
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CEACAM5-AP1G1 (FusionGDB2 ID:15505) |
Fusion Gene Summary for CEACAM5-AP1G1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CEACAM5-AP1G1 | Fusion gene ID: 15505 | Hgene | Tgene | Gene symbol | CEACAM5 | AP1G1 | Gene ID | 1048 | 164 |
| Gene name | CEA cell adhesion molecule 5 | adaptor related protein complex 1 subunit gamma 1 | |
| Synonyms | CD66e|CEA | ADTG|CLAPG1 | |
| Cytomap | 19q13.2 | 16q22.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | carcinoembryonic antigen-related cell adhesion molecule 5carcinoembryonic antigen related cell adhesion molecule 5meconium antigen 100 | AP-1 complex subunit gamma-1adapter-related protein complex 1 subunit gamma-1adaptor protein complex AP-1 subunit gamma-1adaptor related protein complex 1 gamma 1 subunitclathrin assembly protein complex 1 gamma large chainclathrin assembly protein c | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P06731 | O43747 | |
| Ensembl transtripts involved in fusion gene | ENST00000221992, ENST00000398599, ENST00000405816, | ENST00000564155, ENST00000570297, ENST00000299980, ENST00000393512, ENST00000423132, ENST00000433195, ENST00000569748, | |
| Fusion gene scores | * DoF score | 10 X 11 X 4=440 | 12 X 12 X 5=720 |
| # samples | 10 | 14 | |
| ** MAII score | log2(10/440*10)=-2.13750352374993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(14/720*10)=-2.36257007938471 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CEACAM5 [Title/Abstract] AND AP1G1 [Title/Abstract] AND fusion [Title/Abstract] | ||
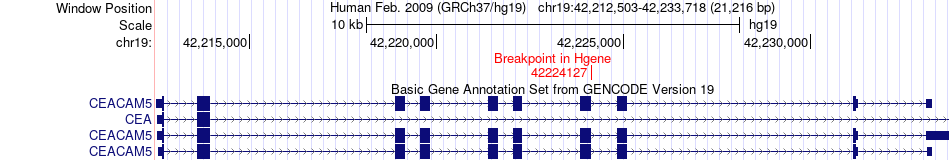
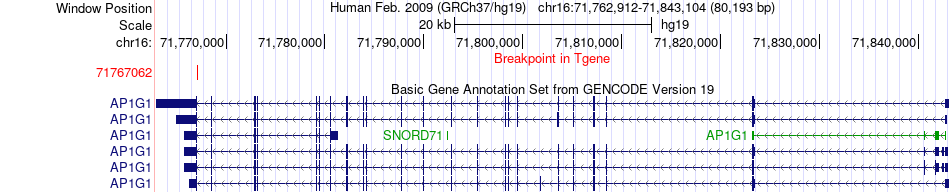
| Most frequent breakpoint | CEACAM5(42224127)-AP1G1(71767062), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CEACAM5-AP1G1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CEACAM5 | GO:0010832 | negative regulation of myotube differentiation | 8408226|10931872 |
| Hgene | CEACAM5 | GO:0034109 | homotypic cell-cell adhesion | 10864933 |
| Hgene | CEACAM5 | GO:0043066 | negative regulation of apoptotic process | 11448920 |
| Hgene | CEACAM5 | GO:2000811 | negative regulation of anoikis | 10910050 |
 Fusion gene breakpoints across CEACAM5 (5'-gene) Fusion gene breakpoints across CEACAM5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across AP1G1 (3'-gene) Fusion gene breakpoints across AP1G1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8683-01A | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
Top |
Fusion Gene ORF analysis for CEACAM5-AP1G1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000221992 | ENST00000564155 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| 5CDS-intron | ENST00000221992 | ENST00000570297 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| 5CDS-intron | ENST00000398599 | ENST00000564155 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| 5CDS-intron | ENST00000398599 | ENST00000570297 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| 5CDS-intron | ENST00000405816 | ENST00000564155 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| 5CDS-intron | ENST00000405816 | ENST00000570297 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000221992 | ENST00000299980 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000221992 | ENST00000393512 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000221992 | ENST00000423132 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000221992 | ENST00000433195 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000398599 | ENST00000299980 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000398599 | ENST00000393512 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000398599 | ENST00000423132 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000398599 | ENST00000433195 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000405816 | ENST00000299980 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000405816 | ENST00000393512 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000405816 | ENST00000423132 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| Frame-shift | ENST00000405816 | ENST00000433195 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| In-frame | ENST00000221992 | ENST00000569748 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| In-frame | ENST00000398599 | ENST00000569748 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
| In-frame | ENST00000405816 | ENST00000569748 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000398599 | CEACAM5 | chr19 | 42224127 | + | ENST00000569748 | AP1G1 | chr16 | 71767062 | - | 3192 | 1915 | 81 | 1949 | 622 |
| ENST00000221992 | CEACAM5 | chr19 | 42224127 | + | ENST00000569748 | AP1G1 | chr16 | 71767062 | - | 3162 | 1885 | 48 | 1919 | 623 |
| ENST00000405816 | CEACAM5 | chr19 | 42224127 | + | ENST00000569748 | AP1G1 | chr16 | 71767062 | - | 3131 | 1854 | 17 | 1888 | 623 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000398599 | ENST00000569748 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - | 0.002155877 | 0.9978441 |
| ENST00000221992 | ENST00000569748 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - | 0.002354644 | 0.9976453 |
| ENST00000405816 | ENST00000569748 | CEACAM5 | chr19 | 42224127 | + | AP1G1 | chr16 | 71767062 | - | 0.00221644 | 0.9977836 |
Top |
Fusion Genomic Features for CEACAM5-AP1G1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
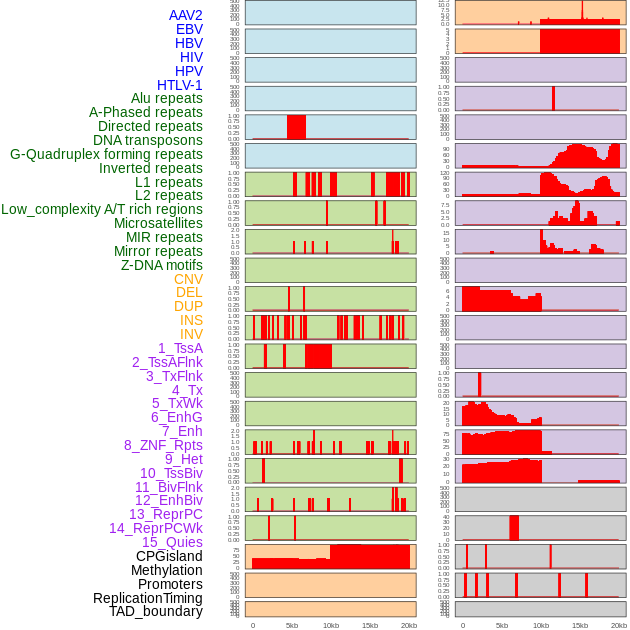 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CEACAM5-AP1G1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:42224127/chr16:71767062) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CEACAM5 | AP1G1 |
| FUNCTION: Cell surface glycoprotein that plays a role in cell adhesion, intracellular signaling and tumor progression (PubMed:2803308, PubMed:10910050, PubMed:10864933). Mediates homophilic and heterophilic cell adhesion with other carcinoembryonic antigen-related cell adhesion molecules, such as CEACAM6 (PubMed:2803308). Plays a role as an oncogene by promoting tumor progression; induces resistance to anoikis of colorectal carcinoma cells (PubMed:10910050). {ECO:0000269|PubMed:10864933, ECO:0000269|PubMed:10910050, ECO:0000269|PubMed:2803308}.; FUNCTION: (Microbial infection) Receptor for E.coli Dr adhesins. Binding of E.coli Dr adhesins leads to dissociation of the homodimer. {ECO:0000269|PubMed:18086185}. | FUNCTION: Subunit of clathrin-associated adaptor protein complex 1 that plays a role in protein sorting in the late-Golgi/trans-Golgi network (TGN) and/or endosomes. The AP complexes mediate both the recruitment of clathrin to membranes and the recognition of sorting signals within the cytosolic tails of transmembrane cargo molecules. In association with AFTPH/aftiphilin in the aftiphilin/p200/gamma-synergin complex, involved in the trafficking of transferrin from early to recycling endosomes, and the membrane trafficking of furin and the lysosomal enzyme cathepsin D between the trans-Golgi network (TGN) and endosomes (PubMed:15758025). {ECO:0000269|PubMed:15758025}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 145_232 | 590 | 848.6666666666666 | Domain | Ig-like C2-type 1 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 240_315 | 590 | 848.6666666666666 | Domain | Ig-like C2-type 2 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 323_410 | 590 | 848.6666666666666 | Domain | Ig-like C2-type 3 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 35_144 | 590 | 848.6666666666666 | Domain | Ig-like V-type |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 418_495 | 590 | 848.6666666666666 | Domain | Ig-like C2-type 4 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 501_588 | 590 | 848.6666666666666 | Domain | Ig-like C2-type 5 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 145_232 | 589 | 685.6666666666666 | Domain | Ig-like C2-type 1 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 240_315 | 589 | 685.6666666666666 | Domain | Ig-like C2-type 2 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 323_410 | 589 | 685.6666666666666 | Domain | Ig-like C2-type 3 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 35_144 | 589 | 685.6666666666666 | Domain | Ig-like V-type |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 418_495 | 589 | 685.6666666666666 | Domain | Ig-like C2-type 4 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 501_588 | 589 | 685.6666666666666 | Domain | Ig-like C2-type 5 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 145_232 | 590 | 696.0 | Domain | Ig-like C2-type 1 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 240_315 | 590 | 696.0 | Domain | Ig-like C2-type 2 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 323_410 | 590 | 696.0 | Domain | Ig-like C2-type 3 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 35_144 | 590 | 696.0 | Domain | Ig-like V-type |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 418_495 | 590 | 696.0 | Domain | Ig-like C2-type 4 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 501_588 | 590 | 696.0 | Domain | Ig-like C2-type 5 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000221992 | + | 7 | 10 | 593_675 | 590 | 848.6666666666666 | Domain | Ig-like C2-type 6 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000398599 | + | 7 | 10 | 593_675 | 589 | 685.6666666666666 | Domain | Ig-like C2-type 6 |
| Hgene | CEACAM5 | chr19:42224127 | chr16:71767062 | ENST00000405816 | + | 7 | 10 | 593_675 | 590 | 696.0 | Domain | Ig-like C2-type 6 |
| Tgene | AP1G1 | chr19:42224127 | chr16:71767062 | ENST00000299980 | 21 | 23 | 702_817 | 789 | 823.0 | Domain | GAE | |
| Tgene | AP1G1 | chr19:42224127 | chr16:71767062 | ENST00000393512 | 22 | 24 | 702_817 | 792 | 826.0 | Domain | GAE | |
| Tgene | AP1G1 | chr19:42224127 | chr16:71767062 | ENST00000569748 | 24 | 26 | 702_817 | 789 | 823.0 | Domain | GAE |
Top |
Fusion Gene Sequence for CEACAM5-AP1G1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >15505_15505_1_CEACAM5-AP1G1_CEACAM5_chr19_42224127_ENST00000221992_AP1G1_chr16_71767062_ENST00000569748_length(transcript)=3162nt_BP=1885nt CTCAGGGCAGAGGGAGGAAGGACAGCAGACCAGACAGTCACAGCAGCCTTGACAAAACGTTCCTGGAACTCAAGCTCTTCTCCACAGAGG AGGACAGAGCAGACAGCAGAGACCATGGAGTCTCCCTCGGCCCCTCCCCACAGATGGTGCATCCCCTGGCAGAGGCTCCTGCTCACAGCC TCACTTCTAACCTTCTGGAACCCGCCCACCACTGCCAAGCTCACTATTGAATCCACGCCGTTCAATGTCGCAGAGGGGAAGGAGGTGCTT CTACTTGTCCACAATCTGCCCCAGCATCTTTTTGGCTACAGCTGGTACAAAGGTGAAAGAGTGGATGGCAACCGTCAAATTATAGGATAT GTAATAGGAACTCAACAAGCTACCCCAGGGCCCGCATACAGTGGTCGAGAGATAATATACCCCAATGCATCCCTGCTGATCCAGAACATC ATCCAGAATGACACAGGATTCTACACCCTACACGTCATAAAGTCAGATCTTGTGAATGAAGAAGCAACTGGCCAGTTCCGGGTATACCCG GAGCTGCCCAAGCCCTCCATCTCCAGCAACAACTCCAAACCCGTGGAGGACAAGGATGCTGTGGCCTTCACCTGTGAACCTGAGACTCAG GACGCAACCTACCTGTGGTGGGTAAACAATCAGAGCCTCCCGGTCAGTCCCAGGCTGCAGCTGTCCAATGGCAACAGGACCCTCACTCTA TTCAATGTCACAAGAAATGACACAGCAAGCTACAAATGTGAAACCCAGAACCCAGTGAGTGCCAGGCGCAGTGATTCAGTCATCCTGAAT GTCCTCTATGGCCCGGATGCCCCCACCATTTCCCCTCTAAACACATCTTACAGATCAGGGGAAAATCTGAACCTCTCCTGCCACGCAGCC TCTAACCCACCTGCACAGTACTCTTGGTTTGTCAATGGGACTTTCCAGCAATCCACCCAAGAGCTCTTTATCCCCAACATCACTGTGAAT AATAGTGGATCCTATACGTGCCAAGCCCATAACTCAGACACTGGCCTCAATAGGACCACAGTCACGACGATCACAGTCTATGCAGAGCCA CCCAAACCCTTCATCACCAGCAACAACTCCAACCCCGTGGAGGATGAGGATGCTGTAGCCTTAACCTGTGAACCTGAGATTCAGAACACA ACCTACCTGTGGTGGGTAAATAATCAGAGCCTCCCGGTCAGTCCCAGGCTGCAGCTGTCCAATGACAACAGGACCCTCACTCTACTCAGT GTCACAAGGAATGATGTAGGACCCTATGAGTGTGGAATCCAGAACAAATTAAGTGTTGACCACAGCGACCCAGTCATCCTGAATGTCCTC TATGGCCCAGACGACCCCACCATTTCCCCCTCATACACCTATTACCGTCCAGGGGTGAACCTCAGCCTCTCCTGCCATGCAGCCTCTAAC CCACCTGCACAGTATTCTTGGCTGATTGATGGGAACATCCAGCAACACACACAAGAGCTCTTTATCTCCAACATCACTGAGAAGAACAGC GGACTCTATACCTGCCAGGCCAATAACTCAGCCAGTGGCCACAGCAGGACTACAGTCAAGACAATCACAGTCTCTGCGGAGCTGCCCAAG CCCTCCATCTCCAGCAACAACTCCAAACCCGTGGAGGACAAGGATGCTGTGGCCTTCACCTGTGAACCTGAGGCTCAGAACACAACCTAC CTGTGGTGGGTAAATGGTCAGAGCCTCCCAGTCAGTCCCAGGCTGCAGCTGTCCAATGGCAACAGGACCCTCACTCTATTCAATGTCACA AGAAATGACGCAAGAGCCTATGTATGTGGAATCCAGAACTCAGTGAGTGCAAACCGCAGTGACCCAGTCACCCTGGATGTCCTCTCAACA GCTGCGAATGCGGATCAAGCTTACATATAATCACAAGGGCTCAGCAATGCAAGATCTAGCAGAGGTGAACAACTTTCCCCCTCAGTCCTG GCAATGAGGGTTTGGCACCATTCTCATTCTTTATCCCACTCAATCAAAGGAACTCTGGGAAGGAGGTTGTGATTGCTGGCAAGTCCCCCC CAACTGTACCACGGGCATGAGGAGCTGAAGAGAACTGCTGAGGAGGATTTTCCTAAAGTTACTGCTGACCTTGAAGCATTGTTAAAGACT AATGTCCTCTCCTCCACTGTTGAGGCTGGCTGCTTCTGGAGGCTACTTTGCACTCTTCCTCTTCTCCTTTTTCCGCACTTCTCCACCCCT CCCACATTTACAGCCAGAATCAACATTCCCTGGGCCCCTGAGGAAATAAGCAGCTGGTCTGGAGGAGAGGACTGCAATCCATGGCGAAAA AACACTCACTTTGTCTCTGCAGCAAAGAGTTGCCCCTTCTTTCTACTGTTGTTTCTCTGTGGACTGGGCAAGGTGGGGTATTTATTCCTC ACTAGCTGGGTTACCATCTTCAGGCACTTTTAACATCTGGCATTCGGAATGGAAATGTAATAATGGACATTAGGGAGCCCTGCCTTTTTC TACTGGTTCCCCCAATGTTTGAAAGAGGCATTAGGCTCCTGGTAGCCTTTTCTGTGCATTGCTGTATACACACAGACACACACATGTATG TTTGTTACCAAGAACTGGTCAGACCTTGCGAGTTTATTTGTAAACACTGGACAGATGGAGTTAAAAAGAGCTTTTGTTGAGATTTGGCAT GAAGGATATGGTGCTCTATTTGTAATAGAAACTTCCAAGGCTCTTCCAGCTCCCCTTTCTCGCCATTCTTTAGCTGTAGTCATGAATAGT CTCCATGATTTTCAAAATTGATTCCCTTTAAAGTGCAAAATGGTCACCTTCTAAAAGATATATTCATAGTTATTAATGACCCTATTCCCA CCACAAATTTTAAAGTGCTCCTAAGCCCATAACTTGCCTGTTTGAACTATGGTAATGGGTGGAAGAGGAGTTCACCAGTTTCAAAGATCA GACTCTGTATCAAAAGTACCTTTGCCCTTAGGAAGAGTGAGTATTGGAGTCATCTTATCTATTACTCCAAACCTCCCTTTTTATTTCTTG AGCCTGGCTTGGACCTTGGCATTCCGTTTGAATTCCTTCTAACTGGAACATTTGTGTTGTATCTGTAACACTGGCACTGAAATAAAGACC >15505_15505_1_CEACAM5-AP1G1_CEACAM5_chr19_42224127_ENST00000221992_AP1G1_chr16_71767062_ENST00000569748_length(amino acids)=623AA_BP= MTKRSWNSSSSPQRRTEQTAETMESPSAPPHRWCIPWQRLLLTASLLTFWNPPTTAKLTIESTPFNVAEGKEVLLLVHNLPQHLFGYSWY KGERVDGNRQIIGYVIGTQQATPGPAYSGREIIYPNASLLIQNIIQNDTGFYTLHVIKSDLVNEEATGQFRVYPELPKPSISSNNSKPVE DKDAVAFTCEPETQDATYLWWVNNQSLPVSPRLQLSNGNRTLTLFNVTRNDTASYKCETQNPVSARRSDSVILNVLYGPDAPTISPLNTS YRSGENLNLSCHAASNPPAQYSWFVNGTFQQSTQELFIPNITVNNSGSYTCQAHNSDTGLNRTTVTTITVYAEPPKPFITSNNSNPVEDE DAVALTCEPEIQNTTYLWWVNNQSLPVSPRLQLSNDNRTLTLLSVTRNDVGPYECGIQNKLSVDHSDPVILNVLYGPDDPTISPSYTYYR PGVNLSLSCHAASNPPAQYSWLIDGNIQQHTQELFISNITEKNSGLYTCQANNSASGHSRTTVKTITVSAELPKPSISSNNSKPVEDKDA -------------------------------------------------------------- >15505_15505_2_CEACAM5-AP1G1_CEACAM5_chr19_42224127_ENST00000398599_AP1G1_chr16_71767062_ENST00000569748_length(transcript)=3192nt_BP=1915nt GATGCTGAGAAGTACTCCTGCCCTAGGAAGAGACTCAGGGCAGAGGGAGGAAGGACAGCAGACCAGACAGTCACAGCAGCCTTGACAAAA CGTTCCTGGAACTCAAGCTCTTCTCCACAGAGGAGGACAGAGCAGACAGCAGAGACCATGGAGTCTCCCTCGGCCCCTCCCCACAGATGG TGCATCCCCTGGCAGAGGCTCCTGCTCACAGCCTCACTTCTAACCTTCTGGAACCCGCCCACCACTGCCAAGCTCACTATTGAATCCACG CCGTTCAATGTCGCAGAGGGGAAGGAGGTGCTTCTACTTGTCCACAATCTGCCCCAGCATCTTTTTGGCTACAGCTGGTACAAAGGTGAA AGAGTGGATGGCAACCGTCAAATTATAGGATATGTAATAGGAACTCAACAAGCTACCCCAGGGCCCGCATACAGTGGTCGAGAGATAATA TACCCCAATGCATCCCTGCTGATCCAGAACATCATCCAGAATGACACAGGATTCTACACCCTACACGTCATAAAGTCAGATCTTGTGAAT GAAGAAGCAACTGGCCAGTTCCGGGTATACCCGGAGCTGCCCAAGCCCTCCATCTCCAGCAACAACTCCAAACCCGTGGAGGACAAGGAT GCTGTGGCCTTCACCTGTGAACCTGAGACTCAGGACGCAACCTACCTGTGGTGGGTAAACAATCAGAGCCTCCCGGTCAGTCCCAGGCTG CAGCTGTCCAATGGCAACAGGACCCTCACTCTATTCAATGTCACAAGAAATGACACAGCAAGCTACAAATGTGAAACCCAGAACCCAGTG AGTGCCAGGCGCAGTGATTCAGTCATCCTGAATGTCCTCTATGGCCCGGATGCCCCCACCATTTCCCCTCTAAACACATCTTACAGATCA GGGGAAAATCTGAACCTCTCCTGCCACGCAGCCTCTAACCCACCTGCACAGTACTCTTGGTTTGTCAATGGGACTTTCCAGCAATCCACC CAAGAGCTCTTTATCCCCAACATCACTGTGAATAATAGTGGATCCTATACGTGCCAAGCCCATAACTCAGACACTGGCCTCAATAGGACC ACAGTCACGACGATCACAGTCTATGAGCCACCCAAACCCTTCATCACCAGCAACAACTCCAACCCCGTGGAGGATGAGGATGCTGTAGCC TTAACCTGTGAACCTGAGATTCAGAACACAACCTACCTGTGGTGGGTAAATAATCAGAGCCTCCCGGTCAGTCCCAGGCTGCAGCTGTCC AATGACAACAGGACCCTCACTCTACTCAGTGTCACAAGGAATGATGTAGGACCCTATGAGTGTGGAATCCAGAACAAATTAAGTGTTGAC CACAGCGACCCAGTCATCCTGAATGTCCTCTATGGCCCAGACGACCCCACCATTTCCCCCTCATACACCTATTACCGTCCAGGGGTGAAC CTCAGCCTCTCCTGCCATGCAGCCTCTAACCCACCTGCACAGTATTCTTGGCTGATTGATGGGAACATCCAGCAACACACACAAGAGCTC TTTATCTCCAACATCACTGAGAAGAACAGCGGACTCTATACCTGCCAGGCCAATAACTCAGCCAGTGGCCACAGCAGGACTACAGTCAAG ACAATCACAGTCTCTGCGGAGCTGCCCAAGCCCTCCATCTCCAGCAACAACTCCAAACCCGTGGAGGACAAGGATGCTGTGGCCTTCACC TGTGAACCTGAGGCTCAGAACACAACCTACCTGTGGTGGGTAAATGGTCAGAGCCTCCCAGTCAGTCCCAGGCTGCAGCTGTCCAATGGC AACAGGACCCTCACTCTATTCAATGTCACAAGAAATGACGCAAGAGCCTATGTATGTGGAATCCAGAACTCAGTGAGTGCAAACCGCAGT GACCCAGTCACCCTGGATGTCCTCTCAACAGCTGCGAATGCGGATCAAGCTTACATATAATCACAAGGGCTCAGCAATGCAAGATCTAGC AGAGGTGAACAACTTTCCCCCTCAGTCCTGGCAATGAGGGTTTGGCACCATTCTCATTCTTTATCCCACTCAATCAAAGGAACTCTGGGA AGGAGGTTGTGATTGCTGGCAAGTCCCCCCCAACTGTACCACGGGCATGAGGAGCTGAAGAGAACTGCTGAGGAGGATTTTCCTAAAGTT ACTGCTGACCTTGAAGCATTGTTAAAGACTAATGTCCTCTCCTCCACTGTTGAGGCTGGCTGCTTCTGGAGGCTACTTTGCACTCTTCCT CTTCTCCTTTTTCCGCACTTCTCCACCCCTCCCACATTTACAGCCAGAATCAACATTCCCTGGGCCCCTGAGGAAATAAGCAGCTGGTCT GGAGGAGAGGACTGCAATCCATGGCGAAAAAACACTCACTTTGTCTCTGCAGCAAAGAGTTGCCCCTTCTTTCTACTGTTGTTTCTCTGT GGACTGGGCAAGGTGGGGTATTTATTCCTCACTAGCTGGGTTACCATCTTCAGGCACTTTTAACATCTGGCATTCGGAATGGAAATGTAA TAATGGACATTAGGGAGCCCTGCCTTTTTCTACTGGTTCCCCCAATGTTTGAAAGAGGCATTAGGCTCCTGGTAGCCTTTTCTGTGCATT GCTGTATACACACAGACACACACATGTATGTTTGTTACCAAGAACTGGTCAGACCTTGCGAGTTTATTTGTAAACACTGGACAGATGGAG TTAAAAAGAGCTTTTGTTGAGATTTGGCATGAAGGATATGGTGCTCTATTTGTAATAGAAACTTCCAAGGCTCTTCCAGCTCCCCTTTCT CGCCATTCTTTAGCTGTAGTCATGAATAGTCTCCATGATTTTCAAAATTGATTCCCTTTAAAGTGCAAAATGGTCACCTTCTAAAAGATA TATTCATAGTTATTAATGACCCTATTCCCACCACAAATTTTAAAGTGCTCCTAAGCCCATAACTTGCCTGTTTGAACTATGGTAATGGGT GGAAGAGGAGTTCACCAGTTTCAAAGATCAGACTCTGTATCAAAAGTACCTTTGCCCTTAGGAAGAGTGAGTATTGGAGTCATCTTATCT ATTACTCCAAACCTCCCTTTTTATTTCTTGAGCCTGGCTTGGACCTTGGCATTCCGTTTGAATTCCTTCTAACTGGAACATTTGTGTTGT >15505_15505_2_CEACAM5-AP1G1_CEACAM5_chr19_42224127_ENST00000398599_AP1G1_chr16_71767062_ENST00000569748_length(amino acids)=622AA_BP= MTKRSWNSSSSPQRRTEQTAETMESPSAPPHRWCIPWQRLLLTASLLTFWNPPTTAKLTIESTPFNVAEGKEVLLLVHNLPQHLFGYSWY KGERVDGNRQIIGYVIGTQQATPGPAYSGREIIYPNASLLIQNIIQNDTGFYTLHVIKSDLVNEEATGQFRVYPELPKPSISSNNSKPVE DKDAVAFTCEPETQDATYLWWVNNQSLPVSPRLQLSNGNRTLTLFNVTRNDTASYKCETQNPVSARRSDSVILNVLYGPDAPTISPLNTS YRSGENLNLSCHAASNPPAQYSWFVNGTFQQSTQELFIPNITVNNSGSYTCQAHNSDTGLNRTTVTTITVYEPPKPFITSNNSNPVEDED AVALTCEPEIQNTTYLWWVNNQSLPVSPRLQLSNDNRTLTLLSVTRNDVGPYECGIQNKLSVDHSDPVILNVLYGPDDPTISPSYTYYRP GVNLSLSCHAASNPPAQYSWLIDGNIQQHTQELFISNITEKNSGLYTCQANNSASGHSRTTVKTITVSAELPKPSISSNNSKPVEDKDAV -------------------------------------------------------------- >15505_15505_3_CEACAM5-AP1G1_CEACAM5_chr19_42224127_ENST00000405816_AP1G1_chr16_71767062_ENST00000569748_length(transcript)=3131nt_BP=1854nt AGACAGTCACAGCAGCCTTGACAAAACGTTCCTGGAACTCAAGCTCTTCTCCACAGAGGAGGACAGAGCAGACAGCAGAGACCATGGAGT CTCCCTCGGCCCCTCCCCACAGATGGTGCATCCCCTGGCAGAGGCTCCTGCTCACAGCCTCACTTCTAACCTTCTGGAACCCGCCCACCA CTGCCAAGCTCACTATTGAATCCACGCCGTTCAATGTCGCAGAGGGGAAGGAGGTGCTTCTACTTGTCCACAATCTGCCCCAGCATCTTT TTGGCTACAGCTGGTACAAAGGTGAAAGAGTGGATGGCAACCGTCAAATTATAGGATATGTAATAGGAACTCAACAAGCTACCCCAGGGC CCGCATACAGTGGTCGAGAGATAATATACCCCAATGCATCCCTGCTGATCCAGAACATCATCCAGAATGACACAGGATTCTACACCCTAC ACGTCATAAAGTCAGATCTTGTGAATGAAGAAGCAACTGGCCAGTTCCGGGTATACCCGGAGCTGCCCAAGCCCTCCATCTCCAGCAACA ACTCCAAACCCGTGGAGGACAAGGATGCTGTGGCCTTCACCTGTGAACCTGAGACTCAGGACGCAACCTACCTGTGGTGGGTAAACAATC AGAGCCTCCCGGTCAGTCCCAGGCTGCAGCTGTCCAATGGCAACAGGACCCTCACTCTATTCAATGTCACAAGAAATGACACAGCAAGCT ACAAATGTGAAACCCAGAACCCAGTGAGTGCCAGGCGCAGTGATTCAGTCATCCTGAATGTCCTCTATGGCCCGGATGCCCCCACCATTT CCCCTCTAAACACATCTTACAGATCAGGGGAAAATCTGAACCTCTCCTGCCACGCAGCCTCTAACCCACCTGCACAGTACTCTTGGTTTG TCAATGGGACTTTCCAGCAATCCACCCAAGAGCTCTTTATCCCCAACATCACTGTGAATAATAGTGGATCCTATACGTGCCAAGCCCATA ACTCAGACACTGGCCTCAATAGGACCACAGTCACGACGATCACAGTCTATGCAGAGCCACCCAAACCCTTCATCACCAGCAACAACTCCA ACCCCGTGGAGGATGAGGATGCTGTAGCCTTAACCTGTGAACCTGAGATTCAGAACACAACCTACCTGTGGTGGGTAAATAATCAGAGCC TCCCGGTCAGTCCCAGGCTGCAGCTGTCCAATGACAACAGGACCCTCACTCTACTCAGTGTCACAAGGAATGATGTAGGACCCTATGAGT GTGGAATCCAGAACAAATTAAGTGTTGACCACAGCGACCCAGTCATCCTGAATGTCCTCTATGGCCCAGACGACCCCACCATTTCCCCCT CATACACCTATTACCGTCCAGGGGTGAACCTCAGCCTCTCCTGCCATGCAGCCTCTAACCCACCTGCACAGTATTCTTGGCTGATTGATG GGAACATCCAGCAACACACACAAGAGCTCTTTATCTCCAACATCACTGAGAAGAACAGCGGACTCTATACCTGCCAGGCCAATAACTCAG CCAGTGGCCACAGCAGGACTACAGTCAAGACAATCACAGTCTCTGCGGAGCTGCCCAAGCCCTCCATCTCCAGCAACAACTCCAAACCCG TGGAGGACAAGGATGCTGTGGCCTTCACCTGTGAACCTGAGGCTCAGAACACAACCTACCTGTGGTGGGTAAATGGTCAGAGCCTCCCAG TCAGTCCCAGGCTGCAGCTGTCCAATGGCAACAGGACCCTCACTCTATTCAATGTCACAAGAAATGACGCAAGAGCCTATGTATGTGGAA TCCAGAACTCAGTGAGTGCAAACCGCAGTGACCCAGTCACCCTGGATGTCCTCTCAACAGCTGCGAATGCGGATCAAGCTTACATATAAT CACAAGGGCTCAGCAATGCAAGATCTAGCAGAGGTGAACAACTTTCCCCCTCAGTCCTGGCAATGAGGGTTTGGCACCATTCTCATTCTT TATCCCACTCAATCAAAGGAACTCTGGGAAGGAGGTTGTGATTGCTGGCAAGTCCCCCCCAACTGTACCACGGGCATGAGGAGCTGAAGA GAACTGCTGAGGAGGATTTTCCTAAAGTTACTGCTGACCTTGAAGCATTGTTAAAGACTAATGTCCTCTCCTCCACTGTTGAGGCTGGCT GCTTCTGGAGGCTACTTTGCACTCTTCCTCTTCTCCTTTTTCCGCACTTCTCCACCCCTCCCACATTTACAGCCAGAATCAACATTCCCT GGGCCCCTGAGGAAATAAGCAGCTGGTCTGGAGGAGAGGACTGCAATCCATGGCGAAAAAACACTCACTTTGTCTCTGCAGCAAAGAGTT GCCCCTTCTTTCTACTGTTGTTTCTCTGTGGACTGGGCAAGGTGGGGTATTTATTCCTCACTAGCTGGGTTACCATCTTCAGGCACTTTT AACATCTGGCATTCGGAATGGAAATGTAATAATGGACATTAGGGAGCCCTGCCTTTTTCTACTGGTTCCCCCAATGTTTGAAAGAGGCAT TAGGCTCCTGGTAGCCTTTTCTGTGCATTGCTGTATACACACAGACACACACATGTATGTTTGTTACCAAGAACTGGTCAGACCTTGCGA GTTTATTTGTAAACACTGGACAGATGGAGTTAAAAAGAGCTTTTGTTGAGATTTGGCATGAAGGATATGGTGCTCTATTTGTAATAGAAA CTTCCAAGGCTCTTCCAGCTCCCCTTTCTCGCCATTCTTTAGCTGTAGTCATGAATAGTCTCCATGATTTTCAAAATTGATTCCCTTTAA AGTGCAAAATGGTCACCTTCTAAAAGATATATTCATAGTTATTAATGACCCTATTCCCACCACAAATTTTAAAGTGCTCCTAAGCCCATA ACTTGCCTGTTTGAACTATGGTAATGGGTGGAAGAGGAGTTCACCAGTTTCAAAGATCAGACTCTGTATCAAAAGTACCTTTGCCCTTAG GAAGAGTGAGTATTGGAGTCATCTTATCTATTACTCCAAACCTCCCTTTTTATTTCTTGAGCCTGGCTTGGACCTTGGCATTCCGTTTGA >15505_15505_3_CEACAM5-AP1G1_CEACAM5_chr19_42224127_ENST00000405816_AP1G1_chr16_71767062_ENST00000569748_length(amino acids)=623AA_BP= MTKRSWNSSSSPQRRTEQTAETMESPSAPPHRWCIPWQRLLLTASLLTFWNPPTTAKLTIESTPFNVAEGKEVLLLVHNLPQHLFGYSWY KGERVDGNRQIIGYVIGTQQATPGPAYSGREIIYPNASLLIQNIIQNDTGFYTLHVIKSDLVNEEATGQFRVYPELPKPSISSNNSKPVE DKDAVAFTCEPETQDATYLWWVNNQSLPVSPRLQLSNGNRTLTLFNVTRNDTASYKCETQNPVSARRSDSVILNVLYGPDAPTISPLNTS YRSGENLNLSCHAASNPPAQYSWFVNGTFQQSTQELFIPNITVNNSGSYTCQAHNSDTGLNRTTVTTITVYAEPPKPFITSNNSNPVEDE DAVALTCEPEIQNTTYLWWVNNQSLPVSPRLQLSNDNRTLTLLSVTRNDVGPYECGIQNKLSVDHSDPVILNVLYGPDDPTISPSYTYYR PGVNLSLSCHAASNPPAQYSWLIDGNIQQHTQELFISNITEKNSGLYTCQANNSASGHSRTTVKTITVSAELPKPSISSNNSKPVEDKDA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CEACAM5-AP1G1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CEACAM5-AP1G1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CEACAM5-AP1G1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
