
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CD47-CBLB (FusionGDB2 ID:14539) |
Fusion Gene Summary for CD47-CBLB |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CD47-CBLB | Fusion gene ID: 14539 | Hgene | Tgene | Gene symbol | CD47 | CBLB | Gene ID | 961 | 868 |
| Gene name | CD47 molecule | Cbl proto-oncogene B | |
| Synonyms | IAP|MER6|OA3 | Cbl-b|Nbla00127|RNF56 | |
| Cytomap | 3q13.12 | 3q13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | leukocyte surface antigen CD47CD47 antigen (Rh-related antigen, integrin-associated signal transducer)CD47 glycoproteinRh-related antigenantigen identified by monoclonal antibody 1D8antigenic surface determinant protein OA3integrin associated protei | E3 ubiquitin-protein ligase CBL-BCas-Br-M (murine) ecotropic retroviral transforming sequence bCbl proto-oncogene B, E3 ubiquitin protein ligaseCbl proto-oncogene, E3 ubiquitin protein ligase BRING finger protein 56RING-type E3 ubiquitin transferase | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q08722 | Q13191 | |
| Ensembl transtripts involved in fusion gene | ENST00000355354, ENST00000361309, ENST00000471694, | ENST00000403724, ENST00000405772, ENST00000545639, ENST00000394027, ENST00000407712, ENST00000264122, | |
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 13 X 14 X 7=1274 |
| # samples | 3 | 16 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(16/1274*10)=-2.99322146736894 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CD47 [Title/Abstract] AND CBLB [Title/Abstract] AND fusion [Title/Abstract] | ||
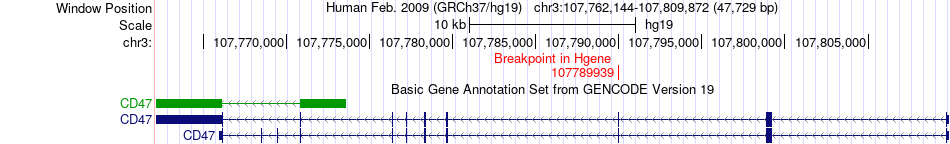
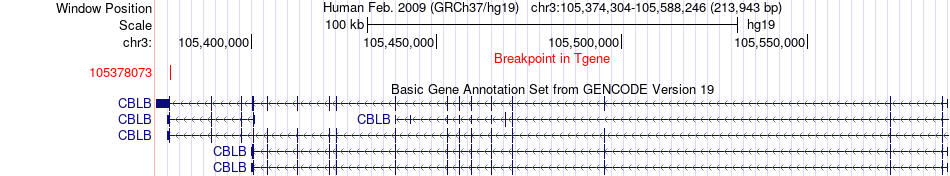
| Most frequent breakpoint | CD47(107789939)-CBLB(105378073), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CD47-CBLB seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CD47-CBLB seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. CD47-CBLB seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CD47 | GO:0008284 | positive regulation of cell proliferation | 15383453 |
| Hgene | CD47 | GO:0022409 | positive regulation of cell-cell adhesion | 15383453 |
| Hgene | CD47 | GO:0050870 | positive regulation of T cell activation | 15383453 |
 Fusion gene breakpoints across CD47 (5'-gene) Fusion gene breakpoints across CD47 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CBLB (3'-gene) Fusion gene breakpoints across CBLB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GX | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
Top |
Fusion Gene ORF analysis for CD47-CBLB |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000355354 | ENST00000403724 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| 5CDS-intron | ENST00000355354 | ENST00000405772 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| 5CDS-intron | ENST00000355354 | ENST00000545639 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| 5CDS-intron | ENST00000361309 | ENST00000403724 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| 5CDS-intron | ENST00000361309 | ENST00000405772 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| 5CDS-intron | ENST00000361309 | ENST00000545639 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| Frame-shift | ENST00000355354 | ENST00000394027 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| Frame-shift | ENST00000355354 | ENST00000407712 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| Frame-shift | ENST00000361309 | ENST00000394027 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| Frame-shift | ENST00000361309 | ENST00000407712 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| In-frame | ENST00000355354 | ENST00000264122 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| In-frame | ENST00000361309 | ENST00000264122 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| intron-3CDS | ENST00000471694 | ENST00000264122 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| intron-3CDS | ENST00000471694 | ENST00000394027 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| intron-3CDS | ENST00000471694 | ENST00000407712 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| intron-intron | ENST00000471694 | ENST00000403724 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| intron-intron | ENST00000471694 | ENST00000405772 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
| intron-intron | ENST00000471694 | ENST00000545639 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000355354 | CD47 | chr3 | 107789939 | - | ENST00000264122 | CBLB | chr3 | 105378073 | - | 4376 | 607 | 42 | 866 | 274 |
| ENST00000361309 | CD47 | chr3 | 107789939 | - | ENST00000264122 | CBLB | chr3 | 105378073 | - | 4365 | 596 | 31 | 855 | 274 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000355354 | ENST00000264122 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - | 0.000289643 | 0.9997104 |
| ENST00000361309 | ENST00000264122 | CD47 | chr3 | 107789939 | - | CBLB | chr3 | 105378073 | - | 0.000290452 | 0.99970955 |
Top |
Fusion Genomic Features for CD47-CBLB |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
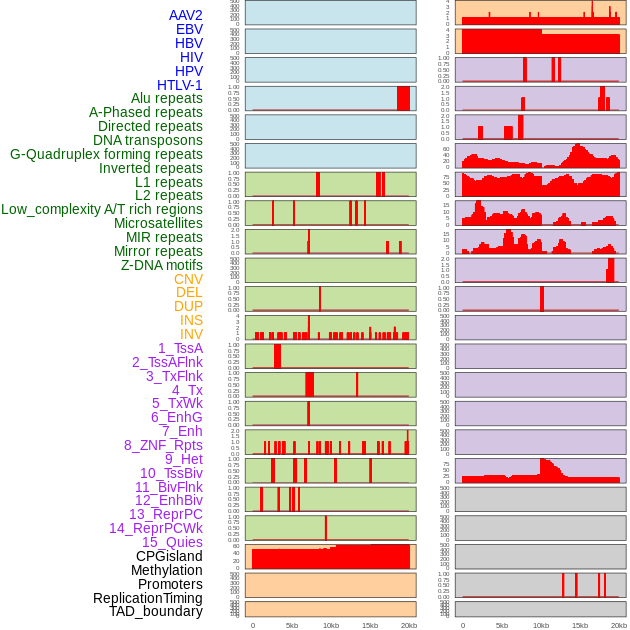
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CD47-CBLB |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:107789939/chr3:105378073) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CD47 | CBLB |
| FUNCTION: Has a role in both cell adhesion by acting as an adhesion receptor for THBS1 on platelets, and in the modulation of integrins. Plays an important role in memory formation and synaptic plasticity in the hippocampus (By similarity). Receptor for SIRPA, binding to which prevents maturation of immature dendritic cells and inhibits cytokine production by mature dendritic cells. Interaction with SIRPG mediates cell-cell adhesion, enhances superantigen-dependent T-cell-mediated proliferation and costimulates T-cell activation. May play a role in membrane transport and/or integrin dependent signal transduction. May prevent premature elimination of red blood cells. May be involved in membrane permeability changes induced following virus infection. {ECO:0000250, ECO:0000269|PubMed:11509594, ECO:0000269|PubMed:15383453, ECO:0000269|PubMed:7691831}. | FUNCTION: E3 ubiquitin-protein ligase which accepts ubiquitin from specific E2 ubiquitin-conjugating enzymes, and transfers it to substrates, generally promoting their degradation by the proteasome. Negatively regulates TCR (T-cell receptor), BCR (B-cell receptor) and FCER1 (high affinity immunoglobulin epsilon receptor) signal transduction pathways. In naive T-cells, inhibits VAV1 activation upon TCR engagement and imposes a requirement for CD28 costimulation for proliferation and IL-2 production. Also acts by promoting PIK3R1/p85 ubiquitination, which impairs its recruitment to the TCR and subsequent activation. In activated T-cells, inhibits PLCG1 activation and calcium mobilization upon restimulation and promotes anergy. In B-cells, acts by ubiquitinating SYK and promoting its proteasomal degradation. Slightly promotes SRC ubiquitination. May be involved in EGFR ubiquitination and internalization. May be functionally coupled with the E2 ubiquitin-protein ligase UB2D3. In association with CBL, required for proper feedback inhibition of ciliary platelet-derived growth factor receptor-alpha (PDGFRA) signaling pathway via ubiquitination and internalization of PDGFRA (By similarity). {ECO:0000250|UniProtKB:Q3TTA7, ECO:0000269|PubMed:10022120, ECO:0000269|PubMed:10086340, ECO:0000269|PubMed:11087752, ECO:0000269|PubMed:11526404, ECO:0000269|PubMed:14661060, ECO:0000269|PubMed:20525694}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 19_127 | 163 | 306.0 | Domain | Note=Ig-like V-type |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 19_127 | 163 | 324.0 | Domain | Note=Ig-like V-type |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 19_141 | 163 | 306.0 | Topological domain | Extracellular |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 19_141 | 163 | 324.0 | Topological domain | Extracellular |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 142_162 | 163 | 306.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 142_162 | 163 | 324.0 | Transmembrane | Helical |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 219_232 | 0 | 771.0 | Calcium binding | Ontology_term=ECO:0000255 | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 477_925 | 0 | 771.0 | Compositional bias | Note=Pro-rich | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 931_970 | 896 | 983.0 | Domain | UBA | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 35_343 | 0 | 771.0 | Domain | Cbl-PTB | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 931_970 | 0 | 771.0 | Domain | UBA | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 168_240 | 0 | 771.0 | Region | Note=EF-hand-like | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 241_343 | 0 | 771.0 | Region | Note=SH2-like | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 344_372 | 0 | 771.0 | Region | Note=Linker | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 35_167 | 0 | 771.0 | Region | Note=4H | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 373_412 | 0 | 771.0 | Zinc finger | RING-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 163_176 | 163 | 306.0 | Topological domain | Cytoplasmic |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 198_207 | 163 | 306.0 | Topological domain | Extracellular |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 229_235 | 163 | 306.0 | Topological domain | Cytoplasmic |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 257_268 | 163 | 306.0 | Topological domain | Extracellular |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 290_323 | 163 | 306.0 | Topological domain | Cytoplasmic |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 163_176 | 163 | 324.0 | Topological domain | Cytoplasmic |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 198_207 | 163 | 324.0 | Topological domain | Extracellular |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 229_235 | 163 | 324.0 | Topological domain | Cytoplasmic |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 257_268 | 163 | 324.0 | Topological domain | Extracellular |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 290_323 | 163 | 324.0 | Topological domain | Cytoplasmic |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 177_197 | 163 | 306.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 208_228 | 163 | 306.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 236_256 | 163 | 306.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000355354 | - | 3 | 9 | 269_289 | 163 | 306.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 177_197 | 163 | 324.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 208_228 | 163 | 324.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 236_256 | 163 | 324.0 | Transmembrane | Helical |
| Hgene | CD47 | chr3:107789939 | chr3:105378073 | ENST00000361309 | - | 3 | 11 | 269_289 | 163 | 324.0 | Transmembrane | Helical |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 219_232 | 896 | 983.0 | Calcium binding | Ontology_term=ECO:0000255 | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 477_925 | 896 | 983.0 | Compositional bias | Note=Pro-rich | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 35_343 | 896 | 983.0 | Domain | Cbl-PTB | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 168_240 | 896 | 983.0 | Region | Note=EF-hand-like | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 241_343 | 896 | 983.0 | Region | Note=SH2-like | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 344_372 | 896 | 983.0 | Region | Note=Linker | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 35_167 | 896 | 983.0 | Region | Note=4H | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 373_412 | 896 | 983.0 | Zinc finger | RING-type |
Top |
Fusion Gene Sequence for CD47-CBLB |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >14539_14539_1_CD47-CBLB_CD47_chr3_107789939_ENST00000355354_CBLB_chr3_105378073_ENST00000264122_length(transcript)=4376nt_BP=607nt TGGGCAGTGGGTCCTGCCTGTGACGCGCGGCGGCGGTCGGTCCTGCCTGTAACGGCGGCGGCGGCTGCTGCTCCGGACACCTGCGGCGGC GGCGGCGACCCCGCGGCGGGCGCGGAGATGTGGCCCCTGGTAGCGGCGCTGTTGCTGGGCTCGGCGTGCTGCGGATCAGCTCAGCTACTA TTTAATAAAACAAAATCTGTAGAATTCACGTTTTGTAATGACACTGTCGTCATTCCATGCTTTGTTACTAATATGGAGGCACAAAACACT ACTGAAGTATACGTAAAGTGGAAATTTAAAGGAAGAGATATTTACACCTTTGATGGAGCTCTAAACAAGTCCACTGTCCCCACTGACTTT AGTAGTGCAAAAATTGAAGTCTCACAATTACTAAAAGGAGATGCCTCTTTGAAGATGGATAAGAGTGATGCTGTCTCACACACAGGAAAC TACACTTGTGAAGTAACAGAATTAACCAGAGAAGGTGAAACGATCATCGAGCTAAAATATCGTGTTGTTTCATGGTTTTCTCCAAATGAA AATATTCTTATTGTTATTTTCCCAATTTTTGCTATACTCCTGTTCTGGGGACAGTTTGGTATTAAAAATGGTTCACAGGCACCAGCCAGA CCCCCTAAACCACGACCGCGCAGGACTGCACCAGAAATTCACCACAGAAAACCCCATGGGCCTGAGGCGGCATTGGAAAATGTCGATGCA AAAATTGCAAAACTCATGGGAGAGGGTTATGCCTTTGAAGAGGTGAAGAGAGCCTTAGAGATAGCCCAGAATAATGTCGAAGTTGCCCGG AGCATCCTCCGAGAATTTGCCTTCCCTCCTCCAGTATCCCCACGTCTAAATCTATAGCAGCCAGAACTGTAGACACCAAAATGGAAAGCA ATCGATGTATTCCAAGAGTGTGGAAATAAAGAGAACTGAGATGGAATTCAAGAGAGAAGTGTCTCCTCCTCGTGTAGCAGCTTGAGAAGA GGCTTGGGAGTGCAGCTTCTCAAAGGAGACCGATGCTTGCTCAGGATGTCGACAGCTGTGGCTTCCTTGTTTTTGCTAGCCATATTTTTA AATCAGGGTTGAACTGACAAAAATAATTTAAAGACGTTTACTTCCCTTGAACTTTGAACCTGTGAAATGCTTTACCTTGTTTACAGTTTG GCAAAGTTGCAGTTTGTTCTTGTTTTTAGTTTAGTTTTGTTTTGGTGTTTTGATACCTGTACTGTGTTCTTCACAGACCCTTTGTAGCGT GGTCAGGTCTGCTGTAACATTTCCCACCAACTCTCTTGCTGTCCACATCAACAGCTAAATCATTTATTCATATGGATCTCTACCATCCCC ATGCCTTGCCCAGGTCCAGTTCCATTTCTCTCATTCACAAGATGCTTTGAAGGTTCTGATTTTCAACTGATCAAACTAATGCAAAAAAAA AAAAGTATGTATTCTTCACTACTGAGTTTCTTCTTTGGAAACCATCACTATTGAGAGATGGGAAAAACCTGAATGTATAAAGCATTTATT TGTCAATAAACTGCCTTTTGTAAGGGGTTTTCACATAACATAGAGGAGCTTCCCTTTTTTGTTTAAGTTTTGTAACCTTTAATCCTCCAT ATTCTCATGTCTGTCATCCCAGGGGTGTCACAACTGTAAAAATCTACAATATTAGAAAGCAGCTACTACAGATGTGGAAGAGAGAACACT TGTATAAGACAATGCTGTACTGAAGTTATATAACAAGGTCCCCTGATACTTATGCCTGCATTACTTTGAGGGATGCTGAATGAGAGGACC ATCTCCCTGAAATATATTAATAATTTTCAGATAAAATTATGAACATAACATACCTTTTCTCAAGTGGAATTAGAGTTCATTTCCCATTTA AGTGATTACACTCTATCTCCTAAGAGCTGTGGGAACCAGTTCATATTGCAGTGGAAACATTAACTCTTCGTTGCACTGTTGAGTGTCAAG GCTACTGTGGCAATGTTTTTTGCAAAGTTCTCAGTAATTTTCTCTCTGCAATCAAATTAGAGTTCATATTTAATTTGCAGATTCTCTCAT TCATTGCTCCACACTGCTTTATCAAATCAGGTGAAAGAAATTAATGTAGTTTTGCCTATTTTACAAAATGGTGGTTGGTTCTTTAAACAC CATGTTATTACTCTTAATAAATCTACAAATATTTATTGAGCATCTATGATATGCTTGGTGCTGTTAGGCACTGTGTTTTTAAAGCACAAA CTCTCAAAAGCTGATTCCAGCGTAATGTTTTATTTTTGTTTTTCTGTGTTTTTTATGACAAAGTCTGTCTGATAGACAAAGCCTGGAAGG TTTCCCCCAGGGCTTTAGAATTTAGAATTAGCTGGTTTTCATTTCAAATGGGAAATTAATGGAGAAATGAGATAAAATCATCTCTGTGTA AATGCTTGAACCCACAGAAAGTTTTCTAAATATGGACTAATTGTGGGTTTGTGCATATTTCTGTGTCTCCTCTTTTCACTTCACACCCAC CATGATATAAAAAGATGCATCAGAGAGTGAGCCTGCCTTCAGGACCTGAAAGGACATGACTGAACATCTAGTAGATCTGGTGGAAATTAA TGTTATACAAAAGACACGTAGACTAAGTTGACCCTGGGAGAGGAGTTAATACATTCCTTTGCAAAGTGACATGTGCCCTTTTTTTGACAG TCTGGCTTTCATGGTATGAGATGAGGACTCACAGGCCTTGGGTAATTTCTGCTTTTGTATGAATTTTTTCAGATGGTTCTGAGCTGGTTA TGTCCCTGGAGGGGAATTTTTCATTTGCCTTAGATGTTCTTCCCATGAGTAGGTACATGAGTGAAGTTTCTTACACTTTTAAAATTGCAT TGGCAAATTTTCTTTTGCTTTATGAACCCCTGTTTTGTCATTATGATTTTTCACTTGTTTTAACAGTTTCAGATGTCACCTTGGAATAGC ACTGTTTATTTTGGCCTTTTTATGATTGCTGATGATAGGTACAGTCTAAAGGCCAAAAGTTGCCACTTCATTGTGATAACATTGTTGCAA CATTCCTAATATATTAGAGAATGATGTAAAATAAGAATGTTAGAGAACATAGCACATTCTCATTCAGTTTATAAACAATATTTGAAAATT TCCAGTGAGAATTAAAAAAGGACTAGTGATGATATAGCAAACCTGGACCTGAAATTTGGGGGTGGGGGGTGGGGGGATATCTGTTTTGCA GAGAAAATTTACCAGCCTCCAGCAAAATCTCTGCATCTTGTACTCAATAGCAACCATTAGTATTGTCATTTAAAAAGAATGAATCAAAAA TTCATACGTAATGGAATGAAAAGAGGAAAACTTTCTATTAAACACTGTCAAGAAAGACCCATTGCTATCTATCACCTATGAAATTAAACC TAAAAAAATTTTTTTTTATTGTAAAAGCGACTGCTCGTTGATGTCTGGAAGTGATCCAGACTTTGTTTCACTGTGATCCAAAAAGTTAGA CATTCCTTCCATTTCTCAAGATTTGGTAACAATTTTTTTTTAAGTCAGCATTTCATATAATACATAAAATTAAGACAAATTAACTTACTG ATATGTGGCCAATATCAACCCTTCTCACCCAACCCAAACATGTTCTGGAGATAGAAGGACTTATCCTCAATTATATTCCAGTAAATTTAT CTCCATAATCTGTTTTATCCCCTAATGTATGATATATCTCACCAAATTTTTCTAGAATTGAATAAAAGCAGTTTTTGCTTAACTTGAATT TCCTTCCACAAATACAACCTTCAGCTCTTCCCTCTCTGTTTCCCTTTTGCCCACATTGCAAATAGTCTTCATAATAGAACCCTCACTGCA ACTTAAAAGGGTCCCATTTCTTAACTCAGCATTTCATATACCAAGAACACTTGCAAAGATTTCTTCATAATGTGATTTTTTAATTTTTCA GTTCCAGAAGCATTTCCTACAGTTCGAACGTTATTTTTTCTCTCAGATTTTCAACTTGAAGCTTTGATTTGCACTGAATTAACTTGGATA AAATTTTAGTTTCTCTCTTCAGCTAACAGATTTGGAAGCAAATTACAGTATACTTGAATAATTCAGGTACTACCTGTTGACCTGGAATTT TAGTGATTCTGGGAAAGTTTTCCAGACAAAGACACCAATAGAGTGAAGGGTTTTTTCCCCAATTACTATTTTATTGTTTTTTAAAATGCC TTTTTAAAACTTGTGTGATGTGGCAAAATAGTCATATGTTCAATTAATTTATATTTTATTCATTCCTTTTCAATTCTGGTTATTATGTAA >14539_14539_1_CD47-CBLB_CD47_chr3_107789939_ENST00000355354_CBLB_chr3_105378073_ENST00000264122_length(amino acids)=274AA_BP=1 MPVTAAAAAAPDTCGGGGDPAAGAEMWPLVAALLLGSACCGSAQLLFNKTKSVEFTFCNDTVVIPCFVTNMEAQNTTEVYVKWKFKGRDI YTFDGALNKSTVPTDFSSAKIEVSQLLKGDASLKMDKSDAVSHTGNYTCEVTELTREGETIIELKYRVVSWFSPNENILIVIFPIFAILL FWGQFGIKNGSQAPARPPKPRPRRTAPEIHHRKPHGPEAALENVDAKIAKLMGEGYAFEEVKRALEIAQNNVEVARSILREFAFPPPVSP -------------------------------------------------------------- >14539_14539_2_CD47-CBLB_CD47_chr3_107789939_ENST00000361309_CBLB_chr3_105378073_ENST00000264122_length(transcript)=4365nt_BP=596nt TCCTGCCTGTGACGCGCGGCGGCGGTCGGTCCTGCCTGTAACGGCGGCGGCGGCTGCTGCTCCGGACACCTGCGGCGGCGGCGGCGACCC CGCGGCGGGCGCGGAGATGTGGCCCCTGGTAGCGGCGCTGTTGCTGGGCTCGGCGTGCTGCGGATCAGCTCAGCTACTATTTAATAAAAC AAAATCTGTAGAATTCACGTTTTGTAATGACACTGTCGTCATTCCATGCTTTGTTACTAATATGGAGGCACAAAACACTACTGAAGTATA CGTAAAGTGGAAATTTAAAGGAAGAGATATTTACACCTTTGATGGAGCTCTAAACAAGTCCACTGTCCCCACTGACTTTAGTAGTGCAAA AATTGAAGTCTCACAATTACTAAAAGGAGATGCCTCTTTGAAGATGGATAAGAGTGATGCTGTCTCACACACAGGAAACTACACTTGTGA AGTAACAGAATTAACCAGAGAAGGTGAAACGATCATCGAGCTAAAATATCGTGTTGTTTCATGGTTTTCTCCAAATGAAAATATTCTTAT TGTTATTTTCCCAATTTTTGCTATACTCCTGTTCTGGGGACAGTTTGGTATTAAAAATGGTTCACAGGCACCAGCCAGACCCCCTAAACC ACGACCGCGCAGGACTGCACCAGAAATTCACCACAGAAAACCCCATGGGCCTGAGGCGGCATTGGAAAATGTCGATGCAAAAATTGCAAA ACTCATGGGAGAGGGTTATGCCTTTGAAGAGGTGAAGAGAGCCTTAGAGATAGCCCAGAATAATGTCGAAGTTGCCCGGAGCATCCTCCG AGAATTTGCCTTCCCTCCTCCAGTATCCCCACGTCTAAATCTATAGCAGCCAGAACTGTAGACACCAAAATGGAAAGCAATCGATGTATT CCAAGAGTGTGGAAATAAAGAGAACTGAGATGGAATTCAAGAGAGAAGTGTCTCCTCCTCGTGTAGCAGCTTGAGAAGAGGCTTGGGAGT GCAGCTTCTCAAAGGAGACCGATGCTTGCTCAGGATGTCGACAGCTGTGGCTTCCTTGTTTTTGCTAGCCATATTTTTAAATCAGGGTTG AACTGACAAAAATAATTTAAAGACGTTTACTTCCCTTGAACTTTGAACCTGTGAAATGCTTTACCTTGTTTACAGTTTGGCAAAGTTGCA GTTTGTTCTTGTTTTTAGTTTAGTTTTGTTTTGGTGTTTTGATACCTGTACTGTGTTCTTCACAGACCCTTTGTAGCGTGGTCAGGTCTG CTGTAACATTTCCCACCAACTCTCTTGCTGTCCACATCAACAGCTAAATCATTTATTCATATGGATCTCTACCATCCCCATGCCTTGCCC AGGTCCAGTTCCATTTCTCTCATTCACAAGATGCTTTGAAGGTTCTGATTTTCAACTGATCAAACTAATGCAAAAAAAAAAAAGTATGTA TTCTTCACTACTGAGTTTCTTCTTTGGAAACCATCACTATTGAGAGATGGGAAAAACCTGAATGTATAAAGCATTTATTTGTCAATAAAC TGCCTTTTGTAAGGGGTTTTCACATAACATAGAGGAGCTTCCCTTTTTTGTTTAAGTTTTGTAACCTTTAATCCTCCATATTCTCATGTC TGTCATCCCAGGGGTGTCACAACTGTAAAAATCTACAATATTAGAAAGCAGCTACTACAGATGTGGAAGAGAGAACACTTGTATAAGACA ATGCTGTACTGAAGTTATATAACAAGGTCCCCTGATACTTATGCCTGCATTACTTTGAGGGATGCTGAATGAGAGGACCATCTCCCTGAA ATATATTAATAATTTTCAGATAAAATTATGAACATAACATACCTTTTCTCAAGTGGAATTAGAGTTCATTTCCCATTTAAGTGATTACAC TCTATCTCCTAAGAGCTGTGGGAACCAGTTCATATTGCAGTGGAAACATTAACTCTTCGTTGCACTGTTGAGTGTCAAGGCTACTGTGGC AATGTTTTTTGCAAAGTTCTCAGTAATTTTCTCTCTGCAATCAAATTAGAGTTCATATTTAATTTGCAGATTCTCTCATTCATTGCTCCA CACTGCTTTATCAAATCAGGTGAAAGAAATTAATGTAGTTTTGCCTATTTTACAAAATGGTGGTTGGTTCTTTAAACACCATGTTATTAC TCTTAATAAATCTACAAATATTTATTGAGCATCTATGATATGCTTGGTGCTGTTAGGCACTGTGTTTTTAAAGCACAAACTCTCAAAAGC TGATTCCAGCGTAATGTTTTATTTTTGTTTTTCTGTGTTTTTTATGACAAAGTCTGTCTGATAGACAAAGCCTGGAAGGTTTCCCCCAGG GCTTTAGAATTTAGAATTAGCTGGTTTTCATTTCAAATGGGAAATTAATGGAGAAATGAGATAAAATCATCTCTGTGTAAATGCTTGAAC CCACAGAAAGTTTTCTAAATATGGACTAATTGTGGGTTTGTGCATATTTCTGTGTCTCCTCTTTTCACTTCACACCCACCATGATATAAA AAGATGCATCAGAGAGTGAGCCTGCCTTCAGGACCTGAAAGGACATGACTGAACATCTAGTAGATCTGGTGGAAATTAATGTTATACAAA AGACACGTAGACTAAGTTGACCCTGGGAGAGGAGTTAATACATTCCTTTGCAAAGTGACATGTGCCCTTTTTTTGACAGTCTGGCTTTCA TGGTATGAGATGAGGACTCACAGGCCTTGGGTAATTTCTGCTTTTGTATGAATTTTTTCAGATGGTTCTGAGCTGGTTATGTCCCTGGAG GGGAATTTTTCATTTGCCTTAGATGTTCTTCCCATGAGTAGGTACATGAGTGAAGTTTCTTACACTTTTAAAATTGCATTGGCAAATTTT CTTTTGCTTTATGAACCCCTGTTTTGTCATTATGATTTTTCACTTGTTTTAACAGTTTCAGATGTCACCTTGGAATAGCACTGTTTATTT TGGCCTTTTTATGATTGCTGATGATAGGTACAGTCTAAAGGCCAAAAGTTGCCACTTCATTGTGATAACATTGTTGCAACATTCCTAATA TATTAGAGAATGATGTAAAATAAGAATGTTAGAGAACATAGCACATTCTCATTCAGTTTATAAACAATATTTGAAAATTTCCAGTGAGAA TTAAAAAAGGACTAGTGATGATATAGCAAACCTGGACCTGAAATTTGGGGGTGGGGGGTGGGGGGATATCTGTTTTGCAGAGAAAATTTA CCAGCCTCCAGCAAAATCTCTGCATCTTGTACTCAATAGCAACCATTAGTATTGTCATTTAAAAAGAATGAATCAAAAATTCATACGTAA TGGAATGAAAAGAGGAAAACTTTCTATTAAACACTGTCAAGAAAGACCCATTGCTATCTATCACCTATGAAATTAAACCTAAAAAAATTT TTTTTTATTGTAAAAGCGACTGCTCGTTGATGTCTGGAAGTGATCCAGACTTTGTTTCACTGTGATCCAAAAAGTTAGACATTCCTTCCA TTTCTCAAGATTTGGTAACAATTTTTTTTTAAGTCAGCATTTCATATAATACATAAAATTAAGACAAATTAACTTACTGATATGTGGCCA ATATCAACCCTTCTCACCCAACCCAAACATGTTCTGGAGATAGAAGGACTTATCCTCAATTATATTCCAGTAAATTTATCTCCATAATCT GTTTTATCCCCTAATGTATGATATATCTCACCAAATTTTTCTAGAATTGAATAAAAGCAGTTTTTGCTTAACTTGAATTTCCTTCCACAA ATACAACCTTCAGCTCTTCCCTCTCTGTTTCCCTTTTGCCCACATTGCAAATAGTCTTCATAATAGAACCCTCACTGCAACTTAAAAGGG TCCCATTTCTTAACTCAGCATTTCATATACCAAGAACACTTGCAAAGATTTCTTCATAATGTGATTTTTTAATTTTTCAGTTCCAGAAGC ATTTCCTACAGTTCGAACGTTATTTTTTCTCTCAGATTTTCAACTTGAAGCTTTGATTTGCACTGAATTAACTTGGATAAAATTTTAGTT TCTCTCTTCAGCTAACAGATTTGGAAGCAAATTACAGTATACTTGAATAATTCAGGTACTACCTGTTGACCTGGAATTTTAGTGATTCTG GGAAAGTTTTCCAGACAAAGACACCAATAGAGTGAAGGGTTTTTTCCCCAATTACTATTTTATTGTTTTTTAAAATGCCTTTTTAAAACT TGTGTGATGTGGCAAAATAGTCATATGTTCAATTAATTTATATTTTATTCATTCCTTTTCAATTCTGGTTATTATGTAACCTCAAGTACT >14539_14539_2_CD47-CBLB_CD47_chr3_107789939_ENST00000361309_CBLB_chr3_105378073_ENST00000264122_length(amino acids)=274AA_BP=1 MPVTAAAAAAPDTCGGGGDPAAGAEMWPLVAALLLGSACCGSAQLLFNKTKSVEFTFCNDTVVIPCFVTNMEAQNTTEVYVKWKFKGRDI YTFDGALNKSTVPTDFSSAKIEVSQLLKGDASLKMDKSDAVSHTGNYTCEVTELTREGETIIELKYRVVSWFSPNENILIVIFPIFAILL FWGQFGIKNGSQAPARPPKPRPRRTAPEIHHRKPHGPEAALENVDAKIAKLMGEGYAFEEVKRALEIAQNNVEVARSILREFAFPPPVSP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CD47-CBLB |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 891_927 | 0 | 771.0 | SH3KBP1 | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000403724 | 0 | 15 | 543_568 | 0 | 771.0 | VAV1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 891_927 | 896.3333333333334 | 983.0 | SH3KBP1 | |
| Tgene | CBLB | chr3:107789939 | chr3:105378073 | ENST00000264122 | 17 | 19 | 543_568 | 896.3333333333334 | 983.0 | VAV1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CD47-CBLB |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CD47-CBLB |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
