
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CD63-COL1A2 (FusionGDB2 ID:14583) |
Fusion Gene Summary for CD63-COL1A2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CD63-COL1A2 | Fusion gene ID: 14583 | Hgene | Tgene | Gene symbol | CD63 | COL1A2 | Gene ID | 967 | 1278 |
| Gene name | CD63 molecule | collagen type I alpha 2 chain | |
| Synonyms | LAMP-3|ME491|MLA1|OMA81H|TSPAN30 | EDSARTH2|EDSCV|OI4 | |
| Cytomap | 12q13.2 | 7q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CD63 antigenCD63 antigen (melanoma 1 antigen)granulophysinlysosomal-associated membrane protein 3lysosome-associated membrane glycoprotein 3melanoma-associated antigen ME491melanoma-associated antigen MLA1ocular melanoma-associated antigentetraspa | collagen alpha-2(I) chainalpha 2 type I procollagenalpha 2(I) procollagenalpha 2(I)-collagenalpha-2 type I collagencollagen I, alpha-2 polypeptidecollagen of skin, tendon and bone, alpha-2 chaincollagen, type I, alpha 2epididymis secretory sperm b | |
| Modification date | 20200327 | 20200322 | |
| UniProtAcc | P08962 | P08123 | |
| Ensembl transtripts involved in fusion gene | ENST00000546939, ENST00000549117, ENST00000550776, ENST00000552067, ENST00000257857, ENST00000420846, ENST00000548160, ENST00000548898, ENST00000552692, ENST00000552754, | ENST00000297268, | |
| Fusion gene scores | * DoF score | 23 X 19 X 9=3933 | 43 X 46 X 7=13846 |
| # samples | 23 | 54 | |
| ** MAII score | log2(23/3933*10)=-4.09592441999854 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(54/13846*10)=-4.68036603576588 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CD63 [Title/Abstract] AND COL1A2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CD63(56120484)-COL1A2(94055062), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CD63-COL1A2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CD63-COL1A2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CD63 | GO:0034613 | cellular protein localization | 15647390 |
| Hgene | CD63 | GO:1901379 | regulation of potassium ion transmembrane transport | 15647390 |
| Tgene | COL1A2 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 17217948 |
| Tgene | COL1A2 | GO:0007266 | Rho protein signal transduction | 17217948 |
 Fusion gene breakpoints across CD63 (5'-gene) Fusion gene breakpoints across CD63 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
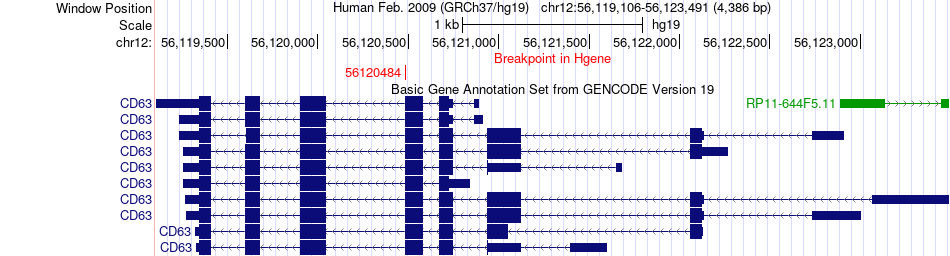 |
 Fusion gene breakpoints across COL1A2 (3'-gene) Fusion gene breakpoints across COL1A2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
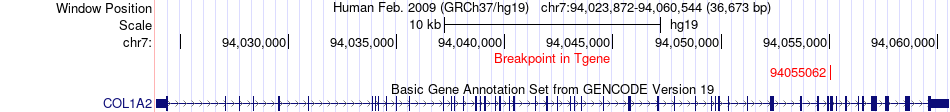 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A3UA-01A | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
Top |
Fusion Gene ORF analysis for CD63-COL1A2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000546939 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| Frame-shift | ENST00000549117 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| Frame-shift | ENST00000550776 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| Frame-shift | ENST00000552067 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| In-frame | ENST00000257857 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| In-frame | ENST00000420846 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| In-frame | ENST00000548160 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| In-frame | ENST00000548898 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| In-frame | ENST00000552692 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
| In-frame | ENST00000552754 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000548898 | CD63 | chr12 | 56120484 | - | ENST00000297268 | COL1A2 | chr7 | 94055062 | + | 2303 | 198 | 39 | 1463 | 474 |
| ENST00000420846 | CD63 | chr12 | 56120484 | - | ENST00000297268 | COL1A2 | chr7 | 94055062 | + | 2716 | 611 | 161 | 1876 | 571 |
| ENST00000548160 | CD63 | chr12 | 56120484 | - | ENST00000297268 | COL1A2 | chr7 | 94055062 | + | 2371 | 266 | 83 | 1531 | 482 |
| ENST00000552692 | CD63 | chr12 | 56120484 | - | ENST00000297268 | COL1A2 | chr7 | 94055062 | + | 2672 | 567 | 141 | 1832 | 563 |
| ENST00000257857 | CD63 | chr12 | 56120484 | - | ENST00000297268 | COL1A2 | chr7 | 94055062 | + | 2810 | 705 | 255 | 1970 | 571 |
| ENST00000552754 | CD63 | chr12 | 56120484 | - | ENST00000297268 | COL1A2 | chr7 | 94055062 | + | 2470 | 365 | 8 | 1630 | 540 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000548898 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + | 0.000465075 | 0.99953496 |
| ENST00000420846 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + | 0.00046495 | 0.9995351 |
| ENST00000548160 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + | 0.000452121 | 0.9995479 |
| ENST00000552692 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + | 0.000383161 | 0.99961686 |
| ENST00000257857 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + | 0.000468287 | 0.99953175 |
| ENST00000552754 | ENST00000297268 | CD63 | chr12 | 56120484 | - | COL1A2 | chr7 | 94055062 | + | 0.000309279 | 0.9996908 |
Top |
Fusion Genomic Features for CD63-COL1A2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CD63 | chr12 | 56120483 | - | COL1A2 | chr7 | 94055061 | + | 0.000315592 | 0.99968445 |
| CD63 | chr12 | 56120483 | - | COL1A2 | chr7 | 94055061 | + | 0.000315592 | 0.99968445 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
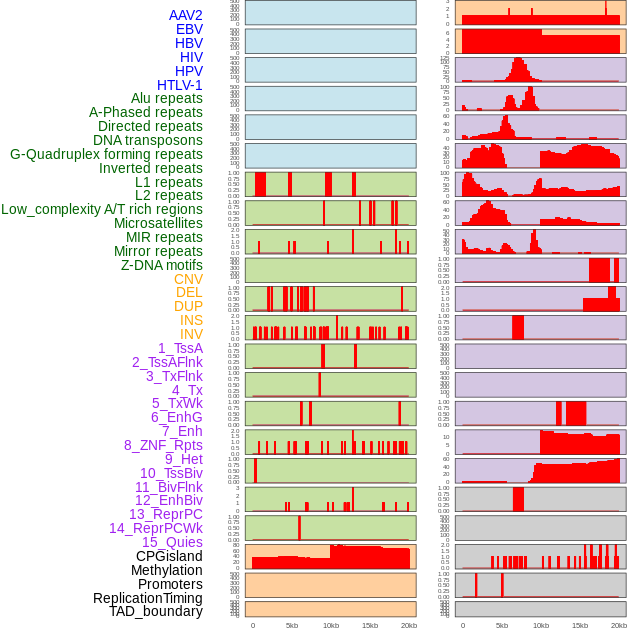 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
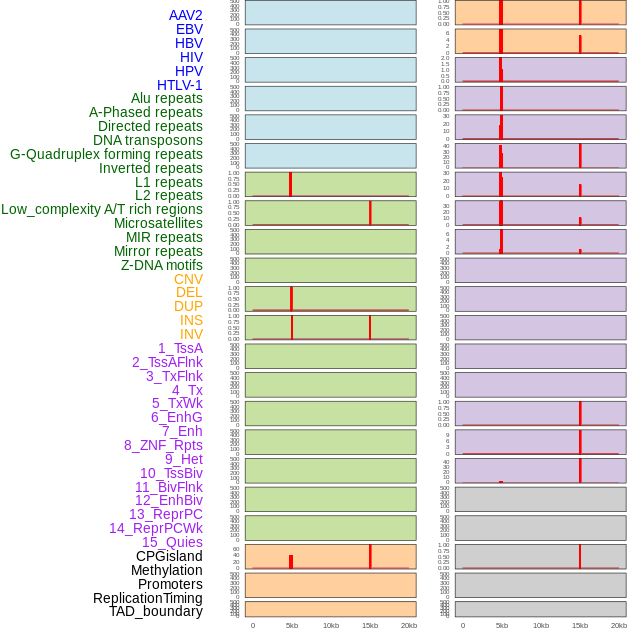 |
Top |
Fusion Protein Features for CD63-COL1A2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:56120484/chr7:94055062) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CD63 | COL1A2 |
| FUNCTION: Functions as cell surface receptor for TIMP1 and plays a role in the activation of cellular signaling cascades. Plays a role in the activation of ITGB1 and integrin signaling, leading to the activation of AKT, FAK/PTK2 and MAP kinases. Promotes cell survival, reorganization of the actin cytoskeleton, cell adhesion, spreading and migration, via its role in the activation of AKT and FAK/PTK2. Plays a role in VEGFA signaling via its role in regulating the internalization of KDR/VEGFR2. Plays a role in intracellular vesicular transport processes, and is required for normal trafficking of the PMEL luminal domain that is essential for the development and maturation of melanocytes. Plays a role in the adhesion of leukocytes onto endothelial cells via its role in the regulation of SELP trafficking. May play a role in mast cell degranulation in response to Ms4a2/FceRI stimulation, but not in mast cell degranulation in response to other stimuli. {ECO:0000269|PubMed:16917503, ECO:0000269|PubMed:21803846, ECO:0000269|PubMed:21962903, ECO:0000269|PubMed:23632027, ECO:0000269|PubMed:24635319}. | FUNCTION: Type I collagen is a member of group I collagen (fibrillar forming collagen). |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 2_11 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 33_51 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 73_81 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 2_11 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 33_51 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 2_11 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 33_51 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 73_81 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 2_11 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 33_51 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 2_11 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 33_51 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 73_81 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 2_11 | 119 | 216.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 33_51 | 119 | 216.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 73_81 | 119 | 216.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 12_32 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 52_72 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 82_102 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 12_32 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 12_32 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 52_72 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 82_102 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 12_32 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 12_32 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 52_72 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 82_102 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 12_32 | 119 | 216.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 52_72 | 119 | 216.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 82_102 | 119 | 216.0 | Transmembrane | Helical |
| Tgene | COL1A2 | chr12:56120484 | chr7:94055062 | ENST00000297268 | 42 | 52 | 1133_1366 | 945 | 1367.0 | Domain | Fibrillar collagen NC1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 234_238 | 142 | 239.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 234_238 | 60 | 157.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 234_238 | 142 | 239.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 234_238 | 60 | 157.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 234_238 | 142 | 239.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 234_238 | 119 | 216.0 | Motif | Lysosomal targeting motif |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 103_203 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 225_238 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 103_203 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 225_238 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 73_81 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 103_203 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 225_238 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 103_203 | 60 | 157.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 225_238 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 73_81 | 60 | 157.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 103_203 | 142 | 239.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 225_238 | 142 | 239.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 103_203 | 119 | 216.0 | Topological domain | Extracellular |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 225_238 | 119 | 216.0 | Topological domain | Cytoplasmic |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000257857 | - | 5 | 8 | 204_224 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 204_224 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 52_72 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000546939 | - | 4 | 7 | 82_102 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000549117 | - | 5 | 8 | 204_224 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 204_224 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 52_72 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000550776 | - | 4 | 7 | 82_102 | 60 | 157.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552692 | - | 4 | 7 | 204_224 | 142 | 239.0 | Transmembrane | Helical |
| Hgene | CD63 | chr12:56120484 | chr7:94055062 | ENST00000552754 | - | 4 | 7 | 204_224 | 119 | 216.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CD63-COL1A2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >14583_14583_1_CD63-COL1A2_CD63_chr12_56120484_ENST00000257857_COL1A2_chr7_94055062_ENST00000297268_length(transcript)=2810nt_BP=705nt CGGGGGTGGGCGCCTTCTTCCGGGTAGGGGCCACGTGGCCCTGGCCGGGCGGGGGGCTCGGCCCACCCCGCGCCGGGCCCAGTGACTCAG GCCGCAGCTGTTACCGCGTCACATGAGGGAGGCCGGCGGCCACTCGGCGGGGGAGGGGACCGTGGCTGGAGCCCGGGGCGGGGCCGCGCG GCAGGCGGGGCGGGAGCCGGGGGGCGCAGCTAGAGAGCCCCGGAGCCGCGGCGGGAGAGGAACGCGCAGCCAGCCTTGGGAAGCCCAGGC CCGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGA CTGATTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATAATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATC ATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCC ATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAAC AACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGGAGAGCGCGGTTAC CCTGGCAATATTGGTCCCGTTGGTGCTGCAGGTGCACCTGGTCCTCATGGCCCCGTGGGTCCTGCTGGCAAACATGGAAACCGTGGTGAA ACTGGTCCTTCTGGTCCTGTTGGTCCTGCTGGTGCTGTTGGCCCAAGAGGTCCTAGTGGCCCACAAGGCATTCGTGGCGATAAGGGAGAG CCCGGTGAAAAGGGGCCCAGAGGTCTTCCTGGCTTAAAGGGACACAATGGATTGCAAGGTCTGCCTGGTATCGCTGGTCACCATGGTGAT CAAGGTGCTCCTGGCTCCGTGGGTCCTGCTGGTCCTAGGGGCCCTGCTGGTCCTTCTGGCCCTGCTGGAAAAGATGGTCGCACTGGACAT CCTGGTACAGTTGGACCTGCTGGCATTCGAGGCCCTCAGGGTCACCAAGGCCCTGCTGGCCCCCCTGGTCCCCCTGGCCCTCCTGGACCT CCAGGTGTAAGCGGTGGTGGTTATGACTTTGGTTACGATGGAGACTTCTACAGGGCTGACCAGCCTCGCTCAGCACCTTCTCTCAGACCC AAGGACTATGAAGTTGATGCTACTCTGAAGTCTCTCAACAACCAGATTGAGACCCTTCTTACTCCTGAAGGCTCTAGAAAGAACCCAGCT CGCACATGCCGTGACTTGAGACTCAGCCACCCAGAGTGGAGCAGTGGTTACTACTGGATTGACCCTAACCAAGGATGCACTATGGATGCT ATCAAAGTATACTGTGATTTCTCTACTGGCGAAACCTGTATCCGGGCCCAACCTGAAAACATCCCAGCCAAGAACTGGTATAGGAGCTCC AAGGACAAGAAACACGTCTGGCTAGGAGAAACTATCAATGCTGGCAGCCAGTTTGAATATAATGTAGAAGGAGTGACTTCCAAGGAAATG GCTACCCAACTTGCCTTCATGCGCCTGCTGGCCAACTATGCCTCTCAGAACATCACCTACCACTGCAAGAACAGCATTGCATACATGGAT GAGGAGACTGGCAACCTGAAAAAGGCTGTCATTCTACAGGGCTCTAATGATGTTGAACTTGTTGCTGAGGGCAACAGCAGGTTCACTTAC ACTGTTCTTGTAGATGGCTGCTCTAAAAAGACAAATGAATGGGGAAAGACAATCATTGAATACAAAACAAATAAGCCATCACGCCTGCCC TTCCTTGATATTGCACCTTTGGACATCGGTGGTGCTGACCAGGAATTCTTTGTGGACATTGGCCCAGTCTGTTTCAAATAAATGAACTCA ATCTAAATTAAAAAAGAAAGAAATTTGAAAAAACTTTCTCTTTGCCATTTCTTCTTCTTCTTTTTTAACTGAAAGCTGAATCCTTCCATT TCTTCTGCACATCTACTTGCTTAAATTGTGGGCAAAAGAGAAAAAGAAGGATTGATCAGAGCATTGTGCAATACAGTTTCATTAACTCCT TCCCCCGCTCCCCCAAAAATTTGAATTTTTTTTTCAACACTCTTACACCTGTTATGGAAAATGTCAACCTTTGTAAGAAAACCAAAATAA AAATTGAAAAATAAAAACCATAAACATTTGCACCACTTGTGGCTTTTGAATATCTTCCACAGAGGGAAGTTTAAAACCCAAACTTCCAAA GGTTTAAACTACCTCAAAACACTTTCCCATGAGTGTGATCCACATTGTTAGGTGCTGACCTAGACAGAGATGAACTGAGGTCCTTGTTTT GTTTTGTTCATAATACAAAGGTGCTAATTAATAGTATTTCAGATACTTGAAGAATGTTGATGGTGCTAGAAGAATTTGAGAAGAAATACT CCTGTATTGAGTTGTATCGTGTGGTGTATTTTTTAAAAAATTTGATTTAGCATTCATATTTTCCATCTTATTCCCAATTAAAAGTATGCA GATTATTTGCCCAAATCTTCTTCAGATTCAGCATTTGTTCTTTGCCAGTCTCATTTTCATCTTCTTCCATGGTTCCACAGAAGCTTTGTT TCTTGGGCAAGCAGAAAAATTAAATTGTACCTATTTTGTATATGTGAGATGTTTAAATAAATTGTGAAAAAAATGAAATAAAGCATGTTT >14583_14583_1_CD63-COL1A2_CD63_chr12_56120484_ENST00000257857_COL1A2_chr7_94055062_ENST00000297268_length(amino acids)=571AA_BP=165 MGSPGPAAMAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCL MITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADGERGYPGNIGPVGAAGAPGPHGPVGPAGKH GNRGETGPSGPVGPAGAVGPRGPSGPQGIRGDKGEPGEKGPRGLPGLKGHNGLQGLPGIAGHHGDQGAPGSVGPAGPRGPAGPSGPAGKD GRTGHPGTVGPAGIRGPQGHQGPAGPPGPPGPPGPPGVSGGGYDFGYDGDFYRADQPRSAPSLRPKDYEVDATLKSLNNQIETLLTPEGS RKNPARTCRDLRLSHPEWSSGYYWIDPNQGCTMDAIKVYCDFSTGETCIRAQPENIPAKNWYRSSKDKKHVWLGETINAGSQFEYNVEGV TSKEMATQLAFMRLLANYASQNITYHCKNSIAYMDEETGNLKKAVILQGSNDVELVAEGNSRFTYTVLVDGCSKKTNEWGKTIIEYKTNK -------------------------------------------------------------- >14583_14583_2_CD63-COL1A2_CD63_chr12_56120484_ENST00000420846_COL1A2_chr7_94055062_ENST00000297268_length(transcript)=2716nt_BP=611nt CAGCTGTTACCGCGTCACATGAGGGAGGCCGGCGGCCACTCGGCGGGGGAGGGGACCGTGGCTGGAGCCCGGGGCGGGGCCGCGCGGCAG GCGGGGCGGGAGCCGGGGGGCGCAGCTAGAGAGCCCCGGAGCCGCGGCGGGAGAGGAACGCGCAGCCAGCCTTGGGAAGCCCAGGCCCGG CAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGACTGA TTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATAATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATCATCG CAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCCATCT TTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACT TCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGGAGAGCGCGGTTACCCTG GCAATATTGGTCCCGTTGGTGCTGCAGGTGCACCTGGTCCTCATGGCCCCGTGGGTCCTGCTGGCAAACATGGAAACCGTGGTGAAACTG GTCCTTCTGGTCCTGTTGGTCCTGCTGGTGCTGTTGGCCCAAGAGGTCCTAGTGGCCCACAAGGCATTCGTGGCGATAAGGGAGAGCCCG GTGAAAAGGGGCCCAGAGGTCTTCCTGGCTTAAAGGGACACAATGGATTGCAAGGTCTGCCTGGTATCGCTGGTCACCATGGTGATCAAG GTGCTCCTGGCTCCGTGGGTCCTGCTGGTCCTAGGGGCCCTGCTGGTCCTTCTGGCCCTGCTGGAAAAGATGGTCGCACTGGACATCCTG GTACAGTTGGACCTGCTGGCATTCGAGGCCCTCAGGGTCACCAAGGCCCTGCTGGCCCCCCTGGTCCCCCTGGCCCTCCTGGACCTCCAG GTGTAAGCGGTGGTGGTTATGACTTTGGTTACGATGGAGACTTCTACAGGGCTGACCAGCCTCGCTCAGCACCTTCTCTCAGACCCAAGG ACTATGAAGTTGATGCTACTCTGAAGTCTCTCAACAACCAGATTGAGACCCTTCTTACTCCTGAAGGCTCTAGAAAGAACCCAGCTCGCA CATGCCGTGACTTGAGACTCAGCCACCCAGAGTGGAGCAGTGGTTACTACTGGATTGACCCTAACCAAGGATGCACTATGGATGCTATCA AAGTATACTGTGATTTCTCTACTGGCGAAACCTGTATCCGGGCCCAACCTGAAAACATCCCAGCCAAGAACTGGTATAGGAGCTCCAAGG ACAAGAAACACGTCTGGCTAGGAGAAACTATCAATGCTGGCAGCCAGTTTGAATATAATGTAGAAGGAGTGACTTCCAAGGAAATGGCTA CCCAACTTGCCTTCATGCGCCTGCTGGCCAACTATGCCTCTCAGAACATCACCTACCACTGCAAGAACAGCATTGCATACATGGATGAGG AGACTGGCAACCTGAAAAAGGCTGTCATTCTACAGGGCTCTAATGATGTTGAACTTGTTGCTGAGGGCAACAGCAGGTTCACTTACACTG TTCTTGTAGATGGCTGCTCTAAAAAGACAAATGAATGGGGAAAGACAATCATTGAATACAAAACAAATAAGCCATCACGCCTGCCCTTCC TTGATATTGCACCTTTGGACATCGGTGGTGCTGACCAGGAATTCTTTGTGGACATTGGCCCAGTCTGTTTCAAATAAATGAACTCAATCT AAATTAAAAAAGAAAGAAATTTGAAAAAACTTTCTCTTTGCCATTTCTTCTTCTTCTTTTTTAACTGAAAGCTGAATCCTTCCATTTCTT CTGCACATCTACTTGCTTAAATTGTGGGCAAAAGAGAAAAAGAAGGATTGATCAGAGCATTGTGCAATACAGTTTCATTAACTCCTTCCC CCGCTCCCCCAAAAATTTGAATTTTTTTTTCAACACTCTTACACCTGTTATGGAAAATGTCAACCTTTGTAAGAAAACCAAAATAAAAAT TGAAAAATAAAAACCATAAACATTTGCACCACTTGTGGCTTTTGAATATCTTCCACAGAGGGAAGTTTAAAACCCAAACTTCCAAAGGTT TAAACTACCTCAAAACACTTTCCCATGAGTGTGATCCACATTGTTAGGTGCTGACCTAGACAGAGATGAACTGAGGTCCTTGTTTTGTTT TGTTCATAATACAAAGGTGCTAATTAATAGTATTTCAGATACTTGAAGAATGTTGATGGTGCTAGAAGAATTTGAGAAGAAATACTCCTG TATTGAGTTGTATCGTGTGGTGTATTTTTTAAAAAATTTGATTTAGCATTCATATTTTCCATCTTATTCCCAATTAAAAGTATGCAGATT ATTTGCCCAAATCTTCTTCAGATTCAGCATTTGTTCTTTGCCAGTCTCATTTTCATCTTCTTCCATGGTTCCACAGAAGCTTTGTTTCTT GGGCAAGCAGAAAAATTAAATTGTACCTATTTTGTATATGTGAGATGTTTAAATAAATTGTGAAAAAAATGAAATAAAGCATGTTTGGTT >14583_14583_2_CD63-COL1A2_CD63_chr12_56120484_ENST00000420846_COL1A2_chr7_94055062_ENST00000297268_length(amino acids)=571AA_BP=165 MGSPGPAAMAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCL MITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADGERGYPGNIGPVGAAGAPGPHGPVGPAGKH GNRGETGPSGPVGPAGAVGPRGPSGPQGIRGDKGEPGEKGPRGLPGLKGHNGLQGLPGIAGHHGDQGAPGSVGPAGPRGPAGPSGPAGKD GRTGHPGTVGPAGIRGPQGHQGPAGPPGPPGPPGPPGVSGGGYDFGYDGDFYRADQPRSAPSLRPKDYEVDATLKSLNNQIETLLTPEGS RKNPARTCRDLRLSHPEWSSGYYWIDPNQGCTMDAIKVYCDFSTGETCIRAQPENIPAKNWYRSSKDKKHVWLGETINAGSQFEYNVEGV TSKEMATQLAFMRLLANYASQNITYHCKNSIAYMDEETGNLKKAVILQGSNDVELVAEGNSRFTYTVLVDGCSKKTNEWGKTIIEYKTNK -------------------------------------------------------------- >14583_14583_3_CD63-COL1A2_CD63_chr12_56120484_ENST00000548160_COL1A2_chr7_94055062_ENST00000297268_length(transcript)=2371nt_BP=266nt GAAGACCTTTGTGAGTGTCCAGGGACCGGCTGTGCCAAAACGGGCCCCTGTAATGCATAGACTCCAAACTTGACTTCTGTCCTTTGCTCC TGCAGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGGTGATGTCAG AGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGGCAGATGGAG AGCGCGGTTACCCTGGCAATATTGGTCCCGTTGGTGCTGCAGGTGCACCTGGTCCTCATGGCCCCGTGGGTCCTGCTGGCAAACATGGAA ACCGTGGTGAAACTGGTCCTTCTGGTCCTGTTGGTCCTGCTGGTGCTGTTGGCCCAAGAGGTCCTAGTGGCCCACAAGGCATTCGTGGCG ATAAGGGAGAGCCCGGTGAAAAGGGGCCCAGAGGTCTTCCTGGCTTAAAGGGACACAATGGATTGCAAGGTCTGCCTGGTATCGCTGGTC ACCATGGTGATCAAGGTGCTCCTGGCTCCGTGGGTCCTGCTGGTCCTAGGGGCCCTGCTGGTCCTTCTGGCCCTGCTGGAAAAGATGGTC GCACTGGACATCCTGGTACAGTTGGACCTGCTGGCATTCGAGGCCCTCAGGGTCACCAAGGCCCTGCTGGCCCCCCTGGTCCCCCTGGCC CTCCTGGACCTCCAGGTGTAAGCGGTGGTGGTTATGACTTTGGTTACGATGGAGACTTCTACAGGGCTGACCAGCCTCGCTCAGCACCTT CTCTCAGACCCAAGGACTATGAAGTTGATGCTACTCTGAAGTCTCTCAACAACCAGATTGAGACCCTTCTTACTCCTGAAGGCTCTAGAA AGAACCCAGCTCGCACATGCCGTGACTTGAGACTCAGCCACCCAGAGTGGAGCAGTGGTTACTACTGGATTGACCCTAACCAAGGATGCA CTATGGATGCTATCAAAGTATACTGTGATTTCTCTACTGGCGAAACCTGTATCCGGGCCCAACCTGAAAACATCCCAGCCAAGAACTGGT ATAGGAGCTCCAAGGACAAGAAACACGTCTGGCTAGGAGAAACTATCAATGCTGGCAGCCAGTTTGAATATAATGTAGAAGGAGTGACTT CCAAGGAAATGGCTACCCAACTTGCCTTCATGCGCCTGCTGGCCAACTATGCCTCTCAGAACATCACCTACCACTGCAAGAACAGCATTG CATACATGGATGAGGAGACTGGCAACCTGAAAAAGGCTGTCATTCTACAGGGCTCTAATGATGTTGAACTTGTTGCTGAGGGCAACAGCA GGTTCACTTACACTGTTCTTGTAGATGGCTGCTCTAAAAAGACAAATGAATGGGGAAAGACAATCATTGAATACAAAACAAATAAGCCAT CACGCCTGCCCTTCCTTGATATTGCACCTTTGGACATCGGTGGTGCTGACCAGGAATTCTTTGTGGACATTGGCCCAGTCTGTTTCAAAT AAATGAACTCAATCTAAATTAAAAAAGAAAGAAATTTGAAAAAACTTTCTCTTTGCCATTTCTTCTTCTTCTTTTTTAACTGAAAGCTGA ATCCTTCCATTTCTTCTGCACATCTACTTGCTTAAATTGTGGGCAAAAGAGAAAAAGAAGGATTGATCAGAGCATTGTGCAATACAGTTT CATTAACTCCTTCCCCCGCTCCCCCAAAAATTTGAATTTTTTTTTCAACACTCTTACACCTGTTATGGAAAATGTCAACCTTTGTAAGAA AACCAAAATAAAAATTGAAAAATAAAAACCATAAACATTTGCACCACTTGTGGCTTTTGAATATCTTCCACAGAGGGAAGTTTAAAACCC AAACTTCCAAAGGTTTAAACTACCTCAAAACACTTTCCCATGAGTGTGATCCACATTGTTAGGTGCTGACCTAGACAGAGATGAACTGAG GTCCTTGTTTTGTTTTGTTCATAATACAAAGGTGCTAATTAATAGTATTTCAGATACTTGAAGAATGTTGATGGTGCTAGAAGAATTTGA GAAGAAATACTCCTGTATTGAGTTGTATCGTGTGGTGTATTTTTTAAAAAATTTGATTTAGCATTCATATTTTCCATCTTATTCCCAATT AAAAGTATGCAGATTATTTGCCCAAATCTTCTTCAGATTCAGCATTTGTTCTTTGCCAGTCTCATTTTCATCTTCTTCCATGGTTCCACA GAAGCTTTGTTTCTTGGGCAAGCAGAAAAATTAAATTGTACCTATTTTGTATATGTGAGATGTTTAAATAAATTGTGAAAAAAATGAAAT >14583_14583_3_CD63-COL1A2_CD63_chr12_56120484_ENST00000548160_COL1A2_chr7_94055062_ENST00000297268_length(amino acids)=482AA_BP=76 MLLQFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADGERGYPGNIGPVGAAGAPGPHGPVGPAGK HGNRGETGPSGPVGPAGAVGPRGPSGPQGIRGDKGEPGEKGPRGLPGLKGHNGLQGLPGIAGHHGDQGAPGSVGPAGPRGPAGPSGPAGK DGRTGHPGTVGPAGIRGPQGHQGPAGPPGPPGPPGPPGVSGGGYDFGYDGDFYRADQPRSAPSLRPKDYEVDATLKSLNNQIETLLTPEG SRKNPARTCRDLRLSHPEWSSGYYWIDPNQGCTMDAIKVYCDFSTGETCIRAQPENIPAKNWYRSSKDKKHVWLGETINAGSQFEYNVEG VTSKEMATQLAFMRLLANYASQNITYHCKNSIAYMDEETGNLKKAVILQGSNDVELVAEGNSRFTYTVLVDGCSKKTNEWGKTIIEYKTN -------------------------------------------------------------- >14583_14583_4_CD63-COL1A2_CD63_chr12_56120484_ENST00000548898_COL1A2_chr7_94055062_ENST00000297268_length(transcript)=2303nt_BP=198nt AGAGCAGCTTGGTTCGGGCTCAGGAAGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTG TTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTG GACAGGATGCAGGCAGATGGAGAGCGCGGTTACCCTGGCAATATTGGTCCCGTTGGTGCTGCAGGTGCACCTGGTCCTCATGGCCCCGTG GGTCCTGCTGGCAAACATGGAAACCGTGGTGAAACTGGTCCTTCTGGTCCTGTTGGTCCTGCTGGTGCTGTTGGCCCAAGAGGTCCTAGT GGCCCACAAGGCATTCGTGGCGATAAGGGAGAGCCCGGTGAAAAGGGGCCCAGAGGTCTTCCTGGCTTAAAGGGACACAATGGATTGCAA GGTCTGCCTGGTATCGCTGGTCACCATGGTGATCAAGGTGCTCCTGGCTCCGTGGGTCCTGCTGGTCCTAGGGGCCCTGCTGGTCCTTCT GGCCCTGCTGGAAAAGATGGTCGCACTGGACATCCTGGTACAGTTGGACCTGCTGGCATTCGAGGCCCTCAGGGTCACCAAGGCCCTGCT GGCCCCCCTGGTCCCCCTGGCCCTCCTGGACCTCCAGGTGTAAGCGGTGGTGGTTATGACTTTGGTTACGATGGAGACTTCTACAGGGCT GACCAGCCTCGCTCAGCACCTTCTCTCAGACCCAAGGACTATGAAGTTGATGCTACTCTGAAGTCTCTCAACAACCAGATTGAGACCCTT CTTACTCCTGAAGGCTCTAGAAAGAACCCAGCTCGCACATGCCGTGACTTGAGACTCAGCCACCCAGAGTGGAGCAGTGGTTACTACTGG ATTGACCCTAACCAAGGATGCACTATGGATGCTATCAAAGTATACTGTGATTTCTCTACTGGCGAAACCTGTATCCGGGCCCAACCTGAA AACATCCCAGCCAAGAACTGGTATAGGAGCTCCAAGGACAAGAAACACGTCTGGCTAGGAGAAACTATCAATGCTGGCAGCCAGTTTGAA TATAATGTAGAAGGAGTGACTTCCAAGGAAATGGCTACCCAACTTGCCTTCATGCGCCTGCTGGCCAACTATGCCTCTCAGAACATCACC TACCACTGCAAGAACAGCATTGCATACATGGATGAGGAGACTGGCAACCTGAAAAAGGCTGTCATTCTACAGGGCTCTAATGATGTTGAA CTTGTTGCTGAGGGCAACAGCAGGTTCACTTACACTGTTCTTGTAGATGGCTGCTCTAAAAAGACAAATGAATGGGGAAAGACAATCATT GAATACAAAACAAATAAGCCATCACGCCTGCCCTTCCTTGATATTGCACCTTTGGACATCGGTGGTGCTGACCAGGAATTCTTTGTGGAC ATTGGCCCAGTCTGTTTCAAATAAATGAACTCAATCTAAATTAAAAAAGAAAGAAATTTGAAAAAACTTTCTCTTTGCCATTTCTTCTTC TTCTTTTTTAACTGAAAGCTGAATCCTTCCATTTCTTCTGCACATCTACTTGCTTAAATTGTGGGCAAAAGAGAAAAAGAAGGATTGATC AGAGCATTGTGCAATACAGTTTCATTAACTCCTTCCCCCGCTCCCCCAAAAATTTGAATTTTTTTTTCAACACTCTTACACCTGTTATGG AAAATGTCAACCTTTGTAAGAAAACCAAAATAAAAATTGAAAAATAAAAACCATAAACATTTGCACCACTTGTGGCTTTTGAATATCTTC CACAGAGGGAAGTTTAAAACCCAAACTTCCAAAGGTTTAAACTACCTCAAAACACTTTCCCATGAGTGTGATCCACATTGTTAGGTGCTG ACCTAGACAGAGATGAACTGAGGTCCTTGTTTTGTTTTGTTCATAATACAAAGGTGCTAATTAATAGTATTTCAGATACTTGAAGAATGT TGATGGTGCTAGAAGAATTTGAGAAGAAATACTCCTGTATTGAGTTGTATCGTGTGGTGTATTTTTTAAAAAATTTGATTTAGCATTCAT ATTTTCCATCTTATTCCCAATTAAAAGTATGCAGATTATTTGCCCAAATCTTCTTCAGATTCAGCATTTGTTCTTTGCCAGTCTCATTTT CATCTTCTTCCATGGTTCCACAGAAGCTTTGTTTCTTGGGCAAGCAGAAAAATTAAATTGTACCTATTTTGTATATGTGAGATGTTTAAA >14583_14583_4_CD63-COL1A2_CD63_chr12_56120484_ENST00000548898_COL1A2_chr7_94055062_ENST00000297268_length(amino acids)=474AA_BP=68 MSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADGERGYPGNIGPVGAAGAPGPHGPVGPAGKHGNRGETG PSGPVGPAGAVGPRGPSGPQGIRGDKGEPGEKGPRGLPGLKGHNGLQGLPGIAGHHGDQGAPGSVGPAGPRGPAGPSGPAGKDGRTGHPG TVGPAGIRGPQGHQGPAGPPGPPGPPGPPGVSGGGYDFGYDGDFYRADQPRSAPSLRPKDYEVDATLKSLNNQIETLLTPEGSRKNPART CRDLRLSHPEWSSGYYWIDPNQGCTMDAIKVYCDFSTGETCIRAQPENIPAKNWYRSSKDKKHVWLGETINAGSQFEYNVEGVTSKEMAT QLAFMRLLANYASQNITYHCKNSIAYMDEETGNLKKAVILQGSNDVELVAEGNSRFTYTVLVDGCSKKTNEWGKTIIEYKTNKPSRLPFL -------------------------------------------------------------- >14583_14583_5_CD63-COL1A2_CD63_chr12_56120484_ENST00000552692_COL1A2_chr7_94055062_ENST00000297268_length(transcript)=2672nt_BP=567nt GCGGGTTTCCGGGTGGGAGGGGCCGGGAATCGGGGCCGTGAAGGGAAGCCCTCGAGGGCCGGCCGGGAGCCCGGAAACCGAGGTCCCCCT TCGGCCAGTTTTAATCCCCCCGCCCTCCCCTCTGCCCCAGGCCCGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTG CTCTACGTCCTCCTGCTGGCCTTTTGCGCCTGTGCAGTGGGACTGATTGCCGTGGGTGTCGGGGCACAGCTTGTCCTGAGTCAGACCATA ATCCAGGGGGCTACCCCTGGCTCTCTGTTGCCAGTGGTCATCATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGC GGGGCCTGCAAGGAGAACTATTGTCTTATGATCACGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCT GGCTATGTGTTTAGAGATAAGGTGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCT TCGATCCTGGACAGGATGCAGGCAGATGGAGAGCGCGGTTACCCTGGCAATATTGGTCCCGTTGGTGCTGCAGGTGCACCTGGTCCTCAT GGCCCCGTGGGTCCTGCTGGCAAACATGGAAACCGTGGTGAAACTGGTCCTTCTGGTCCTGTTGGTCCTGCTGGTGCTGTTGGCCCAAGA GGTCCTAGTGGCCCACAAGGCATTCGTGGCGATAAGGGAGAGCCCGGTGAAAAGGGGCCCAGAGGTCTTCCTGGCTTAAAGGGACACAAT GGATTGCAAGGTCTGCCTGGTATCGCTGGTCACCATGGTGATCAAGGTGCTCCTGGCTCCGTGGGTCCTGCTGGTCCTAGGGGCCCTGCT GGTCCTTCTGGCCCTGCTGGAAAAGATGGTCGCACTGGACATCCTGGTACAGTTGGACCTGCTGGCATTCGAGGCCCTCAGGGTCACCAA GGCCCTGCTGGCCCCCCTGGTCCCCCTGGCCCTCCTGGACCTCCAGGTGTAAGCGGTGGTGGTTATGACTTTGGTTACGATGGAGACTTC TACAGGGCTGACCAGCCTCGCTCAGCACCTTCTCTCAGACCCAAGGACTATGAAGTTGATGCTACTCTGAAGTCTCTCAACAACCAGATT GAGACCCTTCTTACTCCTGAAGGCTCTAGAAAGAACCCAGCTCGCACATGCCGTGACTTGAGACTCAGCCACCCAGAGTGGAGCAGTGGT TACTACTGGATTGACCCTAACCAAGGATGCACTATGGATGCTATCAAAGTATACTGTGATTTCTCTACTGGCGAAACCTGTATCCGGGCC CAACCTGAAAACATCCCAGCCAAGAACTGGTATAGGAGCTCCAAGGACAAGAAACACGTCTGGCTAGGAGAAACTATCAATGCTGGCAGC CAGTTTGAATATAATGTAGAAGGAGTGACTTCCAAGGAAATGGCTACCCAACTTGCCTTCATGCGCCTGCTGGCCAACTATGCCTCTCAG AACATCACCTACCACTGCAAGAACAGCATTGCATACATGGATGAGGAGACTGGCAACCTGAAAAAGGCTGTCATTCTACAGGGCTCTAAT GATGTTGAACTTGTTGCTGAGGGCAACAGCAGGTTCACTTACACTGTTCTTGTAGATGGCTGCTCTAAAAAGACAAATGAATGGGGAAAG ACAATCATTGAATACAAAACAAATAAGCCATCACGCCTGCCCTTCCTTGATATTGCACCTTTGGACATCGGTGGTGCTGACCAGGAATTC TTTGTGGACATTGGCCCAGTCTGTTTCAAATAAATGAACTCAATCTAAATTAAAAAAGAAAGAAATTTGAAAAAACTTTCTCTTTGCCAT TTCTTCTTCTTCTTTTTTAACTGAAAGCTGAATCCTTCCATTTCTTCTGCACATCTACTTGCTTAAATTGTGGGCAAAAGAGAAAAAGAA GGATTGATCAGAGCATTGTGCAATACAGTTTCATTAACTCCTTCCCCCGCTCCCCCAAAAATTTGAATTTTTTTTTCAACACTCTTACAC CTGTTATGGAAAATGTCAACCTTTGTAAGAAAACCAAAATAAAAATTGAAAAATAAAAACCATAAACATTTGCACCACTTGTGGCTTTTG AATATCTTCCACAGAGGGAAGTTTAAAACCCAAACTTCCAAAGGTTTAAACTACCTCAAAACACTTTCCCATGAGTGTGATCCACATTGT TAGGTGCTGACCTAGACAGAGATGAACTGAGGTCCTTGTTTTGTTTTGTTCATAATACAAAGGTGCTAATTAATAGTATTTCAGATACTT GAAGAATGTTGATGGTGCTAGAAGAATTTGAGAAGAAATACTCCTGTATTGAGTTGTATCGTGTGGTGTATTTTTTAAAAAATTTGATTT AGCATTCATATTTTCCATCTTATTCCCAATTAAAAGTATGCAGATTATTTGCCCAAATCTTCTTCAGATTCAGCATTTGTTCTTTGCCAG TCTCATTTTCATCTTCTTCCATGGTTCCACAGAAGCTTTGTTTCTTGGGCAAGCAGAAAAATTAAATTGTACCTATTTTGTATATGTGAG >14583_14583_5_CD63-COL1A2_CD63_chr12_56120484_ENST00000552692_COL1A2_chr7_94055062_ENST00000297268_length(amino acids)=563AA_BP=157 MAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFL SLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADGERGYPGNIGPVGAAGAPGPHGPVGPAGKHGNRGETGP SGPVGPAGAVGPRGPSGPQGIRGDKGEPGEKGPRGLPGLKGHNGLQGLPGIAGHHGDQGAPGSVGPAGPRGPAGPSGPAGKDGRTGHPGT VGPAGIRGPQGHQGPAGPPGPPGPPGPPGVSGGGYDFGYDGDFYRADQPRSAPSLRPKDYEVDATLKSLNNQIETLLTPEGSRKNPARTC RDLRLSHPEWSSGYYWIDPNQGCTMDAIKVYCDFSTGETCIRAQPENIPAKNWYRSSKDKKHVWLGETINAGSQFEYNVEGVTSKEMATQ LAFMRLLANYASQNITYHCKNSIAYMDEETGNLKKAVILQGSNDVELVAEGNSRFTYTVLVDGCSKKTNEWGKTIIEYKTNKPSRLPFLD -------------------------------------------------------------- >14583_14583_6_CD63-COL1A2_CD63_chr12_56120484_ENST00000552754_COL1A2_chr7_94055062_ENST00000297268_length(transcript)=2470nt_BP=365nt CGGCAGCCATGGCGGTGGAAGGAGGAATGAAATGTGTGAAGTTCTTGCTCTACGTCCTCCTGCTGGCCTTTTGCGGGGCTACCCCTGGCT CTCTGTTGCCAGTGGTCATCATCGCAGTGGGTGTCTTCCTCTTCCTGGTGGCTTTTGTGGGCTGCTGCGGGGCCTGCAAGGAGAACTATT GTCTTATGATCACGTTTGCCATCTTTCTGTCTCTTATCATGTTGGTGGAGGTGGCCGCAGCCATTGCTGGCTATGTGTTTAGAGATAAGG TGATGTCAGAGTTTAATAACAACTTCCGGCAGCAGATGGAGAATTACCCGAAAAACAACCACACTGCTTCGATCCTGGACAGGATGCAGG CAGATGGAGAGCGCGGTTACCCTGGCAATATTGGTCCCGTTGGTGCTGCAGGTGCACCTGGTCCTCATGGCCCCGTGGGTCCTGCTGGCA AACATGGAAACCGTGGTGAAACTGGTCCTTCTGGTCCTGTTGGTCCTGCTGGTGCTGTTGGCCCAAGAGGTCCTAGTGGCCCACAAGGCA TTCGTGGCGATAAGGGAGAGCCCGGTGAAAAGGGGCCCAGAGGTCTTCCTGGCTTAAAGGGACACAATGGATTGCAAGGTCTGCCTGGTA TCGCTGGTCACCATGGTGATCAAGGTGCTCCTGGCTCCGTGGGTCCTGCTGGTCCTAGGGGCCCTGCTGGTCCTTCTGGCCCTGCTGGAA AAGATGGTCGCACTGGACATCCTGGTACAGTTGGACCTGCTGGCATTCGAGGCCCTCAGGGTCACCAAGGCCCTGCTGGCCCCCCTGGTC CCCCTGGCCCTCCTGGACCTCCAGGTGTAAGCGGTGGTGGTTATGACTTTGGTTACGATGGAGACTTCTACAGGGCTGACCAGCCTCGCT CAGCACCTTCTCTCAGACCCAAGGACTATGAAGTTGATGCTACTCTGAAGTCTCTCAACAACCAGATTGAGACCCTTCTTACTCCTGAAG GCTCTAGAAAGAACCCAGCTCGCACATGCCGTGACTTGAGACTCAGCCACCCAGAGTGGAGCAGTGGTTACTACTGGATTGACCCTAACC AAGGATGCACTATGGATGCTATCAAAGTATACTGTGATTTCTCTACTGGCGAAACCTGTATCCGGGCCCAACCTGAAAACATCCCAGCCA AGAACTGGTATAGGAGCTCCAAGGACAAGAAACACGTCTGGCTAGGAGAAACTATCAATGCTGGCAGCCAGTTTGAATATAATGTAGAAG GAGTGACTTCCAAGGAAATGGCTACCCAACTTGCCTTCATGCGCCTGCTGGCCAACTATGCCTCTCAGAACATCACCTACCACTGCAAGA ACAGCATTGCATACATGGATGAGGAGACTGGCAACCTGAAAAAGGCTGTCATTCTACAGGGCTCTAATGATGTTGAACTTGTTGCTGAGG GCAACAGCAGGTTCACTTACACTGTTCTTGTAGATGGCTGCTCTAAAAAGACAAATGAATGGGGAAAGACAATCATTGAATACAAAACAA ATAAGCCATCACGCCTGCCCTTCCTTGATATTGCACCTTTGGACATCGGTGGTGCTGACCAGGAATTCTTTGTGGACATTGGCCCAGTCT GTTTCAAATAAATGAACTCAATCTAAATTAAAAAAGAAAGAAATTTGAAAAAACTTTCTCTTTGCCATTTCTTCTTCTTCTTTTTTAACT GAAAGCTGAATCCTTCCATTTCTTCTGCACATCTACTTGCTTAAATTGTGGGCAAAAGAGAAAAAGAAGGATTGATCAGAGCATTGTGCA ATACAGTTTCATTAACTCCTTCCCCCGCTCCCCCAAAAATTTGAATTTTTTTTTCAACACTCTTACACCTGTTATGGAAAATGTCAACCT TTGTAAGAAAACCAAAATAAAAATTGAAAAATAAAAACCATAAACATTTGCACCACTTGTGGCTTTTGAATATCTTCCACAGAGGGAAGT TTAAAACCCAAACTTCCAAAGGTTTAAACTACCTCAAAACACTTTCCCATGAGTGTGATCCACATTGTTAGGTGCTGACCTAGACAGAGA TGAACTGAGGTCCTTGTTTTGTTTTGTTCATAATACAAAGGTGCTAATTAATAGTATTTCAGATACTTGAAGAATGTTGATGGTGCTAGA AGAATTTGAGAAGAAATACTCCTGTATTGAGTTGTATCGTGTGGTGTATTTTTTAAAAAATTTGATTTAGCATTCATATTTTCCATCTTA TTCCCAATTAAAAGTATGCAGATTATTTGCCCAAATCTTCTTCAGATTCAGCATTTGTTCTTTGCCAGTCTCATTTTCATCTTCTTCCAT GGTTCCACAGAAGCTTTGTTTCTTGGGCAAGCAGAAAAATTAAATTGTACCTATTTTGTATATGTGAGATGTTTAAATAAATTGTGAAAA >14583_14583_6_CD63-COL1A2_CD63_chr12_56120484_ENST00000552754_COL1A2_chr7_94055062_ENST00000297268_length(amino acids)=540AA_BP=134 MAVEGGMKCVKFLLYVLLLAFCGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFLSLIMLVEVAAAIAGYVFRDKVMS EFNNNFRQQMENYPKNNHTASILDRMQADGERGYPGNIGPVGAAGAPGPHGPVGPAGKHGNRGETGPSGPVGPAGAVGPRGPSGPQGIRG DKGEPGEKGPRGLPGLKGHNGLQGLPGIAGHHGDQGAPGSVGPAGPRGPAGPSGPAGKDGRTGHPGTVGPAGIRGPQGHQGPAGPPGPPG PPGPPGVSGGGYDFGYDGDFYRADQPRSAPSLRPKDYEVDATLKSLNNQIETLLTPEGSRKNPARTCRDLRLSHPEWSSGYYWIDPNQGC TMDAIKVYCDFSTGETCIRAQPENIPAKNWYRSSKDKKHVWLGETINAGSQFEYNVEGVTSKEMATQLAFMRLLANYASQNITYHCKNSI AYMDEETGNLKKAVILQGSNDVELVAEGNSRFTYTVLVDGCSKKTNEWGKTIIEYKTNKPSRLPFLDIAPLDIGGADQEFFVDIGPVCFK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CD63-COL1A2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CD63-COL1A2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CD63-COL1A2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
