
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CDH3-CHKA (FusionGDB2 ID:15072) |
Fusion Gene Summary for CDH3-CHKA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CDH3-CHKA | Fusion gene ID: 15072 | Hgene | Tgene | Gene symbol | CDH3 | CHKA | Gene ID | 1013 | 1119 |
| Gene name | cadherin 15 | choline kinase alpha | |
| Synonyms | CDH14|CDH3|CDHM|MCAD|MRD3 | CHK|CK|CKI|EK | |
| Cytomap | 16q24.3 | 11q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cadherin-15cadherin 15, type 1, M-cadherin (myotubule)cadherin-14cadherin-3muscle-cadherin | choline kinase alphaCHETK-alphaethanolamine kinase | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P22223 | P35790 | |
| Ensembl transtripts involved in fusion gene | ENST00000264012, ENST00000429102, ENST00000581171, ENST00000569117, | ENST00000356135, ENST00000533728, ENST00000265689, | |
| Fusion gene scores | * DoF score | 12 X 8 X 6=576 | 11 X 10 X 7=770 |
| # samples | 15 | 13 | |
| ** MAII score | log2(15/576*10)=-1.94110631094643 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/770*10)=-2.56634682255381 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CDH3 [Title/Abstract] AND CHKA [Title/Abstract] AND fusion [Title/Abstract] | ||
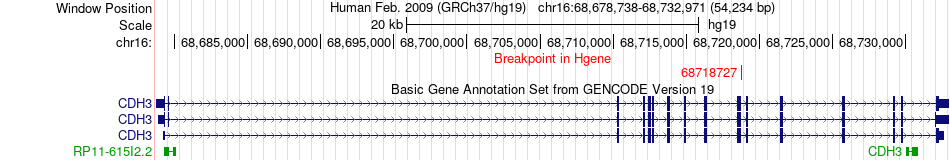
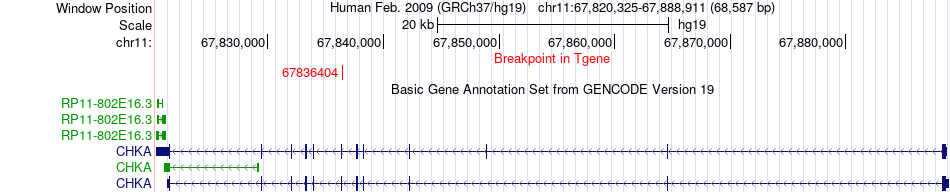
| Most frequent breakpoint | CDH3(68718727)-CHKA(67836404), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CHKA | GO:0006646 | phosphatidylethanolamine biosynthetic process | 19915674 |
| Tgene | CHKA | GO:0006656 | phosphatidylcholine biosynthetic process | 19915674 |
 Fusion gene breakpoints across CDH3 (5'-gene) Fusion gene breakpoints across CDH3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CHKA (3'-gene) Fusion gene breakpoints across CHKA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-DK-A6AV-01A | CDH3 | chr16 | 68718727 | - | CHKA | chr11 | 67836404 | - |
| ChimerDB4 | BLCA | TCGA-DK-A6AV-01A | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| ChimerDB4 | BLCA | TCGA-DK-A6AV | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
Top |
Fusion Gene ORF analysis for CDH3-CHKA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000264012 | ENST00000356135 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| 5CDS-intron | ENST00000264012 | ENST00000533728 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| 5CDS-intron | ENST00000429102 | ENST00000356135 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| 5CDS-intron | ENST00000429102 | ENST00000533728 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| 5CDS-intron | ENST00000581171 | ENST00000356135 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| 5CDS-intron | ENST00000581171 | ENST00000533728 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| In-frame | ENST00000264012 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| In-frame | ENST00000429102 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| In-frame | ENST00000581171 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| intron-3CDS | ENST00000569117 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| intron-intron | ENST00000569117 | ENST00000356135 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
| intron-intron | ENST00000569117 | ENST00000533728 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264012 | CDH3 | chr16 | 68718727 | + | ENST00000265689 | CHKA | chr11 | 67836404 | - | 3602 | 1968 | 346 | 2472 | 708 |
| ENST00000429102 | CDH3 | chr16 | 68718727 | + | ENST00000265689 | CHKA | chr11 | 67836404 | - | 3445 | 1811 | 189 | 2315 | 708 |
| ENST00000581171 | CDH3 | chr16 | 68718727 | + | ENST00000265689 | CHKA | chr11 | 67836404 | - | 3022 | 1388 | 54 | 1892 | 612 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264012 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - | 0.000862223 | 0.99913776 |
| ENST00000429102 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - | 0.000902238 | 0.9990978 |
| ENST00000581171 | ENST00000265689 | CDH3 | chr16 | 68718727 | + | CHKA | chr11 | 67836404 | - | 0.001012625 | 0.9989874 |
Top |
Fusion Genomic Features for CDH3-CHKA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
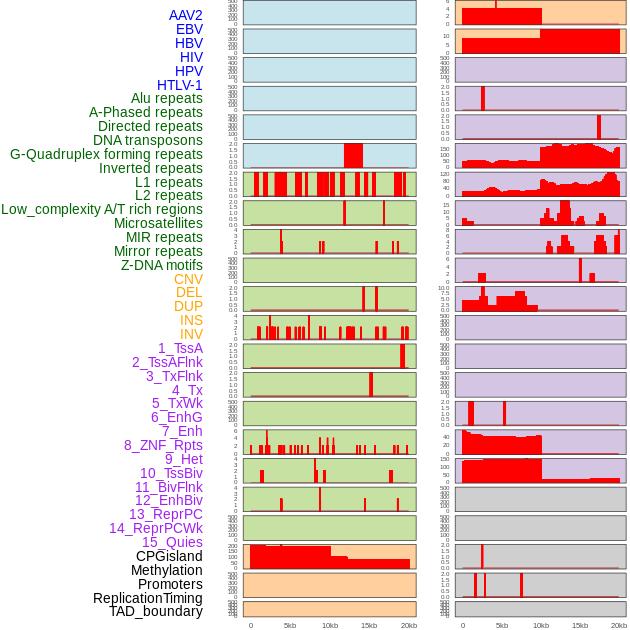 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CDH3-CHKA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:68718727/chr11:67836404) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CDH3 | CHKA |
| FUNCTION: Cadherins are calcium-dependent cell adhesion proteins. They preferentially interact with themselves in a homophilic manner in connecting cells; cadherins may thus contribute to the sorting of heterogeneous cell types. | FUNCTION: Has a key role in phospholipid biosynthesis and may contribute to tumor cell growth. Catalyzes the first step in phosphatidylcholine biosynthesis. Contributes to phosphatidylethanolamine biosynthesis. Phosphorylates choline and ethanolamine. Has higher activity with choline. {ECO:0000269|PubMed:19915674}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 108_215 | 474 | 830.0 | Domain | Cadherin 1 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 216_328 | 474 | 830.0 | Domain | Cadherin 2 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 329_440 | 474 | 830.0 | Domain | Cadherin 3 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 108_215 | 474 | 785.0 | Domain | Cadherin 1 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 216_328 | 474 | 785.0 | Domain | Cadherin 2 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 329_440 | 474 | 785.0 | Domain | Cadherin 3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 785_800 | 474 | 830.0 | Compositional bias | Note=Ser-rich |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 785_800 | 474 | 785.0 | Compositional bias | Note=Ser-rich |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 441_546 | 474 | 830.0 | Domain | Cadherin 4 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 547_650 | 474 | 830.0 | Domain | Cadherin 5 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 441_546 | 474 | 785.0 | Domain | Cadherin 4 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 547_650 | 474 | 785.0 | Domain | Cadherin 5 |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 108_654 | 474 | 830.0 | Topological domain | Extracellular |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 678_829 | 474 | 830.0 | Topological domain | Cytoplasmic |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 108_654 | 474 | 785.0 | Topological domain | Extracellular |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 678_829 | 474 | 785.0 | Topological domain | Cytoplasmic |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000264012 | + | 10 | 16 | 655_677 | 474 | 830.0 | Transmembrane | Helical |
| Hgene | CDH3 | chr16:68718727 | chr11:67836404 | ENST00000429102 | + | 10 | 16 | 655_677 | 474 | 785.0 | Transmembrane | Helical |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000265689 | 5 | 12 | 50_85 | 289 | 458.0 | Compositional bias | Note=Pro-rich | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000356135 | 4 | 11 | 50_85 | 271 | 440.0 | Compositional bias | Note=Pro-rich | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000265689 | 5 | 12 | 117_123 | 289 | 458.0 | Nucleotide binding | Note=ATP | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000265689 | 5 | 12 | 207_213 | 289 | 458.0 | Nucleotide binding | Note=ATP | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000356135 | 4 | 11 | 117_123 | 271 | 440.0 | Nucleotide binding | Note=ATP | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000356135 | 4 | 11 | 207_213 | 271 | 440.0 | Nucleotide binding | Note=ATP | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000265689 | 5 | 12 | 119_121 | 289 | 458.0 | Region | Note=Substrate binding | |
| Tgene | CHKA | chr16:68718727 | chr11:67836404 | ENST00000356135 | 4 | 11 | 119_121 | 271 | 440.0 | Region | Note=Substrate binding |
Top |
Fusion Gene Sequence for CDH3-CHKA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >15072_15072_1_CDH3-CHKA_CDH3_chr16_68718727_ENST00000264012_CHKA_chr11_67836404_ENST00000265689_length(transcript)=3602nt_BP=1968nt GGCTGGCACGGGAGGTGGAGAAAGAGGCTTGGGCGGCCCCGCTGTAGCCGCGTGTGGGAGGACGCACGGGCCTGCTTCAAAGCTTTGGGA TAACAGCGCCTCCGGGGGATAATGAATGCGGAGCCTCCGTTTTCAGTCGACTTCAGATGTGTCTCCACTTTTTTCCGCTGTAGCCGCAAG GCAAGGAAACATTTCTCTTCCCGTACTGAGGAGGCTGAGGAGTGCACTGGGTGTTCTTTTCTCCTCTAACCCAGAACTGCGAGACAGAGG CTGAGTCCCTGTAAAGAACAGCTCCAGAAAAGCCAGGAGAGCGCAGGAGGGCATCCGGGAGGCCAGGAGGGGTTCGCTGGGGCCTCAACC GCACCCACATCGGTCCCACCTGCGAGGGGGCGGGACCTCGTGGCGCTGGACCAATCAGCACCCACCTGCGCTCACCTGGCCTCCTCCCGC TGGCTCCCGGGGGCTGCGGTGCTCAAAGGGGCAAGAGCTGAGCGGAACACCGGCCCGCCGTCGCGGCAGCTGCTTCACCCCTCTCTCTGC AGCCATGGGGCTCCCTCGTGGACCTCTCGCGTCTCTCCTCCTTCTCCAGGTTTGCTGGCTGCAGTGCGCGGCCTCCGAGCCGTGCCGGGC GGTCTTCAGGGAGGCTGAAGTGACCTTGGAGGCGGGAGGCGCGGAGCAGGAGCCCGGCCAGGCGCTGGGGAAAGTATTCATGGGCTGCCC TGGGCAAGAGCCAGCTCTGTTTAGCACTGATAATGATGACTTCACTGTGCGGAATGGCGAGACAGTCCAGGAAAGAAGGTCACTGAAGGA AAGGAATCCATTGAAGATCTTCCCATCCAAACGTATCTTACGAAGACACAAGAGAGATTGGGTGGTTGCTCCAATATCTGTCCCTGAAAA TGGCAAGGGTCCCTTCCCCCAGAGACTGAATCAGCTCAAGTCTAATAAAGATAGAGACACCAAGATTTTCTACAGCATCACGGGGCCGGG GGCAGACAGCCCCCCTGAGGGTGTCTTCGCTGTAGAGAAGGAGACAGGCTGGTTGTTGTTGAATAAGCCACTGGACCGGGAGGAGATTGC CAAGTATGAGCTCTTTGGCCACGCTGTGTCAGAGAATGGTGCCTCAGTGGAGGACCCCATGAACATCTCCATCATCGTGACCGACCAGAA TGACCACAAGCCCAAGTTTACCCAGGACACCTTCCGAGGGAGTGTCTTAGAGGGAGTCCTACCAGGTACTTCTGTGATGCAGGTGACAGC CACGGATGAGGATGATGCCATCTACACCTACAATGGGGTGGTTGCTTACTCCATCCATAGCCAAGAACCAAAGGACCCACACGACCTCAT GTTCACCATTCACCGGAGCACAGGCACCATCAGCGTCATCTCCAGTGGCCTGGACCGGGAAAAAGTCCCTGAGTACACACTGACCATCCA GGCCACAGACATGGATGGGGACGGCTCCACCACCACGGCAGTGGCAGTAGTGGAGATCCTTGATGCCAATGACAATGCTCCCATGTTTGA CCCCCAGAAGTACGAGGCCCATGTGCCTGAGAATGCAGTGGGCCATGAGGTGCAGAGGCTGACGGTCACTGATCTGGACGCCCCCAACTC ACCAGCGTGGCGTGCCACCTACCTTATCATGGGCGGTGACGACGGGGACCATTTTACCATCACCACCCACCCTGAGAGCAACCAGGGCAT CCTGACAACCAGGAAGGGTTTGGATTTTGAGGCCAAAAACCAGCACACCCTGTACGTTGAAGTGACCAACGAGGCCCCTTTTGTGCTGAA GCTCCCAACCTCCACAGCCACCATAGTGGTCCACGTGGAGGATGTGAATGAGGCACCTGTGTTTGTCCCACCCTCCAAAGTCGTTGAGGT CCAGGAGGGCATCCCCACTGGGGAGCCTGTGTGTGTCTACACTGCAGAAGACCCTGACAAGGAGAATCAAAAGATCAGATCATTGCTTGA ATCTACTCCATCTCCAGTTGTATTTTGTCATAATGACTGTCAAGAAGGTAATATCTTGTTGCTGGAAGGCCGAGAGAATTCTGAAAAACA GAAACTGATGCTCATTGATTTCGAATACAGCAGTTACAATTACAGGGGATTCGACATTGGAAATCACTTCTGTGAGTGGATGTATGATTA TAGCTATGAAAAATACCCTTTTTTCAGAGCAAACATCCGGAAGTATCCCACCAAGAAACAACAGCTCCATTTTATTTCCAGTTACTTGCC TGCATTCCAAAATGACTTTGAAAACCTCAGTACTGAAGAAAAATCCATTATAAAAGAAGAAATGTTGCTTGAAGTTAATAGGTTTGCCCT TGCATCTCATTTCCTCTGGGGACTGTGGTCCATTGTACAAGCCAAGATTTCATCTATTGAATTTGGGTACATGGACTACGCCCAAGCAAG GTTTGATGCCTATTTCCACCAGAAGAGGAAGCTTGGGGTGTGACTGTGGGGAGGACTCCATCCACCTCATCACTGGACTGCATGGGGAGG CAGCAGAGCGGGGTCCCCTCTGTGCTTCGACTACTGCTCCTGTGGCAGGAGGCTTTGGGTGGCTCACTACTGAACACATGTGTATGATAC TAAAGACGGTATTAAAATGGAGCGACGTTTATTTCATCTCTTGTTTACGATTTCACTAGGACTCAGAAACGAGATCGGGAAGCAGAAATA TAGTGCAATAGTGCAACATCTCTGAATCCTTTTAATCTAGAGAAGGCATTTCATATTTGGGGGCTAAGGTTTCCAGTCAGATGAGGCAAA CAGCAAGAGTAAGCAGTGTTACTTGCAGGTACTTTGGTTAATGTTGATTTAAATTTTCATGAATGTGCTGGTGAACACTGTGACCAGGCT TTTGTAGATGGCGATGTGTTATAGACGGTGCTCACTCCCAAGGGACAGCAAGTGAGCAGAGATGTACTGCAAAGTCGCCAGTCACTGCTG CAAGGTGGCCTCTGCCTGGGGCCTCCAGAAGCTGCTCCTTTACCCTCTTGGTCCCATGGCTGAAGCTGGAGCAGCGGATTGCTCTGGAGC AGCCAAGGCCGCCAGCGTGTGGAGCAGAGCTCTCCCCTCCTGCTGGGCGTGTGTGACACTGATGAGTTTCACTGTACTGCATGTGACTTC TCCCCTGCCCTTCCTCCTGATGGAGTGTGCAGACAGCCATGCGTGGCCACGGGGGCAGTGTGAGGACCTCCCTGTCTCCCGGCTCCCCTC CCAGGGGAGCCAGCTGCTTGACCTAGCTCTTTGGGCCTCTCCTGCCCTCTGCTCTGCCTGGAGTGTCGGATCCTGTGAGTAGGCTGGGCC TCCCCTGGGCAGGGTTCTCCAAGGGCCCGGTTTCCCGGCCCTTACCTTTCCTGATGCCCCTGACATCATCATTCTTGTGGGAGACAGCAG CCTGTATGTGGTGTGGGGCGTGGATCGAGTGTAGCTGTGAAATCCATATATATGAAATGTCCTGCGGGATACAGTCTTAGCTGACTTTTT TTTACTCTGAACTCTTATTTGAATTGTTTTTTGTGCATATATTTCTGCTACCACAGAGATTGTACTATACAAATAAAAAAATAAAAACCC >15072_15072_1_CDH3-CHKA_CDH3_chr16_68718727_ENST00000264012_CHKA_chr11_67836404_ENST00000265689_length(amino acids)=708AA_BP=540 MGPQPHPHRSHLRGGGTSWRWTNQHPPALTWPPPAGSRGLRCSKGQELSGTPARRRGSCFTPLSAAMGLPRGPLASLLLLQVCWLQCAAS EPCRAVFREAEVTLEAGGAEQEPGQALGKVFMGCPGQEPALFSTDNDDFTVRNGETVQERRSLKERNPLKIFPSKRILRRHKRDWVVAPI SVPENGKGPFPQRLNQLKSNKDRDTKIFYSITGPGADSPPEGVFAVEKETGWLLLNKPLDREEIAKYELFGHAVSENGASVEDPMNISII VTDQNDHKPKFTQDTFRGSVLEGVLPGTSVMQVTATDEDDAIYTYNGVVAYSIHSQEPKDPHDLMFTIHRSTGTISVISSGLDREKVPEY TLTIQATDMDGDGSTTTAVAVVEILDANDNAPMFDPQKYEAHVPENAVGHEVQRLTVTDLDAPNSPAWRATYLIMGGDDGDHFTITTHPE SNQGILTTRKGLDFEAKNQHTLYVEVTNEAPFVLKLPTSTATIVVHVEDVNEAPVFVPPSKVVEVQEGIPTGEPVCVYTAEDPDKENQKI RSLLESTPSPVVFCHNDCQEGNILLLEGRENSEKQKLMLIDFEYSSYNYRGFDIGNHFCEWMYDYSYEKYPFFRANIRKYPTKKQQLHFI -------------------------------------------------------------- >15072_15072_2_CDH3-CHKA_CDH3_chr16_68718727_ENST00000429102_CHKA_chr11_67836404_ENST00000265689_length(transcript)=3445nt_BP=1811nt CTTTTTTCCGCTGTAGCCGCAAGGCAAGGAAACATTTCTCTTCCCGTACTGAGGAGGCTGAGGAGTGCACTGGGTGTTCTTTTCTCCTCT AACCCAGAACTGCGAGACAGAGGCTGAGTCCCTGTAAAGAACAGCTCCAGAAAAGCCAGGAGAGCGCAGGAGGGCATCCGGGAGGCCAGG AGGGGTTCGCTGGGGCCTCAACCGCACCCACATCGGTCCCACCTGCGAGGGGGCGGGACCTCGTGGCGCTGGACCAATCAGCACCCACCT GCGCTCACCTGGCCTCCTCCCGCTGGCTCCCGGGGGCTGCGGTGCTCAAAGGGGCAAGAGCTGAGCGGAACACCGGCCCGCCGTCGCGGC AGCTGCTTCACCCCTCTCTCTGCAGCCATGGGGCTCCCTCGTGGACCTCTCGCGTCTCTCCTCCTTCTCCAGGTTTGCTGGCTGCAGTGC GCGGCCTCCGAGCCGTGCCGGGCGGTCTTCAGGGAGGCTGAAGTGACCTTGGAGGCGGGAGGCGCGGAGCAGGAGCCCGGCCAGGCGCTG GGGAAAGTATTCATGGGCTGCCCTGGGCAAGAGCCAGCTCTGTTTAGCACTGATAATGATGACTTCACTGTGCGGAATGGCGAGACAGTC CAGGAAAGAAGGTCACTGAAGGAAAGGAATCCATTGAAGATCTTCCCATCCAAACGTATCTTACGAAGACACAAGAGAGATTGGGTGGTT GCTCCAATATCTGTCCCTGAAAATGGCAAGGGTCCCTTCCCCCAGAGACTGAATCAGCTCAAGTCTAATAAAGATAGAGACACCAAGATT TTCTACAGCATCACGGGGCCGGGGGCAGACAGCCCCCCTGAGGGTGTCTTCGCTGTAGAGAAGGAGACAGGCTGGTTGTTGTTGAATAAG CCACTGGACCGGGAGGAGATTGCCAAGTATGAGCTCTTTGGCCACGCTGTGTCAGAGAATGGTGCCTCAGTGGAGGACCCCATGAACATC TCCATCATCGTGACCGACCAGAATGACCACAAGCCCAAGTTTACCCAGGACACCTTCCGAGGGAGTGTCTTAGAGGGAGTCCTACCAGGT ACTTCTGTGATGCAGGTGACAGCCACGGATGAGGATGATGCCATCTACACCTACAATGGGGTGGTTGCTTACTCCATCCATAGCCAAGAA CCAAAGGACCCACACGACCTCATGTTCACCATTCACCGGAGCACAGGCACCATCAGCGTCATCTCCAGTGGCCTGGACCGGGAAAAAGTC CCTGAGTACACACTGACCATCCAGGCCACAGACATGGATGGGGACGGCTCCACCACCACGGCAGTGGCAGTAGTGGAGATCCTTGATGCC AATGACAATGCTCCCATGTTTGACCCCCAGAAGTACGAGGCCCATGTGCCTGAGAATGCAGTGGGCCATGAGGTGCAGAGGCTGACGGTC ACTGATCTGGACGCCCCCAACTCACCAGCGTGGCGTGCCACCTACCTTATCATGGGCGGTGACGACGGGGACCATTTTACCATCACCACC CACCCTGAGAGCAACCAGGGCATCCTGACAACCAGGAAGGGTTTGGATTTTGAGGCCAAAAACCAGCACACCCTGTACGTTGAAGTGACC AACGAGGCCCCTTTTGTGCTGAAGCTCCCAACCTCCACAGCCACCATAGTGGTCCACGTGGAGGATGTGAATGAGGCACCTGTGTTTGTC CCACCCTCCAAAGTCGTTGAGGTCCAGGAGGGCATCCCCACTGGGGAGCCTGTGTGTGTCTACACTGCAGAAGACCCTGACAAGGAGAAT CAAAAGATCAGATCATTGCTTGAATCTACTCCATCTCCAGTTGTATTTTGTCATAATGACTGTCAAGAAGGTAATATCTTGTTGCTGGAA GGCCGAGAGAATTCTGAAAAACAGAAACTGATGCTCATTGATTTCGAATACAGCAGTTACAATTACAGGGGATTCGACATTGGAAATCAC TTCTGTGAGTGGATGTATGATTATAGCTATGAAAAATACCCTTTTTTCAGAGCAAACATCCGGAAGTATCCCACCAAGAAACAACAGCTC CATTTTATTTCCAGTTACTTGCCTGCATTCCAAAATGACTTTGAAAACCTCAGTACTGAAGAAAAATCCATTATAAAAGAAGAAATGTTG CTTGAAGTTAATAGGTTTGCCCTTGCATCTCATTTCCTCTGGGGACTGTGGTCCATTGTACAAGCCAAGATTTCATCTATTGAATTTGGG TACATGGACTACGCCCAAGCAAGGTTTGATGCCTATTTCCACCAGAAGAGGAAGCTTGGGGTGTGACTGTGGGGAGGACTCCATCCACCT CATCACTGGACTGCATGGGGAGGCAGCAGAGCGGGGTCCCCTCTGTGCTTCGACTACTGCTCCTGTGGCAGGAGGCTTTGGGTGGCTCAC TACTGAACACATGTGTATGATACTAAAGACGGTATTAAAATGGAGCGACGTTTATTTCATCTCTTGTTTACGATTTCACTAGGACTCAGA AACGAGATCGGGAAGCAGAAATATAGTGCAATAGTGCAACATCTCTGAATCCTTTTAATCTAGAGAAGGCATTTCATATTTGGGGGCTAA GGTTTCCAGTCAGATGAGGCAAACAGCAAGAGTAAGCAGTGTTACTTGCAGGTACTTTGGTTAATGTTGATTTAAATTTTCATGAATGTG CTGGTGAACACTGTGACCAGGCTTTTGTAGATGGCGATGTGTTATAGACGGTGCTCACTCCCAAGGGACAGCAAGTGAGCAGAGATGTAC TGCAAAGTCGCCAGTCACTGCTGCAAGGTGGCCTCTGCCTGGGGCCTCCAGAAGCTGCTCCTTTACCCTCTTGGTCCCATGGCTGAAGCT GGAGCAGCGGATTGCTCTGGAGCAGCCAAGGCCGCCAGCGTGTGGAGCAGAGCTCTCCCCTCCTGCTGGGCGTGTGTGACACTGATGAGT TTCACTGTACTGCATGTGACTTCTCCCCTGCCCTTCCTCCTGATGGAGTGTGCAGACAGCCATGCGTGGCCACGGGGGCAGTGTGAGGAC CTCCCTGTCTCCCGGCTCCCCTCCCAGGGGAGCCAGCTGCTTGACCTAGCTCTTTGGGCCTCTCCTGCCCTCTGCTCTGCCTGGAGTGTC GGATCCTGTGAGTAGGCTGGGCCTCCCCTGGGCAGGGTTCTCCAAGGGCCCGGTTTCCCGGCCCTTACCTTTCCTGATGCCCCTGACATC ATCATTCTTGTGGGAGACAGCAGCCTGTATGTGGTGTGGGGCGTGGATCGAGTGTAGCTGTGAAATCCATATATATGAAATGTCCTGCGG GATACAGTCTTAGCTGACTTTTTTTTACTCTGAACTCTTATTTGAATTGTTTTTTGTGCATATATTTCTGCTACCACAGAGATTGTACTA >15072_15072_2_CDH3-CHKA_CDH3_chr16_68718727_ENST00000429102_CHKA_chr11_67836404_ENST00000265689_length(amino acids)=708AA_BP=540 MGPQPHPHRSHLRGGGTSWRWTNQHPPALTWPPPAGSRGLRCSKGQELSGTPARRRGSCFTPLSAAMGLPRGPLASLLLLQVCWLQCAAS EPCRAVFREAEVTLEAGGAEQEPGQALGKVFMGCPGQEPALFSTDNDDFTVRNGETVQERRSLKERNPLKIFPSKRILRRHKRDWVVAPI SVPENGKGPFPQRLNQLKSNKDRDTKIFYSITGPGADSPPEGVFAVEKETGWLLLNKPLDREEIAKYELFGHAVSENGASVEDPMNISII VTDQNDHKPKFTQDTFRGSVLEGVLPGTSVMQVTATDEDDAIYTYNGVVAYSIHSQEPKDPHDLMFTIHRSTGTISVISSGLDREKVPEY TLTIQATDMDGDGSTTTAVAVVEILDANDNAPMFDPQKYEAHVPENAVGHEVQRLTVTDLDAPNSPAWRATYLIMGGDDGDHFTITTHPE SNQGILTTRKGLDFEAKNQHTLYVEVTNEAPFVLKLPTSTATIVVHVEDVNEAPVFVPPSKVVEVQEGIPTGEPVCVYTAEDPDKENQKI RSLLESTPSPVVFCHNDCQEGNILLLEGRENSEKQKLMLIDFEYSSYNYRGFDIGNHFCEWMYDYSYEKYPFFRANIRKYPTKKQQLHFI -------------------------------------------------------------- >15072_15072_3_CDH3-CHKA_CDH3_chr16_68718727_ENST00000581171_CHKA_chr11_67836404_ENST00000265689_length(transcript)=3022nt_BP=1388nt GCGGTGCTCAAAGGGGCAAGAGCTGAGCGGAACACCGGCCCGCCGTCGCGGCAGCTGCTTCACCCCTCTCTCTGCAGCCATGGGGCTCCC TCGTGGACCTCTCGCGTCTCTCCTCCTTCTCCAGTATTCATGGGCTGCCCTGGGCAAGAGCCAGCTCTGTTTAGCACTGATAATGATGAC TTCACTGTGCGGAATGGCGAGACAGTCCAGGAAAGAAGGTCACTGAAGGAAAGGAATCCATTGAAGATCTTCCCATCCAAACGTATCTTA CGAAGACACAAGAGAGATTGGGTGGTTGCTCCAATATCTGTCCCTGAAAATGGCAAGGGTCCCTTCCCCCAGAGACTGAATCAGCTCAAG TCTAATAAAGATAGAGACACCAAGATTTTCTACAGCATCACGGGGCCGGGGGCAGACAGCCCCCCTGAGGGTGTCTTCGCTGTAGAGAAG GAGACAGGCTGGTTGTTGTTGAATAAGCCACTGGACCGGGAGGAGATTGCCAAGTATGAGCTCTTTGGCCACGCTGTGTCAGAGAATGGT GCCTCAGTGGAGGACCCCATGAACATCTCCATCATCGTGACCGACCAGAATGACCACAAGCCCAAGTTTACCCAGGACACCTTCCGAGGG AGTGTCTTAGAGGGAGTCCTACCAGGTACTTCTGTGATGCAGGTGACAGCCACGGATGAGGATGATGCCATCTACACCTACAATGGGGTG GTTGCTTACTCCATCCATAGCCAAGAACCAAAGGACCCACACGACCTCATGTTCACCATTCACCGGAGCACAGGCACCATCAGCGTCATC TCCAGTGGCCTGGACCGGGAAAAAGTCCCTGAGTACACACTGACCATCCAGGCCACAGACATGGATGGGGACGGCTCCACCACCACGGCA GTGGCAGTAGTGGAGATCCTTGATGCCAATGACAATGCTCCCATGTTTGACCCCCAGAAGTACGAGGCCCATGTGCCTGAGAATGCAGTG GGCCATGAGGTGCAGAGGCTGACGGTCACTGATCTGGACGCCCCCAACTCACCAGCGTGGCGTGCCACCTACCTTATCATGGGCGGTGAC GACGGGGACCATTTTACCATCACCACCCACCCTGAGAGCAACCAGGGCATCCTGACAACCAGGAAGGGTTTGGATTTTGAGGCCAAAAAC CAGCACACCCTGTACGTTGAAGTGACCAACGAGGCCCCTTTTGTGCTGAAGCTCCCAACCTCCACAGCCACCATAGTGGTCCACGTGGAG GATGTGAATGAGGCACCTGTGTTTGTCCCACCCTCCAAAGTCGTTGAGGTCCAGGAGGGCATCCCCACTGGGGAGCCTGTGTGTGTCTAC ACTGCAGAAGACCCTGACAAGGAGAATCAAAAGATCAGATCATTGCTTGAATCTACTCCATCTCCAGTTGTATTTTGTCATAATGACTGT CAAGAAGGTAATATCTTGTTGCTGGAAGGCCGAGAGAATTCTGAAAAACAGAAACTGATGCTCATTGATTTCGAATACAGCAGTTACAAT TACAGGGGATTCGACATTGGAAATCACTTCTGTGAGTGGATGTATGATTATAGCTATGAAAAATACCCTTTTTTCAGAGCAAACATCCGG AAGTATCCCACCAAGAAACAACAGCTCCATTTTATTTCCAGTTACTTGCCTGCATTCCAAAATGACTTTGAAAACCTCAGTACTGAAGAA AAATCCATTATAAAAGAAGAAATGTTGCTTGAAGTTAATAGGTTTGCCCTTGCATCTCATTTCCTCTGGGGACTGTGGTCCATTGTACAA GCCAAGATTTCATCTATTGAATTTGGGTACATGGACTACGCCCAAGCAAGGTTTGATGCCTATTTCCACCAGAAGAGGAAGCTTGGGGTG TGACTGTGGGGAGGACTCCATCCACCTCATCACTGGACTGCATGGGGAGGCAGCAGAGCGGGGTCCCCTCTGTGCTTCGACTACTGCTCC TGTGGCAGGAGGCTTTGGGTGGCTCACTACTGAACACATGTGTATGATACTAAAGACGGTATTAAAATGGAGCGACGTTTATTTCATCTC TTGTTTACGATTTCACTAGGACTCAGAAACGAGATCGGGAAGCAGAAATATAGTGCAATAGTGCAACATCTCTGAATCCTTTTAATCTAG AGAAGGCATTTCATATTTGGGGGCTAAGGTTTCCAGTCAGATGAGGCAAACAGCAAGAGTAAGCAGTGTTACTTGCAGGTACTTTGGTTA ATGTTGATTTAAATTTTCATGAATGTGCTGGTGAACACTGTGACCAGGCTTTTGTAGATGGCGATGTGTTATAGACGGTGCTCACTCCCA AGGGACAGCAAGTGAGCAGAGATGTACTGCAAAGTCGCCAGTCACTGCTGCAAGGTGGCCTCTGCCTGGGGCCTCCAGAAGCTGCTCCTT TACCCTCTTGGTCCCATGGCTGAAGCTGGAGCAGCGGATTGCTCTGGAGCAGCCAAGGCCGCCAGCGTGTGGAGCAGAGCTCTCCCCTCC TGCTGGGCGTGTGTGACACTGATGAGTTTCACTGTACTGCATGTGACTTCTCCCCTGCCCTTCCTCCTGATGGAGTGTGCAGACAGCCAT GCGTGGCCACGGGGGCAGTGTGAGGACCTCCCTGTCTCCCGGCTCCCCTCCCAGGGGAGCCAGCTGCTTGACCTAGCTCTTTGGGCCTCT CCTGCCCTCTGCTCTGCCTGGAGTGTCGGATCCTGTGAGTAGGCTGGGCCTCCCCTGGGCAGGGTTCTCCAAGGGCCCGGTTTCCCGGCC CTTACCTTTCCTGATGCCCCTGACATCATCATTCTTGTGGGAGACAGCAGCCTGTATGTGGTGTGGGGCGTGGATCGAGTGTAGCTGTGA AATCCATATATATGAAATGTCCTGCGGGATACAGTCTTAGCTGACTTTTTTTTACTCTGAACTCTTATTTGAATTGTTTTTTGTGCATAT >15072_15072_3_CDH3-CHKA_CDH3_chr16_68718727_ENST00000581171_CHKA_chr11_67836404_ENST00000265689_length(amino acids)=612AA_BP=444 MLHPSLCSHGAPSWTSRVSPPSPVFMGCPGQEPALFSTDNDDFTVRNGETVQERRSLKERNPLKIFPSKRILRRHKRDWVVAPISVPENG KGPFPQRLNQLKSNKDRDTKIFYSITGPGADSPPEGVFAVEKETGWLLLNKPLDREEIAKYELFGHAVSENGASVEDPMNISIIVTDQND HKPKFTQDTFRGSVLEGVLPGTSVMQVTATDEDDAIYTYNGVVAYSIHSQEPKDPHDLMFTIHRSTGTISVISSGLDREKVPEYTLTIQA TDMDGDGSTTTAVAVVEILDANDNAPMFDPQKYEAHVPENAVGHEVQRLTVTDLDAPNSPAWRATYLIMGGDDGDHFTITTHPESNQGIL TTRKGLDFEAKNQHTLYVEVTNEAPFVLKLPTSTATIVVHVEDVNEAPVFVPPSKVVEVQEGIPTGEPVCVYTAEDPDKENQKIRSLLES TPSPVVFCHNDCQEGNILLLEGRENSEKQKLMLIDFEYSSYNYRGFDIGNHFCEWMYDYSYEKYPFFRANIRKYPTKKQQLHFISSYLPA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CDH3-CHKA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CDH3-CHKA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CDH3-CHKA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
