
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CDK12-FAM20A (FusionGDB2 ID:15132) |
Fusion Gene Summary for CDK12-FAM20A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CDK12-FAM20A | Fusion gene ID: 15132 | Hgene | Tgene | Gene symbol | CDK12 | FAM20A | Gene ID | 51755 | 54757 |
| Gene name | cyclin dependent kinase 12 | FAM20A golgi associated secretory pathway pseudokinase | |
| Synonyms | CRK7|CRKR|CRKRS | AI1G|AIGFS|FP2747 | |
| Cytomap | 17q12 | 17q24.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cyclin-dependent kinase 12CDC2-related protein kinase 7Cdc2-related kinase, arginine/serine-richcell division cycle 2-related protein kinase 7cell division protein kinase 12 | pseudokinase FAM20Afamily with sequence similarity 20, member Aprotein FAM20A | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NYV4 | Q96MK3 | |
| Ensembl transtripts involved in fusion gene | ENST00000430627, ENST00000447079, ENST00000559545, | ENST00000226094, ENST00000592554, | |
| Fusion gene scores | * DoF score | 29 X 13 X 13=4901 | 6 X 5 X 2=60 |
| # samples | 31 | 4 | |
| ** MAII score | log2(31/4901*10)=-3.98273602613552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/60*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CDK12 [Title/Abstract] AND FAM20A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CDK12(37628016)-FAM20A(66551884), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CDK12 | GO:0046777 | protein autophosphorylation | 11683387 |
| Tgene | FAM20A | GO:0001934 | positive regulation of protein phosphorylation | 25789606 |
 Fusion gene breakpoints across CDK12 (5'-gene) Fusion gene breakpoints across CDK12 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
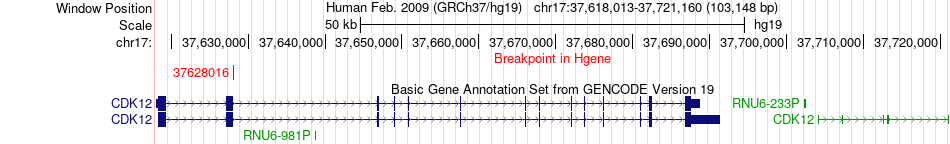 |
 Fusion gene breakpoints across FAM20A (3'-gene) Fusion gene breakpoints across FAM20A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
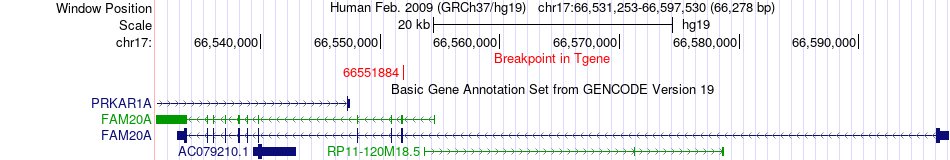 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-3C-AALI-01A | CDK12 | chr17 | 37628016 | - | FAM20A | chr17 | 66551884 | - |
| ChimerDB4 | BRCA | TCGA-3C-AALI-01A | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
Top |
Fusion Gene ORF analysis for CDK12-FAM20A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000430627 | ENST00000226094 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
| 5CDS-5UTR | ENST00000447079 | ENST00000226094 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
| In-frame | ENST00000430627 | ENST00000592554 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
| In-frame | ENST00000447079 | ENST00000592554 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
| intron-3CDS | ENST00000559545 | ENST00000592554 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
| intron-5UTR | ENST00000559545 | ENST00000226094 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000430627 | CDK12 | chr17 | 37628016 | + | ENST00000592554 | FAM20A | chr17 | 66551884 | - | 4070 | 2242 | 185 | 3463 | 1092 |
| ENST00000447079 | CDK12 | chr17 | 37628016 | + | ENST00000592554 | FAM20A | chr17 | 66551884 | - | 3792 | 1964 | 33 | 3185 | 1050 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000430627 | ENST00000592554 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - | 0.000525886 | 0.9994741 |
| ENST00000447079 | ENST00000592554 | CDK12 | chr17 | 37628016 | + | FAM20A | chr17 | 66551884 | - | 0.000441316 | 0.9995586 |
Top |
Fusion Genomic Features for CDK12-FAM20A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
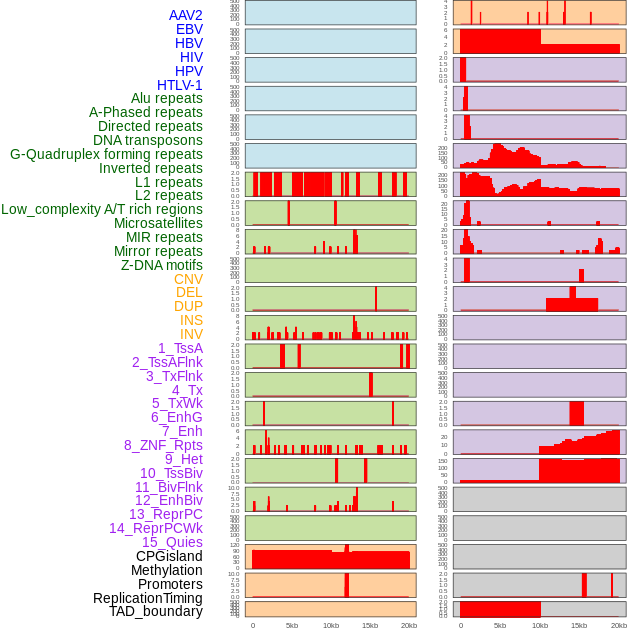 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CDK12-FAM20A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:37628016/chr17:66551884) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CDK12 | FAM20A |
| FUNCTION: Cyclin-dependent kinase that phosphorylates the C-terminal domain (CTD) of the large subunit of RNA polymerase II (POLR2A), thereby acting as a key regulator of transcription elongation. Regulates the expression of genes involved in DNA repair and is required for the maintenance of genomic stability. Preferentially phosphorylates 'Ser-5' in CTD repeats that are already phosphorylated at 'Ser-7', but can also phosphorylate 'Ser-2'. Required for RNA splicing, possibly by phosphorylating SRSF1/SF2. Involved in regulation of MAP kinase activity, possibly leading to affect the response to estrogen inhibitors. {ECO:0000269|PubMed:11683387, ECO:0000269|PubMed:19651820, ECO:0000269|PubMed:20952539, ECO:0000269|PubMed:22012619, ECO:0000269|PubMed:24662513}. | FUNCTION: Pseudokinase that acts as an allosteric activator of the Golgi serine/threonine protein kinase FAM20C and is involved in biomineralization of teeth. Forms a complex with FAM20C and increases the ability of FAM20C to phosphorylate the proteins that form the 'matrix' that guides the deposition of the enamel minerals. {ECO:0000269|PubMed:25789606}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000430627 | + | 2 | 14 | 407_413 | 643 | 1482.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000430627 | + | 2 | 14 | 535_540 | 643 | 1482.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000447079 | + | 2 | 14 | 407_413 | 643 | 1491.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000447079 | + | 2 | 14 | 535_540 | 643 | 1491.0 | Compositional bias | Note=Poly-Pro |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000430627 | + | 2 | 14 | 1266_1280 | 643 | 1482.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000447079 | + | 2 | 14 | 1266_1280 | 643 | 1491.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000430627 | + | 2 | 14 | 727_1020 | 643 | 1482.0 | Domain | Protein kinase |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000447079 | + | 2 | 14 | 727_1020 | 643 | 1491.0 | Domain | Protein kinase |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000430627 | + | 2 | 14 | 733_741 | 643 | 1482.0 | Nucleotide binding | Note=ATP |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000430627 | + | 2 | 14 | 814_819 | 643 | 1482.0 | Nucleotide binding | Note=ATP |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000447079 | + | 2 | 14 | 733_741 | 643 | 1491.0 | Nucleotide binding | Note=ATP |
| Hgene | CDK12 | chr17:37628016 | chr17:66551884 | ENST00000447079 | + | 2 | 14 | 814_819 | 643 | 1491.0 | Nucleotide binding | Note=ATP |
Top |
Fusion Gene Sequence for CDK12-FAM20A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >15132_15132_1_CDK12-FAM20A_CDK12_chr17_37628016_ENST00000430627_FAM20A_chr17_66551884_ENST00000592554_length(transcript)=4070nt_BP=2242nt CCCGGCGCCCGGGCCTGAGGACTGGCTCGGCGGAGGGAGAAGAGGAAACAGACTTGAGCAGCTCCCCGTTGTCTCGCAACTCCACTGCCG AGGAACTCTCATTTCTTCCCTCGCTCCTTCACCCCCCACCTCATGTAGAAGGGTGCTGAGGCGTCGGGAGGGAGGAGGAGCCTGGGCTAC CGTCCCTGCCCTCCCCACCCCCTTCCCGGGGCGCTTTGGTGGGCGTGGAGTTGGGGTTGGGGGGGTGGGTGGGGGTTGCTTTTTGGAGTG CTGGGGAACTTTTTTCCCTTCTTCAGGTCAGGGGAAAGGGAATGCCCAATTCAGAGAGACATGGGGGCAAGAAGGACGGGAGTGGAGGAG CTTCTGGAACTTTGCAGCCGTCATCGGGAGGCGGCAGCTCTAACAGCAGAGAGCGTCACCGCTTGGTATCGAAGCACAAGCGGCATAAGT CCAAACACTCCAAAGACATGGGGTTGGTGACCCCCGAAGCAGCATCCCTGGGCACAGTTATCAAACCTTTGGTGGAGTATGATGATATCA GCTCTGATTCCGACACCTTCTCCGATGACATGGCCTTCAAACTAGACCGAAGGGAGAACGACGAACGTCGTGGATCAGATCGGAGCGACC GCCTGCACAAACATCGTCACCACCAGCACAGGCGTTCCCGGGACTTACTAAAAGCTAAACAGACCGAAAAAGAAAAAAGCCAAGAAGTCT CCAGCAAGTCGGGATCGATGAAGGACCGGATATCGGGAAGTTCAAAGCGTTCGAATGAGGAGACTGATGACTATGGGAAGGCGCAGGTAG CCAAAAGCAGCAGCAAGGAATCCAGGTCATCCAAGCTCCACAAGGAGAAGACCAGGAAAGAACGGGAGCTGAAGTCTGGGCACAAAGACC GGAGTAAAAGTCATCGAAAAAGGGAAACACCCAAAAGTTACAAAACAGTGGACAGCCCAAAACGGAGATCCAGGAGCCCCCACAGGAAGT GGTCTGACAGCTCCAAACAAGATGATAGCCCCTCGGGAGCTTCTTATGGCCAAGATTATGACCTTAGTCCCTCACGATCTCATACCTCGA GCAATTATGACTCCTACAAGAAAAGTCCTGGAAGTACCTCGAGAAGGCAGTCGGTCAGTCCCCCTTACAAGGAGCCTTCGGCCTACCAGT CCAGCACCCGGTCACCGAGCCCCTACAGTAGGCGACAGAGATCTGTCAGTCCCTATAGCAGGAGACGGTCGTCCAGCTACGAAAGAAGTG GCTCTTACAGCGGGCGATCGCCCAGTCCCTATGGTCGAAGGCGGTCCAGCAGCCCTTTCCTGAGCAAGCGGTCTCTGAGTCGGAGTCCAC TCCCCAGTAGGAAATCCATGAAGTCCAGAAGTAGAAGTCCTGCATATTCAAGACATTCATCTTCTCATAGTAAAAAGAAGAGATCCAGTT CACGCAGTCGTCATTCCAGTATCTCACCTGTCAGGCTTCCACTTAATTCCAGTCTGGGAGCTGAACTCAGTAGGAAAAAGAAGGAAAGAG CAGCTGCTGCTGCTGCAGCAAAGATGGATGGAAAGGAGTCCAAGGGTTCACCTGTATTTTTGCCTAGAAAAGAGAACAGTTCAGTAGAGG CTAAGGATTCAGGTTTGGAGTCTAAAAAGTTACCCAGAAGTGTAAAATTGGAAAAATCTGCCCCAGATACTGAACTGGTGAATGTAACAC ATCTAAACACAGAGGTAAAAAATTCTTCAGATACAGGGAAAGTAAAGTTGGATGAGAACTCCGAGAAGCATCTTGTTAAAGATTTGAAAG CACAGGGAACAAGAGACTCTAAACCCATAGCACTGAAAGAGGAGATTGTTACTCCAAAGGAGACAGAAACATCAGAAAAGGAGACCCCTC CACCTCTTCCCACAATTGCTTCTCCCCCACCCCCTCTACCAACTACTACCCCTCCACCTCAGACACCCCCTTTGCCACCTTTGCCTCCAA TACCAGCTCTTCCACAGCAACCACCTCTGCCTCCTTCTCAGCCAGCATTTAGTCAGGTTCCTGCTTCCAGTACTTCAACTTTGCCCCCTT CTACTCACTCAAAGACATCTGCTGTGTCCTCTCAGGCAAATTCTCAGCCCCCTGTACAGGTTTCTGTGAAGACTCAAGTATCTGTAACAG CTGCTATTCCACACCTGAAAACTTCAACGTTGCCTCCTTTGCCCCTCCCACCCTTATTACCTGGAGATGATGACATGGATAGGCGACACA AGATGTACAGAGAGCAGATGAACCTTACCTCCCTGGACCCCCCACTGCAGCTCCGACTCGAGGCCAGCTGGGTCCAGTTCCACCTGGGTA TTAACCGCCATGGGCTCTACTCCCGGTCCAGCCCTGTTGTCAGCAAACTTCTGCAAGACATGAGGCACTTTCCCACCATCAGTGCTGATT ACAGTCAAGATGAGAAAGCCTTGCTGGGGGCATGTGACTGCACCCAGATTGTGAAACCCAGTGGGGTCCACCTCAAGCTGGTGCTGAGGT TCTCGGATTTCGGGAAGGCCATGTTCAAACCCATGAGACAGCAGCGAGATGAGGAGACACCAGTGGACTTCTTCTACTTCATTGACTTTC AGAGACACAATGCTGAGATCGCAGCTTTCCATCTGGACAGGATTCTGGACTTCCGACGGGTGCCGCCAACAGTGGGGAGGATAGTAAATG TCACCAAGGAAATCCTAGAGGTCACCAAGAATGAAATCCTGCAGAGTGTTTTCTTTGTCTCTCCAGCGAGCAACGTGTGCTTCTTCGCCA AGTGTCCATACATGTGCAAGACGGAGTATGCTGTCTGTGGCAACCCACACCTGCTGGAGGGTTCCCTCTCTGCCTTCCTGCCGTCCCTCA ACCTGGCCCCCAGGCTGTCTGTGCCCAACCCCTGGATCCGCTCCTACACACTGGCAGGAAAAGAGGAGTGGGAGGTCAATCCCCTTTACT GTGACACAGTGAAACAGATCTACCCGTACAACAACAGCCAGCGGCTCCTCAATGTCATCGACATGGCCATCTTCGACTTCTTGATAGGGA ATATGGACCGGCACCATTATGAGATGTTCACCAAGTTCGGGGATGATGGGTTCCTTATTCACCTTGACAACGCCAGAGGGTTCGGACGAC ACTCCCATGATGAAATCTCCATCCTCTCGCCTCTCTCCCAGTGCTGCATGATAAAAAAGAAAACACTTTTGCACCTGCAGCTGCTGGCCC AAGCTGACTACAGACTCAGCGATGTGATGCGAGAATCACTGCTGGAAGACCAGCTCAGCCCTGTCCTCACTGAACCCCACCTCCTTGCCC TGGATCGAAGGCTCCAAACCATCCTAAGGACAGTGGAGGGGTGCATAGTGGCCCATGGACAGCAGAGTGTCATAGTCGACGGCCCAGTGG AACAGTTGGCCCCAGACTCTGGCCAGGCTAACTTGACAAGCTAAGGGCTGGCAGAGTCCAGTTTCAGAAAATACGCCTGGAGCCAGAGCA GTCGACTCGAGTGCCGACCCTGCGTCCTCACTCCCACCTGTTACTGCTGGGAGTCAAGTCAGCTAGGAAGGAAGCAGGACATTTTCTCAA ACAGCAAGTGGGGCCCATGGAACTGAATCTTTACTCCTTGGTGCACCGCTTCTGTCGTGCGTTGCCTTGCTCCGTTTTTCCCAAAAAGCA CTGGCTTCATCAAGGCCACCGACGATCTCCTGAGTGCACTGGGAAATCTGGGTATAGGTCAGGCTTGGCAGCCTTGATCCCAGGAGAGTA CTAATGGTAACAAGTCAAATAAAAGGACATCAAGTGGATACCTGACTTCTCAGGATCCTTATTCTAGCTACAAGTCAAAGATAACTCCTG GTCCAGACAAAACACCTGGCCTATCACAAGCTGACTAAAAATCTGCACTTTGGGCCAGCGCAGGCAACAGTAACTCTGACAGGTTCAAAT TAGACCTCACACTTTCTACTCATATTCTAGTCACTGGACCCATCTGAATCAGTAATCCCTACTGCCCGGTCCTGGAGTAACTTCTTAGAG >15132_15132_1_CDK12-FAM20A_CDK12_chr17_37628016_ENST00000430627_FAM20A_chr17_66551884_ENST00000592554_length(amino acids)=1092AA_BP=431 MPSPPPSRGALVGVELGLGGWVGVAFWSAGELFSLLQVRGKGMPNSERHGGKKDGSGGASGTLQPSSGGGSSNSRERHRLVSKHKRHKSK HSKDMGLVTPEAASLGTVIKPLVEYDDISSDSDTFSDDMAFKLDRRENDERRGSDRSDRLHKHRHHQHRRSRDLLKAKQTEKEKSQEVSS KSGSMKDRISGSSKRSNEETDDYGKAQVAKSSSKESRSSKLHKEKTRKERELKSGHKDRSKSHRKRETPKSYKTVDSPKRRSRSPHRKWS DSSKQDDSPSGASYGQDYDLSPSRSHTSSNYDSYKKSPGSTSRRQSVSPPYKEPSAYQSSTRSPSPYSRRQRSVSPYSRRRSSSYERSGS YSGRSPSPYGRRRSSSPFLSKRSLSRSPLPSRKSMKSRSRSPAYSRHSSSHSKKKRSSSRSRHSSISPVRLPLNSSLGAELSRKKKERAA AAAAAKMDGKESKGSPVFLPRKENSSVEAKDSGLESKKLPRSVKLEKSAPDTELVNVTHLNTEVKNSSDTGKVKLDENSEKHLVKDLKAQ GTRDSKPIALKEEIVTPKETETSEKETPPPLPTIASPPPPLPTTTPPPQTPPLPPLPPIPALPQQPPLPPSQPAFSQVPASSTSTLPPST HSKTSAVSSQANSQPPVQVSVKTQVSVTAAIPHLKTSTLPPLPLPPLLPGDDDMDRRHKMYREQMNLTSLDPPLQLRLEASWVQFHLGIN RHGLYSRSSPVVSKLLQDMRHFPTISADYSQDEKALLGACDCTQIVKPSGVHLKLVLRFSDFGKAMFKPMRQQRDEETPVDFFYFIDFQR HNAEIAAFHLDRILDFRRVPPTVGRIVNVTKEILEVTKNEILQSVFFVSPASNVCFFAKCPYMCKTEYAVCGNPHLLEGSLSAFLPSLNL APRLSVPNPWIRSYTLAGKEEWEVNPLYCDTVKQIYPYNNSQRLLNVIDMAIFDFLIGNMDRHHYEMFTKFGDDGFLIHLDNARGFGRHS HDEISILSPLSQCCMIKKKTLLHLQLLAQADYRLSDVMRESLLEDQLSPVLTEPHLLALDRRLQTILRTVEGCIVAHGQQSVIVDGPVEQ -------------------------------------------------------------- >15132_15132_2_CDK12-FAM20A_CDK12_chr17_37628016_ENST00000447079_FAM20A_chr17_66551884_ENST00000592554_length(transcript)=3792nt_BP=1964nt CTTTTTTCCCTTCTTCAGGTCAGGGGAAAGGGAATGCCCAATTCAGAGAGACATGGGGGCAAGAAGGACGGGAGTGGAGGAGCTTCTGGA ACTTTGCAGCCGTCATCGGGAGGCGGCAGCTCTAACAGCAGAGAGCGTCACCGCTTGGTATCGAAGCACAAGCGGCATAAGTCCAAACAC TCCAAAGACATGGGGTTGGTGACCCCCGAAGCAGCATCCCTGGGCACAGTTATCAAACCTTTGGTGGAGTATGATGATATCAGCTCTGAT TCCGACACCTTCTCCGATGACATGGCCTTCAAACTAGACCGAAGGGAGAACGACGAACGTCGTGGATCAGATCGGAGCGACCGCCTGCAC AAACATCGTCACCACCAGCACAGGCGTTCCCGGGACTTACTAAAAGCTAAACAGACCGAAAAAGAAAAAAGCCAAGAAGTCTCCAGCAAG TCGGGATCGATGAAGGACCGGATATCGGGAAGTTCAAAGCGTTCGAATGAGGAGACTGATGACTATGGGAAGGCGCAGGTAGCCAAAAGC AGCAGCAAGGAATCCAGGTCATCCAAGCTCCACAAGGAGAAGACCAGGAAAGAACGGGAGCTGAAGTCTGGGCACAAAGACCGGAGTAAA AGTCATCGAAAAAGGGAAACACCCAAAAGTTACAAAACAGTGGACAGCCCAAAACGGAGATCCAGGAGCCCCCACAGGAAGTGGTCTGAC AGCTCCAAACAAGATGATAGCCCCTCGGGAGCTTCTTATGGCCAAGATTATGACCTTAGTCCCTCACGATCTCATACCTCGAGCAATTAT GACTCCTACAAGAAAAGTCCTGGAAGTACCTCGAGAAGGCAGTCGGTCAGTCCCCCTTACAAGGAGCCTTCGGCCTACCAGTCCAGCACC CGGTCACCGAGCCCCTACAGTAGGCGACAGAGATCTGTCAGTCCCTATAGCAGGAGACGGTCGTCCAGCTACGAAAGAAGTGGCTCTTAC AGCGGGCGATCGCCCAGTCCCTATGGTCGAAGGCGGTCCAGCAGCCCTTTCCTGAGCAAGCGGTCTCTGAGTCGGAGTCCACTCCCCAGT AGGAAATCCATGAAGTCCAGAAGTAGAAGTCCTGCATATTCAAGACATTCATCTTCTCATAGTAAAAAGAAGAGATCCAGTTCACGCAGT CGTCATTCCAGTATCTCACCTGTCAGGCTTCCACTTAATTCCAGTCTGGGAGCTGAACTCAGTAGGAAAAAGAAGGAAAGAGCAGCTGCT GCTGCTGCAGCAAAGATGGATGGAAAGGAGTCCAAGGGTTCACCTGTATTTTTGCCTAGAAAAGAGAACAGTTCAGTAGAGGCTAAGGAT TCAGGTTTGGAGTCTAAAAAGTTACCCAGAAGTGTAAAATTGGAAAAATCTGCCCCAGATACTGAACTGGTGAATGTAACACATCTAAAC ACAGAGGTAAAAAATTCTTCAGATACAGGGAAAGTAAAGTTGGATGAGAACTCCGAGAAGCATCTTGTTAAAGATTTGAAAGCACAGGGA ACAAGAGACTCTAAACCCATAGCACTGAAAGAGGAGATTGTTACTCCAAAGGAGACAGAAACATCAGAAAAGGAGACCCCTCCACCTCTT CCCACAATTGCTTCTCCCCCACCCCCTCTACCAACTACTACCCCTCCACCTCAGACACCCCCTTTGCCACCTTTGCCTCCAATACCAGCT CTTCCACAGCAACCACCTCTGCCTCCTTCTCAGCCAGCATTTAGTCAGGTTCCTGCTTCCAGTACTTCAACTTTGCCCCCTTCTACTCAC TCAAAGACATCTGCTGTGTCCTCTCAGGCAAATTCTCAGCCCCCTGTACAGGTTTCTGTGAAGACTCAAGTATCTGTAACAGCTGCTATT CCACACCTGAAAACTTCAACGTTGCCTCCTTTGCCCCTCCCACCCTTATTACCTGGAGATGATGACATGGATAGGCGACACAAGATGTAC AGAGAGCAGATGAACCTTACCTCCCTGGACCCCCCACTGCAGCTCCGACTCGAGGCCAGCTGGGTCCAGTTCCACCTGGGTATTAACCGC CATGGGCTCTACTCCCGGTCCAGCCCTGTTGTCAGCAAACTTCTGCAAGACATGAGGCACTTTCCCACCATCAGTGCTGATTACAGTCAA GATGAGAAAGCCTTGCTGGGGGCATGTGACTGCACCCAGATTGTGAAACCCAGTGGGGTCCACCTCAAGCTGGTGCTGAGGTTCTCGGAT TTCGGGAAGGCCATGTTCAAACCCATGAGACAGCAGCGAGATGAGGAGACACCAGTGGACTTCTTCTACTTCATTGACTTTCAGAGACAC AATGCTGAGATCGCAGCTTTCCATCTGGACAGGATTCTGGACTTCCGACGGGTGCCGCCAACAGTGGGGAGGATAGTAAATGTCACCAAG GAAATCCTAGAGGTCACCAAGAATGAAATCCTGCAGAGTGTTTTCTTTGTCTCTCCAGCGAGCAACGTGTGCTTCTTCGCCAAGTGTCCA TACATGTGCAAGACGGAGTATGCTGTCTGTGGCAACCCACACCTGCTGGAGGGTTCCCTCTCTGCCTTCCTGCCGTCCCTCAACCTGGCC CCCAGGCTGTCTGTGCCCAACCCCTGGATCCGCTCCTACACACTGGCAGGAAAAGAGGAGTGGGAGGTCAATCCCCTTTACTGTGACACA GTGAAACAGATCTACCCGTACAACAACAGCCAGCGGCTCCTCAATGTCATCGACATGGCCATCTTCGACTTCTTGATAGGGAATATGGAC CGGCACCATTATGAGATGTTCACCAAGTTCGGGGATGATGGGTTCCTTATTCACCTTGACAACGCCAGAGGGTTCGGACGACACTCCCAT GATGAAATCTCCATCCTCTCGCCTCTCTCCCAGTGCTGCATGATAAAAAAGAAAACACTTTTGCACCTGCAGCTGCTGGCCCAAGCTGAC TACAGACTCAGCGATGTGATGCGAGAATCACTGCTGGAAGACCAGCTCAGCCCTGTCCTCACTGAACCCCACCTCCTTGCCCTGGATCGA AGGCTCCAAACCATCCTAAGGACAGTGGAGGGGTGCATAGTGGCCCATGGACAGCAGAGTGTCATAGTCGACGGCCCAGTGGAACAGTTG GCCCCAGACTCTGGCCAGGCTAACTTGACAAGCTAAGGGCTGGCAGAGTCCAGTTTCAGAAAATACGCCTGGAGCCAGAGCAGTCGACTC GAGTGCCGACCCTGCGTCCTCACTCCCACCTGTTACTGCTGGGAGTCAAGTCAGCTAGGAAGGAAGCAGGACATTTTCTCAAACAGCAAG TGGGGCCCATGGAACTGAATCTTTACTCCTTGGTGCACCGCTTCTGTCGTGCGTTGCCTTGCTCCGTTTTTCCCAAAAAGCACTGGCTTC ATCAAGGCCACCGACGATCTCCTGAGTGCACTGGGAAATCTGGGTATAGGTCAGGCTTGGCAGCCTTGATCCCAGGAGAGTACTAATGGT AACAAGTCAAATAAAAGGACATCAAGTGGATACCTGACTTCTCAGGATCCTTATTCTAGCTACAAGTCAAAGATAACTCCTGGTCCAGAC AAAACACCTGGCCTATCACAAGCTGACTAAAAATCTGCACTTTGGGCCAGCGCAGGCAACAGTAACTCTGACAGGTTCAAATTAGACCTC ACACTTTCTACTCATATTCTAGTCACTGGACCCATCTGAATCAGTAATCCCTACTGCCCGGTCCTGGAGTAACTTCTTAGAGATATTATA >15132_15132_2_CDK12-FAM20A_CDK12_chr17_37628016_ENST00000447079_FAM20A_chr17_66551884_ENST00000592554_length(amino acids)=1050AA_BP=389 MPNSERHGGKKDGSGGASGTLQPSSGGGSSNSRERHRLVSKHKRHKSKHSKDMGLVTPEAASLGTVIKPLVEYDDISSDSDTFSDDMAFK LDRRENDERRGSDRSDRLHKHRHHQHRRSRDLLKAKQTEKEKSQEVSSKSGSMKDRISGSSKRSNEETDDYGKAQVAKSSSKESRSSKLH KEKTRKERELKSGHKDRSKSHRKRETPKSYKTVDSPKRRSRSPHRKWSDSSKQDDSPSGASYGQDYDLSPSRSHTSSNYDSYKKSPGSTS RRQSVSPPYKEPSAYQSSTRSPSPYSRRQRSVSPYSRRRSSSYERSGSYSGRSPSPYGRRRSSSPFLSKRSLSRSPLPSRKSMKSRSRSP AYSRHSSSHSKKKRSSSRSRHSSISPVRLPLNSSLGAELSRKKKERAAAAAAAKMDGKESKGSPVFLPRKENSSVEAKDSGLESKKLPRS VKLEKSAPDTELVNVTHLNTEVKNSSDTGKVKLDENSEKHLVKDLKAQGTRDSKPIALKEEIVTPKETETSEKETPPPLPTIASPPPPLP TTTPPPQTPPLPPLPPIPALPQQPPLPPSQPAFSQVPASSTSTLPPSTHSKTSAVSSQANSQPPVQVSVKTQVSVTAAIPHLKTSTLPPL PLPPLLPGDDDMDRRHKMYREQMNLTSLDPPLQLRLEASWVQFHLGINRHGLYSRSSPVVSKLLQDMRHFPTISADYSQDEKALLGACDC TQIVKPSGVHLKLVLRFSDFGKAMFKPMRQQRDEETPVDFFYFIDFQRHNAEIAAFHLDRILDFRRVPPTVGRIVNVTKEILEVTKNEIL QSVFFVSPASNVCFFAKCPYMCKTEYAVCGNPHLLEGSLSAFLPSLNLAPRLSVPNPWIRSYTLAGKEEWEVNPLYCDTVKQIYPYNNSQ RLLNVIDMAIFDFLIGNMDRHHYEMFTKFGDDGFLIHLDNARGFGRHSHDEISILSPLSQCCMIKKKTLLHLQLLAQADYRLSDVMRESL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CDK12-FAM20A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CDK12-FAM20A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CDK12-FAM20A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
