
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CD63-C12orf10 (FusionGDB2 ID:14579) |
Fusion Gene Summary for CD63-C12orf10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CD63-C12orf10 | Fusion gene ID: 14579 | Hgene | Tgene | Gene symbol | CD63 | C12orf10 | Gene ID | 967 | 60314 |
| Gene name | CD63 molecule | MYG1 exonuclease | |
| Synonyms | LAMP-3|ME491|MLA1|OMA81H|TSPAN30 | C12orf10|Gamm1|MST024|MSTP024|MYG | |
| Cytomap | 12q13.2 | 12q13.13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CD63 antigenCD63 antigen (melanoma 1 antigen)granulophysinlysosomal-associated membrane protein 3lysosome-associated membrane glycoprotein 3melanoma-associated antigen ME491melanoma-associated antigen MLA1ocular melanoma-associated antigentetraspa | UPF0160 protein MYG1, mitochondrialmelanocyte proliferating gene 1melanocyte related | |
| Modification date | 20200327 | 20200320 | |
| UniProtAcc | P08962 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000257857, ENST00000420846, ENST00000546939, ENST00000548160, ENST00000548898, ENST00000549117, ENST00000550776, ENST00000552067, ENST00000552692, ENST00000552754, | ENST00000549488, ENST00000267103, ENST00000548632, | |
| Fusion gene scores | * DoF score | 23 X 19 X 9=3933 | 8 X 12 X 5=480 |
| # samples | 23 | 12 | |
| ** MAII score | log2(23/3933*10)=-4.09592441999854 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/480*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CD63 [Title/Abstract] AND C12orf10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CD63(56120484)-C12orf10(53693934), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CD63-C12orf10 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CD63-C12orf10 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CD63 | GO:0034613 | cellular protein localization | 15647390 |
| Hgene | CD63 | GO:1901379 | regulation of potassium ion transmembrane transport | 15647390 |
 Fusion gene breakpoints across CD63 (5'-gene) Fusion gene breakpoints across CD63 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
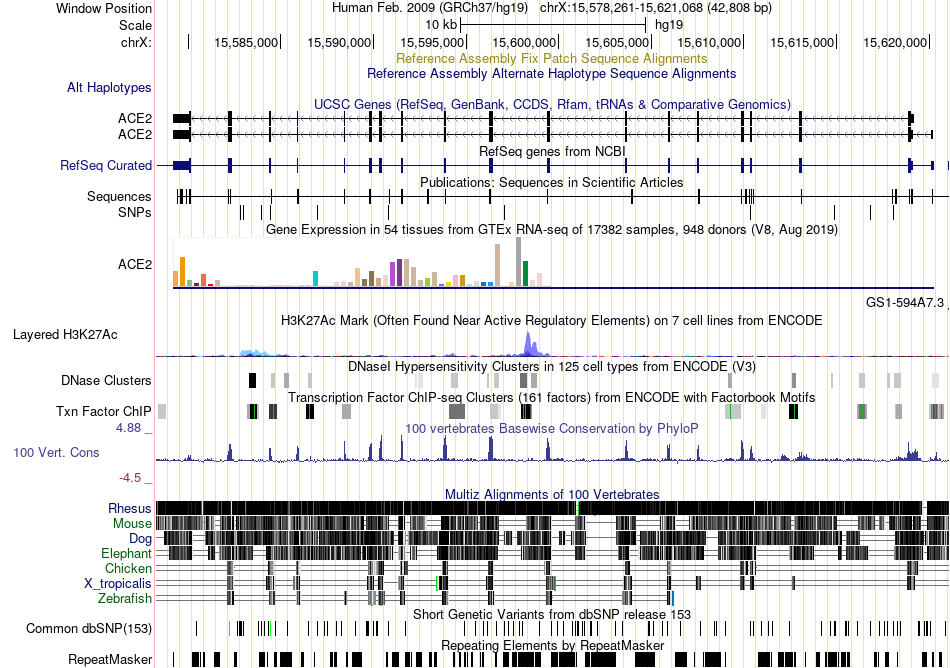 |
 Fusion gene breakpoints across C12orf10 (3'-gene) Fusion gene breakpoints across C12orf10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
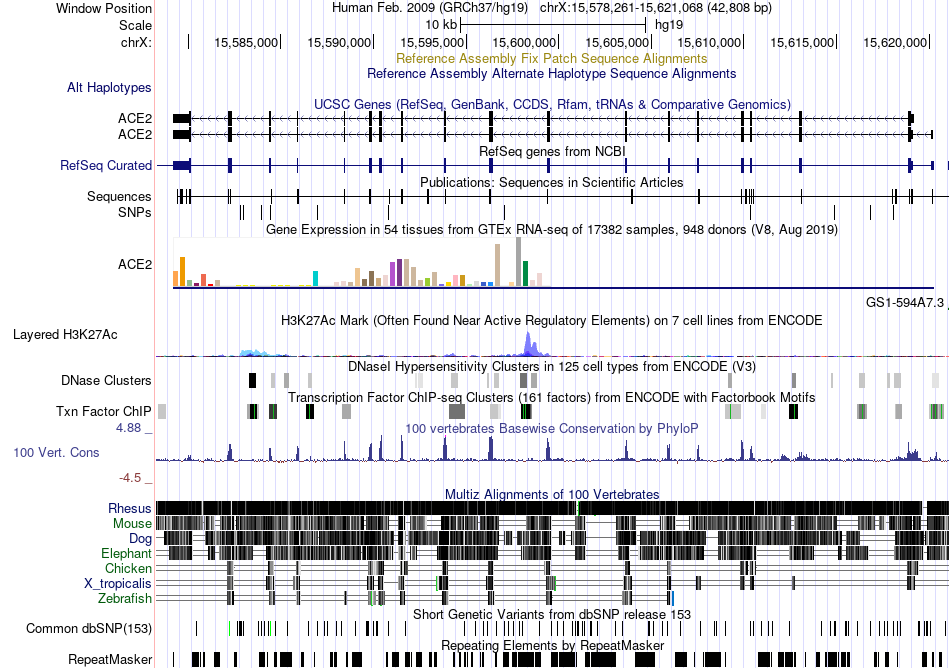 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-D3-A3C1-06A | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
Top |
Fusion Gene ORF analysis for CD63-C12orf10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000257857 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000420846 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000546939 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000548160 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000548898 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000549117 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000550776 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000552067 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000552692 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| 5CDS-5UTR | ENST00000552754 | ENST00000549488 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000546939 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000546939 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000549117 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000549117 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000550776 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000550776 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000552067 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| Frame-shift | ENST00000552067 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000257857 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000257857 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000420846 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000420846 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000548160 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000548160 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000548898 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000548898 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000552692 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000552692 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000552754 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
| In-frame | ENST00000552754 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000548898 | CD63 | chr12 | 56120484 | - | ENST00000267103 | C12orf10 | chr12 | 53693934 | + | 1122 | 198 | 39 | 1112 | 357 |
| ENST00000548898 | CD63 | chr12 | 56120484 | - | ENST00000548632 | C12orf10 | chr12 | 53693934 | + | 982 | 198 | 39 | 959 | 306 |
| ENST00000420846 | CD63 | chr12 | 56120484 | - | ENST00000267103 | C12orf10 | chr12 | 53693934 | + | 1535 | 611 | 161 | 1525 | 454 |
| ENST00000420846 | CD63 | chr12 | 56120484 | - | ENST00000548632 | C12orf10 | chr12 | 53693934 | + | 1395 | 611 | 161 | 1372 | 403 |
| ENST00000548160 | CD63 | chr12 | 56120484 | - | ENST00000267103 | C12orf10 | chr12 | 53693934 | + | 1190 | 266 | 83 | 1180 | 365 |
| ENST00000548160 | CD63 | chr12 | 56120484 | - | ENST00000548632 | C12orf10 | chr12 | 53693934 | + | 1050 | 266 | 83 | 1027 | 314 |
| ENST00000552692 | CD63 | chr12 | 56120484 | - | ENST00000267103 | C12orf10 | chr12 | 53693934 | + | 1491 | 567 | 141 | 1481 | 446 |
| ENST00000552692 | CD63 | chr12 | 56120484 | - | ENST00000548632 | C12orf10 | chr12 | 53693934 | + | 1351 | 567 | 141 | 1328 | 395 |
| ENST00000257857 | CD63 | chr12 | 56120484 | - | ENST00000267103 | C12orf10 | chr12 | 53693934 | + | 1629 | 705 | 255 | 1619 | 454 |
| ENST00000257857 | CD63 | chr12 | 56120484 | - | ENST00000548632 | C12orf10 | chr12 | 53693934 | + | 1489 | 705 | 255 | 1466 | 403 |
| ENST00000552754 | CD63 | chr12 | 56120484 | - | ENST00000267103 | C12orf10 | chr12 | 53693934 | + | 1289 | 365 | 8 | 1279 | 423 |
| ENST00000552754 | CD63 | chr12 | 56120484 | - | ENST00000548632 | C12orf10 | chr12 | 53693934 | + | 1149 | 365 | 8 | 1126 | 372 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000548898 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.006989917 | 0.9930101 |
| ENST00000548898 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.012997316 | 0.9870027 |
| ENST00000420846 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.002596815 | 0.9974032 |
| ENST00000420846 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.001881837 | 0.99811816 |
| ENST00000548160 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.005876635 | 0.9941234 |
| ENST00000548160 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.010889193 | 0.98911077 |
| ENST00000552692 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.002092924 | 0.99790704 |
| ENST00000552692 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.001509516 | 0.99849045 |
| ENST00000257857 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.002546023 | 0.997454 |
| ENST00000257857 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.001791613 | 0.9982084 |
| ENST00000552754 | ENST00000267103 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.001806544 | 0.99819344 |
| ENST00000552754 | ENST00000548632 | CD63 | chr12 | 56120484 | - | C12orf10 | chr12 | 53693934 | + | 0.001932333 | 0.9980677 |
Top |
Fusion Genomic Features for CD63-C12orf10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CD63 | chr12 | 56120483 | - | C12orf10 | chr12 | 53693933 | + | 1.92E-05 | 0.9999808 |
| CD63 | chr12 | 56120483 | - | C12orf10 | chr12 | 53693933 | + | 1.92E-05 | 0.9999808 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
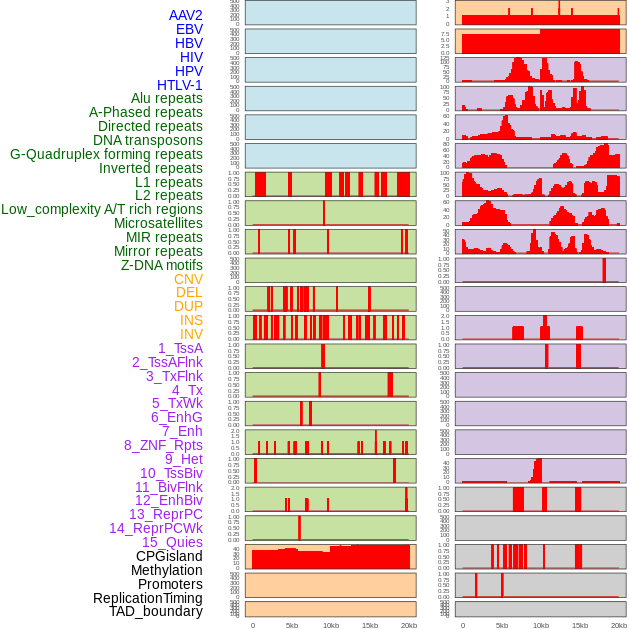 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
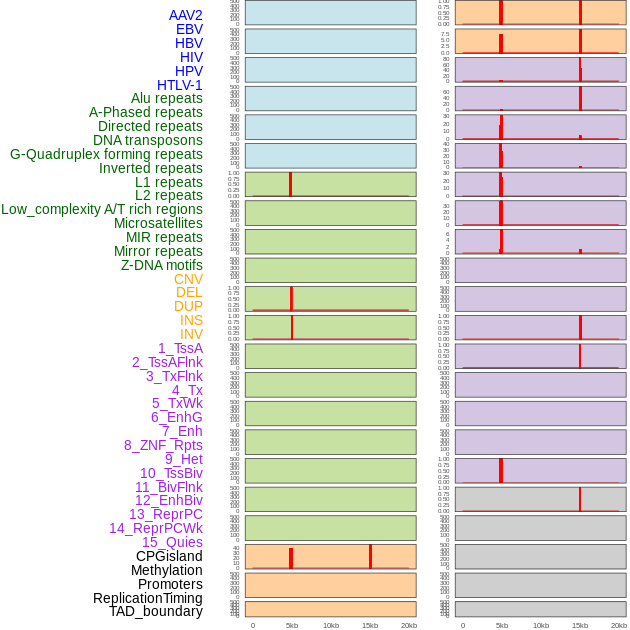 |
Top |
Fusion Protein Features for CD63-C12orf10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:56120484/chr12:53693934) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CD63 | . |
| FUNCTION: Functions as cell surface receptor for TIMP1 and plays a role in the activation of cellular signaling cascades. Plays a role in the activation of ITGB1 and integrin signaling, leading to the activation of AKT, FAK/PTK2 and MAP kinases. Promotes cell survival, reorganization of the actin cytoskeleton, cell adhesion, spreading and migration, via its role in the activation of AKT and FAK/PTK2. Plays a role in VEGFA signaling via its role in regulating the internalization of KDR/VEGFR2. Plays a role in intracellular vesicular transport processes, and is required for normal trafficking of the PMEL luminal domain that is essential for the development and maturation of melanocytes. Plays a role in the adhesion of leukocytes onto endothelial cells via its role in the regulation of SELP trafficking. May play a role in mast cell degranulation in response to Ms4a2/FceRI stimulation, but not in mast cell degranulation in response to other stimuli. {ECO:0000269|PubMed:16917503, ECO:0000269|PubMed:21803846, ECO:0000269|PubMed:21962903, ECO:0000269|PubMed:23632027, ECO:0000269|PubMed:24635319}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 2_11 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 33_51 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 73_81 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 2_11 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 33_51 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 2_11 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 33_51 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 73_81 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 2_11 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 33_51 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 2_11 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 33_51 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 73_81 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 2_11 | 119 | 216.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 33_51 | 119 | 216.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 73_81 | 119 | 216.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 12_32 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 52_72 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 82_102 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 12_32 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 12_32 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 52_72 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 82_102 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 12_32 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 12_32 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 52_72 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 82_102 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 12_32 | 119 | 216.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 52_72 | 119 | 216.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 82_102 | 119 | 216.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 234_238 | 142 | 239.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 234_238 | 60 | 157.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 234_238 | 142 | 239.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 234_238 | 60 | 157.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 234_238 | 142 | 239.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 234_238 | 119 | 216.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 103_203 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 225_238 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 103_203 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 225_238 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 73_81 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 103_203 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 225_238 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 103_203 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 225_238 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 73_81 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 103_203 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 225_238 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 103_203 | 119 | 216.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 225_238 | 119 | 216.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000257857 | - | 5 | 8 | 204_224 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 204_224 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 52_72 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000546939 | - | 4 | 7 | 82_102 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000549117 | - | 5 | 8 | 204_224 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 204_224 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 52_72 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000550776 | - | 4 | 7 | 82_102 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552692 | - | 4 | 7 | 204_224 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr12:53693934 | ENST00000552754 | - | 4 | 7 | 204_224 | 119 | 216.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CD63-C12orf10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >14579_14579_1_CD63-C12orf10_CD63_chr12_56120484_ENST00000257857_C12orf10_chr12_53693934_ENST00000267103_length(transcript)=1629nt_BP=705nt CGGGGGTGGGCGCCTTCTTCCGGGTAGGGGCCACGTGGCCCTGGCCGGGCGGGGGGCTCGGCCCACCCCGCGCCGGGCCCAGTGACTCAG GCCGCAGCTGTTACCGCGTCACATGAGGGAGGCCGGCGGCCACTCGGCGGGGGAGGGGACCGTGGCTGGAGCCCGGGGCGGGGCCGCGCG GCAGGCGGGGCGGGAGCCGGGGGGCGCAGCTAGAGAGCCCCGGAGCCGCGGCGGGAGAGGAACGCGCAGCCAGCCTTGGGAAGCCCAGGC CCGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGA CTGATTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATAATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATC ATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCC ATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAAC AACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGATGCAGAGATTGTG CGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTCGGAGACACCGATATGACCAT CACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCAGTGCGGGACTCATCTATCTG CACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCTATGACAAGATGTATGAGAAC TTTGTGGAGGAGGTGGATGCTGTGGACAATGGGATCTCCCAGTGGGCAGAGGGGGAGCCTCGATATGCACTGACCACTACCCTGAGTGCA CGAGTTGCTCGACTTAATCCTACCTGGAACCACCCCGACCAAGACACTGAGGCAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAG TTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGAC CCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTG GCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTG CCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGC TTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAGCGCTCATACCTCCCACAAATCTCCTAG >14579_14579_1_CD63-C12orf10_CD63_chr12_56120484_ENST00000257857_C12orf10_chr12_53693934_ENST00000267103_length(amino acids)=454AA_BP=150 MGSPGPAAMAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCL MITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRR HRYDHHQRSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKMYENFVEEVDAVDNGISQWAEGEPRYALT TTLSARVARLNPTWNHPDQDTEAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESG LSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQVSGIPGCIFVHASGFTGGHHTREGALSMARATLAQRSYL -------------------------------------------------------------- >14579_14579_2_CD63-C12orf10_CD63_chr12_56120484_ENST00000257857_C12orf10_chr12_53693934_ENST00000548632_length(transcript)=1489nt_BP=705nt CGGGGGTGGGCGCCTTCTTCCGGGTAGGGGCCACGTGGCCCTGGCCGGGCGGGGGGCTCGGCCCACCCCGCGCCGGGCCCAGTGACTCAG GCCGCAGCTGTTACCGCGTCACATGAGGGAGGCCGGCGGCCACTCGGCGGGGGAGGGGACCGTGGCTGGAGCCCGGGGCGGGGCCGCGCG GCAGGCGGGGCGGGAGCCGGGGGGCGCAGCTAGAGAGCCCCGGAGCCGCGGCGGGAGAGGAACGCGCAGCCAGCCTTGGGAAGCCCAGGC CCGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGA CTGATTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATAATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATC ATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCC ATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAAC AACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGATGCAGAGATTGTG CGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTCGGAGACACCGATATGACCAT CACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCAGTGCGGGACTCATCTATCTG CACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCTATGACAAGGCAGGGTTCAAG CGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAG GCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCAC CTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAG GAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCT GGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAG >14579_14579_2_CD63-C12orf10_CD63_chr12_56120484_ENST00000257857_C12orf10_chr12_53693934_ENST00000548632_length(amino acids)=403AA_BP=150 MGSPGPAAMAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCL MITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRR HRYDHHQRSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKAGFKRAMDLVQEEFLQRLDFYQHSWLPAR ALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQ -------------------------------------------------------------- >14579_14579_3_CD63-C12orf10_CD63_chr12_56120484_ENST00000420846_C12orf10_chr12_53693934_ENST00000267103_length(transcript)=1535nt_BP=611nt CAGCTGTTACCGCGTCACATGAGGGAGGCCGGCGGCCACTCGGCGGGGGAGGGGACCGTGGCTGGAGCCCGGGGCGGGGCCGCGCGGCAG GCGGGGCGGGAGCCGGGGGGCGCAGCTAGAGAGCCCCGGAGCCGCGGCGGGAGAGGAACGCGCAGCCAGCCTTGGGAAGCCCAGGCCCGG CAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGACTGA TTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATAATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATCATCG CAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCCATCT TTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACT TCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGATGCAGAGATTGTGCGGA CCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTCGGAGACACCGATATGACCATCACC AGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCAGTGCGGGACTCATCTATCTGCACT TCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCTATGACAAGATGTATGAGAACTTTG TGGAGGAGGTGGATGCTGTGGACAATGGGATCTCCCAGTGGGCAGAGGGGGAGCCTCGATATGCACTGACCACTACCCTGAGTGCACGAG TTGCTCGACTTAATCCTACCTGGAACCACCCCGACCAAGACACTGAGGCAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTC TGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAA GTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCA TCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCC TGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCA CTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAGCGCTCATACCTCCCACAAATCTCCTAGTCTA >14579_14579_3_CD63-C12orf10_CD63_chr12_56120484_ENST00000420846_C12orf10_chr12_53693934_ENST00000267103_length(amino acids)=454AA_BP=150 MGSPGPAAMAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCL MITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRR HRYDHHQRSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKMYENFVEEVDAVDNGISQWAEGEPRYALT TTLSARVARLNPTWNHPDQDTEAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESG LSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQVSGIPGCIFVHASGFTGGHHTREGALSMARATLAQRSYL -------------------------------------------------------------- >14579_14579_4_CD63-C12orf10_CD63_chr12_56120484_ENST00000420846_C12orf10_chr12_53693934_ENST00000548632_length(transcript)=1395nt_BP=611nt CAGCTGTTACCGCGTCACATGAGGGAGGCCGGCGGCCACTCGGCGGGGGAGGGGACCGTGGCTGGAGCCCGGGGCGGGGCCGCGCGGCAG GCGGGGCGGGAGCCGGGGGGCGCAGCTAGAGAGCCCCGGAGCCGCGGCGGGAGAGGAACGCGCAGCCAGCCTTGGGAAGCCCAGGCCCGG CAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGACTGA TTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATAATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATCATCG CAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCCATCT TTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACT TCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGATGCAGAGATTGTGCGGA CCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTCGGAGACACCGATATGACCATCACC AGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCAGTGCGGGACTCATCTATCTGCACT TCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCTATGACAAGGCAGGGTTCAAGCGTG CAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCC TTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGG AATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGC CCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCT GCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAGCGCT >14579_14579_4_CD63-C12orf10_CD63_chr12_56120484_ENST00000420846_C12orf10_chr12_53693934_ENST00000548632_length(amino acids)=403AA_BP=150 MGSPGPAAMAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCL MITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRR HRYDHHQRSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKAGFKRAMDLVQEEFLQRLDFYQHSWLPAR ALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQ -------------------------------------------------------------- >14579_14579_5_CD63-C12orf10_CD63_chr12_56120484_ENST00000548160_C12orf10_chr12_53693934_ENST00000267103_length(transcript)=1190nt_BP=266nt GAAGACCTTTGTGAGTGTCCAGGGACCGGCTGTGCCAAAACGGGCCCCTGTAATGCATAGACTCCAAACTTGACTTCTGTCCTTTGCTCC TGCAGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAG AGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGATG CAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTCGGAGACACC GATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCAGTGCGGGAC TCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCTATGACAAGA TGTATGAGAACTTTGTGGAGGAGGTGGATGCTGTGGACAATGGGATCTCCCAGTGGGCAGAGGGGGAGCCTCGATATGCACTGACCACTA CCCTGAGTGCACGAGTTGCTCGACTTAATCCTACCTGGAACCACCCCGACCAAGACACTGAGGCAGGGTTCAAGCGTGCAATGGATCTGG TTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCCTTGCCCAGCGAT TCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGGAATCTGGGCTGT CCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGCCCCACTCATTCC AAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCTGCATCTTCGTCC ATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAGCGCTCATACCTCCCAC >14579_14579_5_CD63-C12orf10_CD63_chr12_56120484_ENST00000548160_C12orf10_chr12_53693934_ENST00000267103_length(amino acids)=365AA_BP=61 MLLQFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPR RHRYDHHQRSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKMYENFVEEVDAVDNGISQWAEGEPRYAL TTTLSARVARLNPTWNHPDQDTEAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLES GLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQVSGIPGCIFVHASGFTGGHHTREGALSMARATLAQRSY -------------------------------------------------------------- >14579_14579_6_CD63-C12orf10_CD63_chr12_56120484_ENST00000548160_C12orf10_chr12_53693934_ENST00000548632_length(transcript)=1050nt_BP=266nt GAAGACCTTTGTGAGTGTCCAGGGACCGGCTGTGCCAAAACGGGCCCCTGTAATGCATAGACTCCAAACTTGACTTCTGTCCTTTGCTCC TGCAGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAG AGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGATG CAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTCGGAGACACC GATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCAGTGCGGGAC TCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCTATGACAAGG CAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCT TGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGC ATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGT GTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCA GTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCA >14579_14579_6_CD63-C12orf10_CD63_chr12_56120484_ENST00000548160_C12orf10_chr12_53693934_ENST00000548632_length(amino acids)=314AA_BP=61 MLLQFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPR RHRYDHHQRSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKAGFKRAMDLVQEEFLQRLDFYQHSWLPA RALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALD -------------------------------------------------------------- >14579_14579_7_CD63-C12orf10_CD63_chr12_56120484_ENST00000548898_C12orf10_chr12_53693934_ENST00000267103_length(transcript)=1122nt_BP=198nt AGAGCAGCTTGGTTCGGGCTCAGGAAGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTG TTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTG GACAGGATGCAGGCAGATGATGCAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGC GAGTACGACCCTCGGAGACACCGATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAG ACCAAGCTGAGCAGTGCGGGACTCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATG GTGGGCACCCTCTATGACAAGATGTATGAGAACTTTGTGGAGGAGGTGGATGCTGTGGACAATGGGATCTCCCAGTGGGCAGAGGGGGAG CCTCGATATGCACTGACCACTACCCTGAGTGCACGAGTTGCTCGACTTAATCCTACCTGGAACCACCCCGACCAAGACACTGAGGCAGGG TTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTG GAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTC TACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTG CCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGG ATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTG >14579_14579_7_CD63-C12orf10_CD63_chr12_56120484_ENST00000548898_C12orf10_chr12_53693934_ENST00000267103_length(amino acids)=357AA_BP=53 MSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRRHRYDHHQ RSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKMYENFVEEVDAVDNGISQWAEGEPRYALTTTLSARV ARLNPTWNHPDQDTEAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAI -------------------------------------------------------------- >14579_14579_8_CD63-C12orf10_CD63_chr12_56120484_ENST00000548898_C12orf10_chr12_53693934_ENST00000548632_length(transcript)=982nt_BP=198nt AGAGCAGCTTGGTTCGGGCTCAGGAAGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTG TTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTG GACAGGATGCAGGCAGATGATGCAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGC GAGTACGACCCTCGGAGACACCGATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAG ACCAAGCTGAGCAGTGCGGGACTCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATG GTGGGCACCCTCTATGACAAGGCAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACAC AGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAA GGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAG GCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGG GACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGT >14579_14579_8_CD63-C12orf10_CD63_chr12_56120484_ENST00000548898_C12orf10_chr12_53693934_ENST00000548632_length(amino acids)=306AA_BP=53 MSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRRHRYDHHQ RSFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEAL AQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQVSGIPGC -------------------------------------------------------------- >14579_14579_9_CD63-C12orf10_CD63_chr12_56120484_ENST00000552692_C12orf10_chr12_53693934_ENST00000267103_length(transcript)=1491nt_BP=567nt GCGGGTTTCCGGGTGGGAGGGGCCGGGAATCGGGGCCGTGAAGGGAAGCCCTCGAGGGCCGGCCGGGAGCCCGGAAACCGAGGTCCCCCT TCGGCCAGTTTTAATCCCCCCGCCCTCCCCTCTGCCCCAGGCCCGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTG CTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGACTGATTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATA ATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATCATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGC GGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCT GGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCT TCGATCCTGGACAGGATGCAGGCAGATGATGCAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGAC GTGGGGGGCGAGTACGACCCTCGGAGACACCGATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAG CCGTGGCAGACCAAGCTGAGCAGTGCGGGACTCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAG GACAGCATGGTGGGCACCCTCTATGACAAGATGTATGAGAACTTTGTGGAGGAGGTGGATGCTGTGGACAATGGGATCTCCCAGTGGGCA GAGGGGGAGCCTCGATATGCACTGACCACTACCCTGAGTGCACGAGTTGCTCGACTTAATCCTACCTGGAACCACCCCGACCAAGACACT GAGGCAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGG GCCTTGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAG GAGCATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATA CAGTGTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAG GTCAGTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGT >14579_14579_9_CD63-C12orf10_CD63_chr12_56120484_ENST00000552692_C12orf10_chr12_53693934_ENST00000267103_length(amino acids)=446AA_BP=142 MAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFL SLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRRHRYDHHQR SFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKMYENFVEEVDAVDNGISQWAEGEPRYALTTTLSARVA RLNPTWNHPDQDTEAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIF -------------------------------------------------------------- >14579_14579_10_CD63-C12orf10_CD63_chr12_56120484_ENST00000552692_C12orf10_chr12_53693934_ENST00000548632_length(transcript)=1351nt_BP=567nt GCGGGTTTCCGGGTGGGAGGGGCCGGGAATCGGGGCCGTGAAGGGAAGCCCTCGAGGGCCGGCCGGGAGCCCGGAAACCGAGGTCCCCCT TCGGCCAGTTTTAATCCCCCCGCCCTCCCCTCTGCCCCAGGCCCGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTG CTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGACTGATTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATA ATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATCATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGC GGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCT GGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCT TCGATCCTGGACAGGATGCAGGCAGATGATGCAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGAC GTGGGGGGCGAGTACGACCCTCGGAGACACCGATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAG CCGTGGCAGACCAAGCTGAGCAGTGCGGGACTCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAG GACAGCATGGTGGGCACCCTCTATGACAAGGCAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTC TACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAA CTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTAC ACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGG GGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACC CGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAGCGCTCATACCTCCCACAAATCTCCTAGTCTAATAAAACCTTCCATCTC >14579_14579_10_CD63-C12orf10_CD63_chr12_56120484_ENST00000552692_C12orf10_chr12_53693934_ENST00000548632_length(amino acids)=395AA_BP=142 MAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFL SLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRRHRYDHHQR SFTETMSSLSPGKPWQTKLSSAGLIYLHFGHKLLAQLLGTSEEDSMVGTLYDKAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALA QRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQVSGIPGCI -------------------------------------------------------------- >14579_14579_11_CD63-C12orf10_CD63_chr12_56120484_ENST00000552754_C12orf10_chr12_53693934_ENST00000267103_length(transcript)=1289nt_BP=365nt CGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGGGGCTACCCCTGGCT CTCTGTTGCCAGTGGTCATCATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATT GTCTTATGATCACGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGG TGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGG CAGATGATGCAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTC GGAGACACCGATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCA GTGCGGGACTCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCT ATGACAAGATGTATGAGAACTTTGTGGAGGAGGTGGATGCTGTGGACAATGGGATCTCCCAGTGGGCAGAGGGGGAGCCTCGATATGCAC TGACCACTACCCTGAGTGCACGAGTTGCTCGACTTAATCCTACCTGGAACCACCCCGACCAAGACACTGAGGCAGGGTTCAAGCGTGCAA TGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAGCCCGGGCCTTGGTGGAAGAGGCCCTTG CCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCTGGAAGGAGCATCTCTACCACCTGGAAT CTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGCGAATACAGTGTGTGCCCAAGGAGCCCC ACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGGACCAGGTCAGTGGGATCCCTGGCTGCA TCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGGCCCGTGCCACCTTGGCCCAGCGCTCAT >14579_14579_11_CD63-C12orf10_CD63_chr12_56120484_ENST00000552754_C12orf10_chr12_53693934_ENST00000267103_length(amino acids)=423AA_BP=119 MAVEGGMKCVKFLLYVLLLAFCGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFLSLIMLVEVAAAIAGYVFRDKVMS EFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRRHRYDHHQRSFTETMSSLSPGKPWQTKLSSAG LIYLHFGHKLLAQLLGTSEEDSMVGTLYDKMYENFVEEVDAVDNGISQWAEGEPRYALTTTLSARVARLNPTWNHPDQDTEAGFKRAMDL VQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKEHLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSF -------------------------------------------------------------- >14579_14579_12_CD63-C12orf10_CD63_chr12_56120484_ENST00000552754_C12orf10_chr12_53693934_ENST00000548632_length(transcript)=1149nt_BP=365nt CGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGGGGCTACCCCTGGCT CTCTGTTGCCAGTGGTCATCATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATT GTCTTATGATCACGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGG TGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGG CAGATGATGCAGAGATTGTGCGGACCCGGGATCCCGAAAAACTCGCTTCCTGTGACATCGTGGTGGACGTGGGGGGCGAGTACGACCCTC GGAGACACCGATATGACCATCACCAGAGGTCTTTCACAGAGACCATGAGCTCCCTGTCCCCTGGGAAGCCGTGGCAGACCAAGCTGAGCA GTGCGGGACTCATCTATCTGCACTTCGGGCACAAGCTGCTGGCCCAGTTGCTGGGCACTAGTGAAGAGGACAGCATGGTGGGCACCCTCT ATGACAAGGCAGGGTTCAAGCGTGCAATGGATCTGGTTCAAGAGGAGTTTCTGCAGAGATTAGATTTCTACCAACACAGCTGGCTGCCAG CCCGGGCCTTGGTGGAAGAGGCCCTTGCCCAGCGATTCCAGGTGGACCCAAGTGGAGAGATTGTGGAACTGGCGAAAGGTGCATGTCCCT GGAAGGAGCATCTCTACCACCTGGAATCTGGGCTGTCCCCTCCAGTGGCCATCTTCTTTGTTATCTACACTGACCAGGCTGGACAGTGGC GAATACAGTGTGTGCCCAAGGAGCCCCACTCATTCCAAAGCCGGCTGCCCCTGCCAGAGCCATGGCGGGGTCTTCGGGACGAGGCCCTGG ACCAGGTCAGTGGGATCCCTGGCTGCATCTTCGTCCATGCAAGCGGCTTCACTGGCGGTCACCACACCCGAGAGGGTGCCTTGAGCATGG >14579_14579_12_CD63-C12orf10_CD63_chr12_56120484_ENST00000552754_C12orf10_chr12_53693934_ENST00000548632_length(amino acids)=372AA_BP=119 MAVEGGMKCVKFLLYVLLLAFCGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFLSLIMLVEVAAAIAGYVFRDKVMS EFNNNFRQQMENYPKNNHTASILDRMQADDAEIVRTRDPEKLASCDIVVDVGGEYDPRRHRYDHHQRSFTETMSSLSPGKPWQTKLSSAG LIYLHFGHKLLAQLLGTSEEDSMVGTLYDKAGFKRAMDLVQEEFLQRLDFYQHSWLPARALVEEALAQRFQVDPSGEIVELAKGACPWKE HLYHLESGLSPPVAIFFVIYTDQAGQWRIQCVPKEPHSFQSRLPLPEPWRGLRDEALDQVSGIPGCIFVHASGFTGGHHTREGALSMARA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CD63-C12orf10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CD63-C12orf10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CD63-C12orf10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
