCTCTCTGTGGCCA CAGAGCAACCAGAACTGGAAGGACCACGGGAAGGATATGGAGGCTTCAGGGGACAGCGGGAAGGCAGTCGAGGCTTCAGGGGACAGCGGG ACGGAAACAGAAGATTCAGAGGACAGCGGGAAGGCAGTAGAGGCCCGAGAGGACAGCGATCAGGAGGTGGCAACAAAAGTAACAGATCCC AAAACAAAGGCCAGAAGCGGAGTTTCAGTAAAGCATTTGGTCAATAATTAGAAATAGAAGATTTATATAGCAAAAAGAGAATGATGTTTG GCAATATAGAACTGAACATTATTTTTCATGCAAAGTTAAAAGCACATTGTGCCTCCTTTTGACCACTTGCCAAGTCCCTGTCTCTTTCAG ACACAGACAAGCTTCATTTAAATTATTTCATCTGATCATTATCATTTATAACTTTATTGTTACTTCTTCATCAGTTTTTCCTTTTGAAAG GTGTATGAATTCATTACATTTTTATTCTAATGTATTATCTGTAGATTAGAAGATAAAATCAAGCATGTATCTGCCTATACTTTGTGAGTT CACCTGTCTTTATACTCAAAAGTGTCCCTTAATAGTGTCCTTCCCTGAAATAAATACCTAAGGGAGTGTAACAGTCTCTGGAGGACCACT TTGAGCCTTTGGAAGTTAAGGTTTCCTCAGCCACCTGCCGAACAGTTTCTCATGTGGTCCTATTATTTGTCTACTGAGACTTAATACTGA GCAATGTTTTGAAACAAGATTTCAAACTAATCTGGGTTGTAATACAGTTTATACCAGTGTATGCTCTAGACTTGGAAGATGTAGTATGTT TGATGTGGATTACCTATACTTATGTTCGTTTTGATACATTTTTAGCTTCTCATTATAAGGTGATTCATGCTTTAGTGAATTCTTCATAGA TAGTATATATAAAAGTACATTTTAATAGAAAGCCAGGGTTTTAAGGAATTTCACATGTATAAGGTGGCTCCATAGCTTTATTTGTAAGTA GGCTGGATAAATGGTGCTTAAATGGTAATGTACTCCACTTCTTCCTATTGGAAGATTAACATTATTTACCAAGAAGGACTTAAGGGAGTA GGGGGCGCAGATTAGCATTGCTCAAGAGTATGTAAAAAAAAAAAAAAAAAAAAAGAACCAAACCACTGGAAATAATCAAATGCAAAAAGG TAACAAATTCATAACTGGAAAGCAAAGAGAAGAACAAGTATGATTTGGATGATAAAGCATTGTTTTAATGGTGAAAACTTCACAGATCAC TAATGTTTCTAGAGGTTAACTTCAAGTGGGCAAGCTGGGGTTTTTAGGTAGTCAGTGGCCTAGTTCCTAAAGCCACAGTATAGGATCTGT TAAACTGAATGTCTGTTGAAAGTTTGTTTTAGCTGCTTGGAGGCTTCCTTTTAAGACAAACTGTATGTGATTAAGTTGTTTTGAGGGAAC TGAAGAACCTGATGTAGCCCCTGGCCAGATAACTGCCTGATTTCTCAGATATTATTTCTCTGGGAAACATTCTACATAGCACAGGAGCTT AAGAGTGGCATTATCTTCTCGCCTTAATTTCCAGAGATTATTTCTGTACTGAGAATCCTGGAACTACTATGCTAGGAAATTTAAAGCTGC ATGGTCTGTCTTGTTTTCATTTAATTATTGTGAATACCTAGAATCTTTCTTGGTCCTGATTTCTCTTGCTTAATCCAGTCTTTATCTCTA ACTGCCCCTTATTTGATCACCATGTACTAGGAGCTCTGATAGCCAGCTCAGCTCCTAATCCTTGAGGCAACATTCTTTTTCTATTTGAAC TTCAGTTCTGTCCTTGAATCCCGACTAGATATTTCTTGCCCTCTGGTCTCAGAATTCTCTTGGCTTTTATTCCTTGATCCACTTGCCAGT TTTATCACTTTACCCTTGTTCCTCATGGCTTCCCATCAAGCCATGGGTATTAGGTGACAGTGTAATTTATTAGATTCTGGTTTTGCCCAA ATACTGGGCATGCTTTAATAATAACTGAACCATTTCATTATTTGGATAGGCATGGGTACCTTATCAAGCAGATTAAAAGGATATGGTACC CGTCCTTTAGAAAAGAACAGCTAAAACCTTGTTGTGGATTATGGATTTAGCCTAAAGAAAAATAATCTGGCATAAATTAAGAGTAAGAGA GAAGATTAATAGAAATTTCACTTCACATAACTTAAAACATGGCTATTTCAATAAAGGACTAAAGTTTCTCCTGGATCCCAGAATTCAACC TGTATTTATAAATGTATAATGTATTTAGCTACTTTTTGGTTTAAATGAACTTGTTGGGTTAGCTTGGTAAATGTTATAATTTTTACTATT TTCTACAAAGAAAATATTTTCTAATTTAAGTTGGAGCTATCTGTGCAGCAGTTTCTCTACAGTTGTGCATAAATGTTTTTACTATAAAAT >13517_13517_1_CCAR1-DDX21_CCAR1_chr10_70482334_ENST00000265872_DDX21_chr10_70723046_ENST00000354185_length(amino acids)=605AA_BP=18 MAQFGGQKNPPWATQFTATAVSQPGRGVTFLFPIQAKTFHHVYSGKDLIAQARTGTGKTFSFAIPLIEKLHGELQDRKRGRAPQVLVLAP TRELANQVSKDFSDITKKLSVACFYGGTPYGGQFERMRNGIDILVGTPGRIKDHIQNGKLDLTKLKHVVLDEVDQMLDMGFADQVEEILS VAYKKDSEDNPQTLLFSATCPHWVFNVAKKYMKSTYEQVDLIGKKTQKTAITVEHLAIKCHWTQRAAVIGDVIRVYSGHQGRTIIFCETK KEAQELSQNSAIKQDAQSLHGDIPQKQREITLKGFRNGSFGVLVATNVAARGLDIPEVDLVIQSSPPKDVESYIHRSGRTGRAGRTGVCI CFYQHKEEYQLVQVEQKAGIKFKRIGVPSATEIIKASSKDAIRLLDSVPPTAISHFKQSAEKLIEEKGAVEALAAALAHISGATSVDQRS LINSNVGFVTMILQCSIEMPNISYAWKELKEQLGEEIDSKVKGMVFLKGKLGVCFDVPTASVTEIQEKWHDSRRWQLSVATEQPELEGPR -------------------------------------------------------------- >13517_13517_2_CCAR1-DDX21_CCAR1_chr10_70482334_ENST00000535016_DDX21_chr10_70723046_ENST00000354185_length(transcript)=4198nt_BP=192nt GAAGTTGGCGCATGCGCCTAAAGCTGACGGGTTTGAAATGGCTTCGATGTTAGCCGGGACCCGACTCAGATCGATGCTATAGAAGACAAA CAAGGGAAGGTTTTTTTTCCTTTTGCATCATGGCTCAATTTGGAGGACAGAAGAATCCGCCATGGGCTACTCAGTTTACAGCCACTGCAG TATCACAGCCAGGCCGAGGAGTGACCTTCCTATTTCCTATACAAGCAAAGACATTCCATCATGTTTACAGCGGGAAGGACTTAATTGCAC AGGCACGGACAGGAACTGGGAAGACATTCTCCTTTGCCATCCCTTTGATTGAGAAACTTCATGGGGAACTGCAAGACAGGAAGAGAGGCC GTGCCCCTCAGGTACTGGTTCTTGCACCTACAAGAGAGTTGGCAAATCAAGTAAGCAAAGACTTCAGTGACATCACAAAAAAGCTGTCAG TGGCTTGTTTTTATGGTGGAACTCCCTATGGAGGTCAATTTGAACGCATGAGGAATGGGATTGATATCCTGGTTGGAACACCAGGTCGTA TCAAAGACCACATACAGAATGGCAAACTAGATCTCACCAAACTTAAGCATGTTGTCCTGGATGAAGTGGACCAGATGTTGGATATGGGAT TTGCTGATCAAGTGGAAGAGATTTTAAGTGTGGCATACAAGAAAGATTCTGAAGACAATCCCCAAACATTGCTTTTTTCTGCAACTTGCC CTCATTGGGTATTTAATGTTGCCAAGAAATACATGAAATCTACATATGAACAGGTGGACCTGATTGGTAAAAAGACTCAGAAAACGGCAA TAACTGTGGAGCATCTGGCTATTAAGTGCCACTGGACTCAGAGGGCAGCAGTTATTGGGGATGTCATCCGAGTATATAGTGGTCATCAAG GACGCACTATCATCTTTTGTGAAACCAAGAAAGAAGCCCAGGAGCTGTCCCAGAATTCAGCTATAAAGCAGGATGCTCAGTCCTTGCATG GAGACATTCCACAGAAGCAAAGGGAAATCACCCTGAAAGGTTTTAGAAATGGTAGTTTTGGAGTTTTGGTGGCAACCAATGTTGCTGCAC GTGGGTTAGACATCCCTGAGGTTGATTTGGTTATACAAAGCTCTCCACCAAAGGATGTAGAGTCCTACATTCATCGATCCGGGCGGACAG GCAGAGCTGGAAGGACGGGGGTGTGCATCTGCTTTTATCAGCACAAGGAAGAATATCAGTTAGTACAAGTGGAGCAAAAAGCGGGAATTA AGTTCAAACGAATAGGTGTTCCTTCTGCAACAGAAATAATAAAAGCTTCCAGCAAAGATGCCATCAGGCTTTTGGATTCCGTGCCTCCCA CTGCCATTAGTCACTTCAAACAATCAGCTGAGAAGCTGATAGAGGAGAAGGGAGCTGTGGAAGCTCTGGCAGCAGCACTGGCCCATATTT CAGGTGCCACGTCCGTAGACCAGCGCTCCTTGATCAACTCAAATGTGGGTTTTGTGACCATGATCTTGCAGTGCTCAATTGAAATGCCAA ATATTAGTTATGCTTGGAAAGAACTTAAAGAGCAGCTGGGCGAGGAGATTGATTCCAAAGTGAAGGGAATGGTTTTTCTCAAAGGAAAGC TGGGTGTTTGCTTTGATGTACCTACCGCATCAGTAACAGAAATACAGGAGAAATGGCATGATTCACGACGCTGGCAGCTCTCTGTGGCCA CAGAGCAACCAGAACTGGAAGGACCACGGGAAGGATATGGAGGCTTCAGGGGACAGCGGGAAGGCAGTCGAGGCTTCAGGGGACAGCGGG ACGGAAACAGAAGATTCAGAGGACAGCGGGAAGGCAGTAGAGGCCCGAGAGGACAGCGATCAGGAGGTGGCAACAAAAGTAACAGATCCC AAAACAAAGGCCAGAAGCGGAGTTTCAGTAAAGCATTTGGTCAATAATTAGAAATAGAAGATTTATATAGCAAAAAGAGAATGATGTTTG GCAATATAGAACTGAACATTATTTTTCATGCAAAGTTAAAAGCACATTGTGCCTCCTTTTGACCACTTGCCAAGTCCCTGTCTCTTTCAG ACACAGACAAGCTTCATTTAAATTATTTCATCTGATCATTATCATTTATAACTTTATTGTTACTTCTTCATCAGTTTTTCCTTTTGAAAG GTGTATGAATTCATTACATTTTTATTCTAATGTATTATCTGTAGATTAGAAGATAAAATCAAGCATGTATCTGCCTATACTTTGTGAGTT CACCTGTCTTTATACTCAAAAGTGTCCCTTAATAGTGTCCTTCCCTGAAATAAATACCTAAGGGAGTGTAACAGTCTCTGGAGGACCACT TTGAGCCTTTGGAAGTTAAGGTTTCCTCAGCCACCTGCCGAACAGTTTCTCATGTGGTCCTATTATTTGTCTACTGAGACTTAATACTGA GCAATGTTTTGAAACAAGATTTCAAACTAATCTGGGTTGTAATACAGTTTATACCAGTGTATGCTCTAGACTTGGAAGATGTAGTATGTT TGATGTGGATTACCTATACTTATGTTCGTTTTGATACATTTTTAGCTTCTCATTATAAGGTGATTCATGCTTTAGTGAATTCTTCATAGA TAGTATATATAAAAGTACATTTTAATAGAAAGCCAGGGTTTTAAGGAATTTCACATGTATAAGGTGGCTCCATAGCTTTATTTGTAAGTA GGCTGGATAAATGGTGCTTAAATGGTAATGTACTCCACTTCTTCCTATTGGAAGATTAACATTATTTACCAAGAAGGACTTAAGGGAGTA GGGGGCGCAGATTAGCATTGCTCAAGAGTATGTAAAAAAAAAAAAAAAAAAAAAGAACCAAACCACTGGAAATAATCAAATGCAAAAAGG TAACAAATTCATAACTGGAAAGCAAAGAGAAGAACAAGTATGATTTGGATGATAAAGCATTGTTTTAATGGTGAAAACTTCACAGATCAC TAATGTTTCTAGAGGTTAACTTCAAGTGGGCAAGCTGGGGTTTTTAGGTAGTCAGTGGCCTAGTTCCTAAAGCCACAGTATAGGATCTGT TAAACTGAATGTCTGTTGAAAGTTTGTTTTAGCTGCTTGGAGGCTTCCTTTTAAGACAAACTGTATGTGATTAAGTTGTTTTGAGGGAAC TGAAGAACCTGATGTAGCCCCTGGCCAGATAACTGCCTGATTTCTCAGATATTATTTCTCTGGGAAACATTCTACATAGCACAGGAGCTT AAGAGTGGCATTATCTTCTCGCCTTAATTTCCAGAGATTATTTCTGTACTGAGAATCCTGGAACTACTATGCTAGGAAATTTAAAGCTGC ATGGTCTGTCTTGTTTTCATTTAATTATTGTGAATACCTAGAATCTTTCTTGGTCCTGATTTCTCTTGCTTAATCCAGTCTTTATCTCTA ACTGCCCCTTATTTGATCACCATGTACTAGGAGCTCTGATAGCCAGCTCAGCTCCTAATCCTTGAGGCAACATTCTTTTTCTATTTGAAC TTCAGTTCTGTCCTTGAATCCCGACTAGATATTTCTTGCCCTCTGGTCTCAGAATTCTCTTGGCTTTTATTCCTTGATCCACTTGCCAGT TTTATCACTTTACCCTTGTTCCTCATGGCTTCCCATCAAGCCATGGGTATTAGGTGACAGTGTAATTTATTAGATTCTGGTTTTGCCCAA ATACTGGGCATGCTTTAATAATAACTGAACCATTTCATTATTTGGATAGGCATGGGTACCTTATCAAGCAGATTAAAAGGATATGGTACC CGTCCTTTAGAAAAGAACAGCTAAAACCTTGTTGTGGATTATGGATTTAGCCTAAAGAAAAATAATCTGGCATAAATTAAGAGTAAGAGA GAAGATTAATAGAAATTTCACTTCACATAACTTAAAACATGGCTATTTCAATAAAGGACTAAAGTTTCTCCTGGATCCCAGAATTCAACC TGTATTTATAAATGTATAATGTATTTAGCTACTTTTTGGTTTAAATGAACTTGTTGGGTTAGCTTGGTAAATGTTATAATTTTTACTATT TTCTACAAAGAAAATATTTTCTAATTTAAGTTGGAGCTATCTGTGCAGCAGTTTCTCTACAGTTGTGCATAAATGTTTTTACTATAAAAT >13517_13517_2_CCAR1-DDX21_CCAR1_chr10_70482334_ENST00000535016_DDX21_chr10_70723046_ENST00000354185_length(amino acids)=605AA_BP=18 MAQFGGQKNPPWATQFTATAVSQPGRGVTFLFPIQAKTFHHVYSGKDLIAQARTGTGKTFSFAIPLIEKLHGELQDRKRGRAPQVLVLAP TRELANQVSKDFSDITKKLSVACFYGGTPYGGQFERMRNGIDILVGTPGRIKDHIQNGKLDLTKLKHVVLDEVDQMLDMGFADQVEEILS VAYKKDSEDNPQTLLFSATCPHWVFNVAKKYMKSTYEQVDLIGKKTQKTAITVEHLAIKCHWTQRAAVIGDVIRVYSGHQGRTIIFCETK KEAQELSQNSAIKQDAQSLHGDIPQKQREITLKGFRNGSFGVLVATNVAARGLDIPEVDLVIQSSPPKDVESYIHRSGRTGRAGRTGVCI CFYQHKEEYQLVQVEQKAGIKFKRIGVPSATEIIKASSKDAIRLLDSVPPTAISHFKQSAEKLIEEKGAVEALAAALAHISGATSVDQRS LINSNVGFVTMILQCSIEMPNISYAWKELKEQLGEEIDSKVKGMVFLKGKLGVCFDVPTASVTEIQEKWHDSRRWQLSVATEQPELEGPR -------------------------------------------------------------- >13517_13517_3_CCAR1-DDX21_CCAR1_chr10_70482334_ENST00000543719_DDX21_chr10_70723046_ENST00000354185_length(transcript)=4192nt_BP=186nt TGCGCCTAAAGCTGACGGGTTTGAAATGGCTTCGATGTTAGCCGGGACCCGACTCAGGTGAAGATCGATGCTATAGAAGACAAACAAGGG AAGGTTTTTTTTCCTTTTGCATCATGGCTCAATTTGGAGGACAGAAGAATCCGCCATGGGCTACTCAGTTTACAGCCACTGCAGTATCAC AGCCAGGCCGAGGAGTGACCTTCCTATTTCCTATACAAGCAAAGACATTCCATCATGTTTACAGCGGGAAGGACTTAATTGCACAGGCAC GGACAGGAACTGGGAAGACATTCTCCTTTGCCATCCCTTTGATTGAGAAACTTCATGGGGAACTGCAAGACAGGAAGAGAGGCCGTGCCC CTCAGGTACTGGTTCTTGCACCTACAAGAGAGTTGGCAAATCAAGTAAGCAAAGACTTCAGTGACATCACAAAAAAGCTGTCAGTGGCTT GTTTTTATGGTGGAACTCCCTATGGAGGTCAATTTGAACGCATGAGGAATGGGATTGATATCCTGGTTGGAACACCAGGTCGTATCAAAG ACCACATACAGAATGGCAAACTAGATCTCACCAAACTTAAGCATGTTGTCCTGGATGAAGTGGACCAGATGTTGGATATGGGATTTGCTG ATCAAGTGGAAGAGATTTTAAGTGTGGCATACAAGAAAGATTCTGAAGACAATCCCCAAACATTGCTTTTTTCTGCAACTTGCCCTCATT GGGTATTTAATGTTGCCAAGAAATACATGAAATCTACATATGAACAGGTGGACCTGATTGGTAAAAAGACTCAGAAAACGGCAATAACTG TGGAGCATCTGGCTATTAAGTGCCACTGGACTCAGAGGGCAGCAGTTATTGGGGATGTCATCCGAGTATATAGTGGTCATCAAGGACGCA CTATCATCTTTTGTGAAACCAAGAAAGAAGCCCAGGAGCTGTCCCAGAATTCAGCTATAAAGCAGGATGCTCAGTCCTTGCATGGAGACA TTCCACAGAAGCAAAGGGAAATCACCCTGAAAGGTTTTAGAAATGGTAGTTTTGGAGTTTTGGTGGCAACCAATGTTGCTGCACGTGGGT TAGACATCCCTGAGGTTGATTTGGTTATACAAAGCTCTCCACCAAAGGATGTAGAGTCCTACATTCATCGATCCGGGCGGACAGGCAGAG CTGGAAGGACGGGGGTGTGCATCTGCTTTTATCAGCACAAGGAAGAATATCAGTTAGTACAAGTGGAGCAAAAAGCGGGAATTAAGTTCA AACGAATAGGTGTTCCTTCTGCAACAGAAATAATAAAAGCTTCCAGCAAAGATGCCATCAGGCTTTTGGATTCCGTGCCTCCCACTGCCA TTAGTCACTTCAAACAATCAGCTGAGAAGCTGATAGAGGAGAAGGGAGCTGTGGAAGCTCTGGCAGCAGCACTGGCCCATATTTCAGGTG CCACGTCCGTAGACCAGCGCTCCTTGATCAACTCAAATGTGGGTTTTGTGACCATGATCTTGCAGTGCTCAATTGAAATGCCAAATATTA GTTATGCTTGGAAAGAACTTAAAGAGCAGCTGGGCGAGGAGATTGATTCCAAAGTGAAGGGAATGGTTTTTCTCAAAGGAAAGCTGGGTG TTTGCTTTGATGTACCTACCGCATCAGTAACAGAAATACAGGAGAAATGGCATGATTCACGACGCTGGCAGCTCTCTGTGGCCACAGAGC AACCAGAACTGGAAGGACCACGGGAAGGATATGGAGGCTTCAGGGGACAGCGGGAAGGCAGTCGAGGCTTCAGGGGACAGCGGGACGGAA ACAGAAGATTCAGAGGACAGCGGGAAGGCAGTAGAGGCCCGAGAGGACAGCGATCAGGAGGTGGCAACAAAAGTAACAGATCCCAAAACA AAGGCCAGAAGCGGAGTTTCAGTAAAGCATTTGGTCAATAATTAGAAATAGAAGATTTATATAGCAAAAAGAGAATGATGTTTGGCAATA TAGAACTGAACATTATTTTTCATGCAAAGTTAAAAGCACATTGTGCCTCCTTTTGACCACTTGCCAAGTCCCTGTCTCTTTCAGACACAG ACAAGCTTCATTTAAATTATTTCATCTGATCATTATCATTTATAACTTTATTGTTACTTCTTCATCAGTTTTTCCTTTTGAAAGGTGTAT GAATTCATTACATTTTTATTCTAATGTATTATCTGTAGATTAGAAGATAAAATCAAGCATGTATCTGCCTATACTTTGTGAGTTCACCTG TCTTTATACTCAAAAGTGTCCCTTAATAGTGTCCTTCCCTGAAATAAATACCTAAGGGAGTGTAACAGTCTCTGGAGGACCACTTTGAGC CTTTGGAAGTTAAGGTTTCCTCAGCCACCTGCCGAACAGTTTCTCATGTGGTCCTATTATTTGTCTACTGAGACTTAATACTGAGCAATG TTTTGAAACAAGATTTCAAACTAATCTGGGTTGTAATACAGTTTATACCAGTGTATGCTCTAGACTTGGAAGATGTAGTATGTTTGATGT GGATTACCTATACTTATGTTCGTTTTGATACATTTTTAGCTTCTCATTATAAGGTGATTCATGCTTTAGTGAATTCTTCATAGATAGTAT ATATAAAAGTACATTTTAATAGAAAGCCAGGGTTTTAAGGAATTTCACATGTATAAGGTGGCTCCATAGCTTTATTTGTAAGTAGGCTGG ATAAATGGTGCTTAAATGGTAATGTACTCCACTTCTTCCTATTGGAAGATTAACATTATTTACCAAGAAGGACTTAAGGGAGTAGGGGGC GCAGATTAGCATTGCTCAAGAGTATGTAAAAAAAAAAAAAAAAAAAAAGAACCAAACCACTGGAAATAATCAAATGCAAAAAGGTAACAA ATTCATAACTGGAAAGCAAAGAGAAGAACAAGTATGATTTGGATGATAAAGCATTGTTTTAATGGTGAAAACTTCACAGATCACTAATGT TTCTAGAGGTTAACTTCAAGTGGGCAAGCTGGGGTTTTTAGGTAGTCAGTGGCCTAGTTCCTAAAGCCACAGTATAGGATCTGTTAAACT GAATGTCTGTTGAAAGTTTGTTTTAGCTGCTTGGAGGCTTCCTTTTAAGACAAACTGTATGTGATTAAGTTGTTTTGAGGGAACTGAAGA ACCTGATGTAGCCCCTGGCCAGATAACTGCCTGATTTCTCAGATATTATTTCTCTGGGAAACATTCTACATAGCACAGGAGCTTAAGAGT GGCATTATCTTCTCGCCTTAATTTCCAGAGATTATTTCTGTACTGAGAATCCTGGAACTACTATGCTAGGAAATTTAAAGCTGCATGGTC TGTCTTGTTTTCATTTAATTATTGTGAATACCTAGAATCTTTCTTGGTCCTGATTTCTCTTGCTTAATCCAGTCTTTATCTCTAACTGCC CCTTATTTGATCACCATGTACTAGGAGCTCTGATAGCCAGCTCAGCTCCTAATCCTTGAGGCAACATTCTTTTTCTATTTGAACTTCAGT TCTGTCCTTGAATCCCGACTAGATATTTCTTGCCCTCTGGTCTCAGAATTCTCTTGGCTTTTATTCCTTGATCCACTTGCCAGTTTTATC ACTTTACCCTTGTTCCTCATGGCTTCCCATCAAGCCATGGGTATTAGGTGACAGTGTAATTTATTAGATTCTGGTTTTGCCCAAATACTG GGCATGCTTTAATAATAACTGAACCATTTCATTATTTGGATAGGCATGGGTACCTTATCAAGCAGATTAAAAGGATATGGTACCCGTCCT TTAGAAAAGAACAGCTAAAACCTTGTTGTGGATTATGGATTTAGCCTAAAGAAAAATAATCTGGCATAAATTAAGAGTAAGAGAGAAGAT TAATAGAAATTTCACTTCACATAACTTAAAACATGGCTATTTCAATAAAGGACTAAAGTTTCTCCTGGATCCCAGAATTCAACCTGTATT TATAAATGTATAATGTATTTAGCTACTTTTTGGTTTAAATGAACTTGTTGGGTTAGCTTGGTAAATGTTATAATTTTTACTATTTTCTAC AAAGAAAATATTTTCTAATTTAAGTTGGAGCTATCTGTGCAGCAGTTTCTCTACAGTTGTGCATAAATGTTTTTACTATAAAATGAGCTA >13517_13517_3_CCAR1-DDX21_CCAR1_chr10_70482334_ENST00000543719_DDX21_chr10_70723046_ENST00000354185_length(amino acids)=605AA_BP=18 MAQFGGQKNPPWATQFTATAVSQPGRGVTFLFPIQAKTFHHVYSGKDLIAQARTGTGKTFSFAIPLIEKLHGELQDRKRGRAPQVLVLAP TRELANQVSKDFSDITKKLSVACFYGGTPYGGQFERMRNGIDILVGTPGRIKDHIQNGKLDLTKLKHVVLDEVDQMLDMGFADQVEEILS VAYKKDSEDNPQTLLFSATCPHWVFNVAKKYMKSTYEQVDLIGKKTQKTAITVEHLAIKCHWTQRAAVIGDVIRVYSGHQGRTIIFCETK KEAQELSQNSAIKQDAQSLHGDIPQKQREITLKGFRNGSFGVLVATNVAARGLDIPEVDLVIQSSPPKDVESYIHRSGRTGRAGRTGVCI CFYQHKEEYQLVQVEQKAGIKFKRIGVPSATEIIKASSKDAIRLLDSVPPTAISHFKQSAEKLIEEKGAVEALAAALAHISGATSVDQRS LINSNVGFVTMILQCSIEMPNISYAWKELKEQLGEEIDSKVKGMVFLKGKLGVCFDVPTASVTEIQEKWHDSRRWQLSVATEQPELEGPR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CCAR1-DDX21 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 1_249 | 24.333333333333332 | 1151.0 | AR |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 1_249 | 24.333333333333332 | 1136.0 | AR |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 1_249 | 24.333333333333332 | 1136.0 | AR |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 643_1150 | 24.333333333333332 | 1151.0 | GATA1 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 643_1150 | 24.333333333333332 | 1136.0 | GATA1 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 643_1150 | 24.333333333333332 | 1136.0 | GATA1 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000265872 | + | 2 | 25 | 203_660 | 24.333333333333332 | 1151.0 | GATA2 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000535016 | + | 2 | 24 | 203_660 | 24.333333333333332 | 1136.0 | GATA2 |
| Hgene | CCAR1 | chr10:70482334 | chr10:70723046 | ENST00000543719 | + | 2 | 24 | 203_660 | 24.333333333333332 | 1136.0 | GATA2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CCAR1-DDX21 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CCAR1-DDX21 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
