
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CELSR1-BTAF1 (FusionGDB2 ID:15640) |
Fusion Gene Summary for CELSR1-BTAF1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CELSR1-BTAF1 | Fusion gene ID: 15640 | Hgene | Tgene | Gene symbol | CELSR1 | BTAF1 | Gene ID | 9620 | 9044 |
| Gene name | cadherin EGF LAG seven-pass G-type receptor 1 | B-TFIID TATA-box binding protein associated factor 1 | |
| Synonyms | ADGRC1|CDHF9|FMI2|HFMI2|ME2 | MOT1|TAF(II)170|TAF172|TAFII170 | |
| Cytomap | 22q13.31 | 10q23.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cadherin EGF LAG seven-pass G-type receptor 1adhesion G protein-coupled receptor C1cadherin family member 9cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)flamingo homolog 2protocadherin flamingo 2 | TATA-binding protein-associated factor 172ATP-dependent helicase BTAF1B-TFIID transcription factor-associated 170 kDa subunitBTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae)Mot1 homologTBP-associa | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | Q9NYQ6 | O14981 | |
| Ensembl transtripts involved in fusion gene | ENST00000262738, ENST00000395964, ENST00000497509, | ENST00000471217, ENST00000544642, ENST00000265990, | |
| Fusion gene scores | * DoF score | 23 X 19 X 11=4807 | 3 X 3 X 2=18 |
| # samples | 28 | 3 | |
| ** MAII score | log2(28/4807*10)=-4.10163807119293 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: CELSR1 [Title/Abstract] AND BTAF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CELSR1(46829290)-BTAF1(93767880), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BTAF1 | GO:0045892 | negative regulation of transcription, DNA-templated | 9488487 |
 Fusion gene breakpoints across CELSR1 (5'-gene) Fusion gene breakpoints across CELSR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
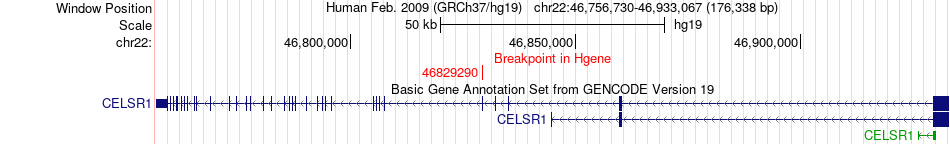 |
 Fusion gene breakpoints across BTAF1 (3'-gene) Fusion gene breakpoints across BTAF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
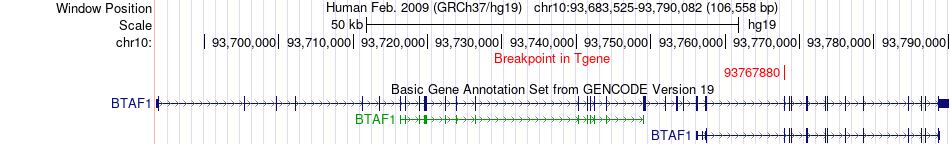 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-A4IY-01A | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
Top |
Fusion Gene ORF analysis for CELSR1-BTAF1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000262738 | ENST00000471217 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| 5CDS-intron | ENST00000262738 | ENST00000544642 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| In-frame | ENST00000262738 | ENST00000265990 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| intron-3CDS | ENST00000395964 | ENST00000265990 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| intron-3CDS | ENST00000497509 | ENST00000265990 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| intron-intron | ENST00000395964 | ENST00000471217 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| intron-intron | ENST00000395964 | ENST00000544642 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| intron-intron | ENST00000497509 | ENST00000471217 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
| intron-intron | ENST00000497509 | ENST00000544642 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262738 | CELSR1 | chr22 | 46829290 | - | ENST00000265990 | BTAF1 | chr10 | 93767880 | + | 7893 | 4611 | 0 | 6500 | 2166 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262738 | ENST00000265990 | CELSR1 | chr22 | 46829290 | - | BTAF1 | chr10 | 93767880 | + | 0.000884119 | 0.9991159 |
Top |
Fusion Genomic Features for CELSR1-BTAF1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CELSR1 | chr22 | 46829289 | - | BTAF1 | chr10 | 93767879 | + | 0.006734146 | 0.99326587 |
| CELSR1 | chr22 | 46829289 | - | BTAF1 | chr10 | 93767879 | + | 0.006734146 | 0.99326587 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
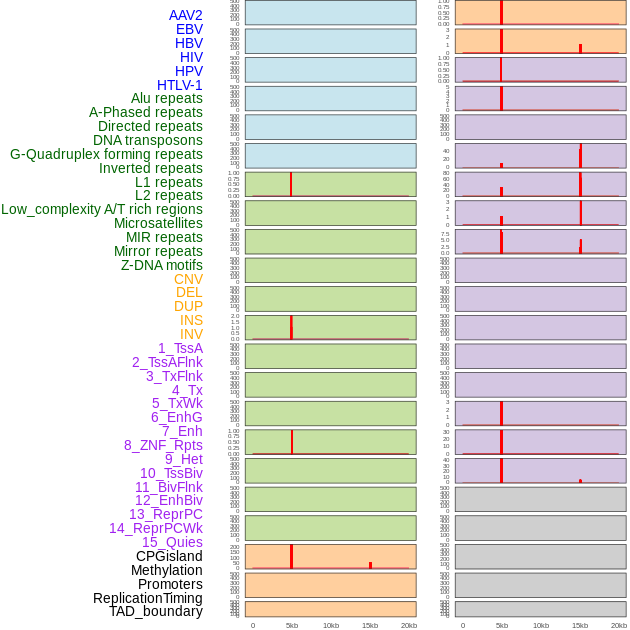 |
Top |
Fusion Protein Features for CELSR1-BTAF1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:46829290/chr10:93767880) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CELSR1 | BTAF1 |
| FUNCTION: Receptor that may have an important role in cell/cell signaling during nervous system formation. | FUNCTION: Regulates transcription in association with TATA binding protein (TBP). Removes TBP from the TATA box in an ATP-dependent manner. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1000_1101 | 1537 | 3015.0 | Domain | Cadherin 8 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1106_1224 | 1537 | 3015.0 | Domain | Cadherin 9 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1303_1361 | 1537 | 3015.0 | Domain | EGF-like 1%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1363_1399 | 1537 | 3015.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1403_1441 | 1537 | 3015.0 | Domain | EGF-like 3%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 246_353 | 1537 | 3015.0 | Domain | Cadherin 1 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 354_459 | 1537 | 3015.0 | Domain | Cadherin 2 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 460_565 | 1537 | 3015.0 | Domain | Cadherin 3 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 566_687 | 1537 | 3015.0 | Domain | Cadherin 4 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 688_789 | 1537 | 3015.0 | Domain | Cadherin 5 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 790_892 | 1537 | 3015.0 | Domain | Cadherin 6 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 893_999 | 1537 | 3015.0 | Domain | Cadherin 7 |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 1278_1453 | 1220 | 1850.0 | Domain | Helicase ATP-binding | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 1636_1790 | 1220 | 1850.0 | Domain | Helicase C-terminal | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 1404_1407 | 1220 | 1850.0 | Motif | Note=DEGH box | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 1291_1298 | 1220 | 1850.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2659_2663 | 1537 | 3015.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1442_1646 | 1537 | 3015.0 | Domain | Laminin G-like 1 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1649_1685 | 1537 | 3015.0 | Domain | EGF-like 4%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1689_1870 | 1537 | 3015.0 | Domain | Laminin G-like 2 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1872_1907 | 1537 | 3015.0 | Domain | EGF-like 5%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1908_1946 | 1537 | 3015.0 | Domain | EGF-like 6%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1947_1979 | 1537 | 3015.0 | Domain | EGF-like 7%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 1981_2016 | 1537 | 3015.0 | Domain | EGF-like 8%3B calcium-binding |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2003_2050 | 1537 | 3015.0 | Domain | Laminin EGF-like |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2408_2460 | 1537 | 3015.0 | Domain | GPS |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 22_2469 | 1537 | 3015.0 | Topological domain | Extracellular |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2491_2501 | 1537 | 3015.0 | Topological domain | Cytoplasmic |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2523_2527 | 1537 | 3015.0 | Topological domain | Extracellular |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2549_2572 | 1537 | 3015.0 | Topological domain | Cytoplasmic |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2594_2611 | 1537 | 3015.0 | Topological domain | Extracellular |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2633_2655 | 1537 | 3015.0 | Topological domain | Cytoplasmic |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2677_2683 | 1537 | 3015.0 | Topological domain | Extracellular |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2705_3014 | 1537 | 3015.0 | Topological domain | Cytoplasmic |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2470_2490 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2502_2522 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2528_2548 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2573_2593 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2612_2632 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2656_2676 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | CELSR1 | chr22:46829290 | chr10:93767880 | ENST00000262738 | - | 5 | 35 | 2684_2704 | 1537 | 3015.0 | Transmembrane | Helical%3B Name%3D7 |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 191_207 | 1220 | 1850.0 | Motif | Nuclear localization signal | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 1102_1139 | 1220 | 1850.0 | Repeat | Note=HEAT 7 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 1182_1219 | 1220 | 1850.0 | Repeat | Note=HEAT 8 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 385_422 | 1220 | 1850.0 | Repeat | Note=HEAT 1 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 426_463 | 1220 | 1850.0 | Repeat | Note=HEAT 2 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 513_550 | 1220 | 1850.0 | Repeat | Note=HEAT 3 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 554_596 | 1220 | 1850.0 | Repeat | Note=HEAT 4 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 818_855 | 1220 | 1850.0 | Repeat | Note=HEAT 5 | |
| Tgene | BTAF1 | chr22:46829290 | chr10:93767880 | ENST00000265990 | 24 | 38 | 872_910 | 1220 | 1850.0 | Repeat | Note=HEAT 6 |
Top |
Fusion Gene Sequence for CELSR1-BTAF1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >15640_15640_1_CELSR1-BTAF1_CELSR1_chr22_46829290_ENST00000262738_BTAF1_chr10_93767880_ENST00000265990_length(transcript)=7893nt_BP=4611nt ATGGCGCCGCCGCCGCCGCCCGTGCTGCCCGTGCTGCTGCTCCTGGCCGCCGCCGCCGCCCTGCCGGCGATGGGGCTGCGAGCGGCCGCC TGGGAGCCGCGCGTACCCGGCGGGACCCGCGCCTTCGCCCTCCGGCCCGGCTGTACCTACGCGGTGGGCGCCGCTTGCACGCCCCGGGCG CCGCGGGAGCTGCTGGACGTGGGCCGCGATGGGCGGCTGGCAGGACGTCGGCGCGTCTCGGGCGCGGGGCGCCCGCTGCCGCTGCAAGTC CGCTTGGTGGCCCGCAGTGCCCCGACGGCGCTGAGCCGCCGCCTGCGGGCGCGCACGCACCTTCCCGGCTGCGGAGCCCGTGCCCGGCTC TGCGGAACCGGTGCCCGGCTCTGCGGGGCGCTCTGCTTCCCCGTCCCCGGCGGCTGCGCGGCCGCGCAGCATTCGGCGCTCGCAGCTCCG ACCACCTTACCCGCCTGCCGCTGCCCGCCGCGCCCCAGGCCCCGCTGTCCCGGCCGTCCCATCTGCCTGCCGCCGGGCGGCTCGGTCCGC CTGCGTCTGCTGTGCGCCCTGCGGCGCGCGGCTGGCGCCGTCCGGGTGGGACTGGCGCTGGAGGCCGCCACCGCGGGGACGCCCTCCGCG TCGCCATCCCCATCGCCGCCCCTGCCGCCGAACTTGCCCGAAGCCCGGGCGGGGCCGGCGCGACGGGCCCGGCGGGGCACGAGCGGCAGA GGGAGCCTGAAGTTTCCGATGCCCAACTACCAGGTGGCGTTGTTTGAGAACGAACCGGCGGGCACCCTCATCCTCCAGCTGCACGCGCAC TACACCATCGAGGGCGAGGAGGAGCGCGTGAGCTATTACATGGAGGGGCTGTTCGACGAGCGCTCCCGGGGCTACTTCCGAATCGACTCT GCCACGGGCGCCGTGAGCACGGACAGCGTACTGGACCGCGAGACCAAGGAGACGCACGTCCTCAGGGTGAAAGCCGTGGACTACAGTACG CCGCCGCGCTCGGCCACCACCTACATCACTGTCTTGGTCAAAGACACCAACGACCACAGCCCGGTCTTCGAGCAGTCGGAGTACCGCGAG CGCGTGCGGGAGAACCTGGAGGTGGGCTACGAGGTGCTGACCATCCGCGCCAGCGACCGCGACTCGCCCATCAACGCCAACTTGCGTTAC CGCGTGTTGGGGGGCGCGTGGGACGTCTTCCAGCTCAACGAGAGCTCTGGCGTGGTGAGCACACGGGCGGTGCTGGACCGGGAGGAGGCG GCCGAGTACCAGCTCCTGGTGGAGGCCAACGACCAGGGGCGCAATCCGGGCCCGCTCAGTGCCACGGCCACCGTGTACATCGAGGTGGAG GACGAGAACGACAACTACCCCCAGTTCAGCGAGCAGAACTACGTGGTCCAGGTGCCCGAGGACGTGGGGCTCAACACGGCTGTGCTGCGA GTGCAGGCCACGGACCGGGACCAGGGCCAGAACGCGGCCATTCACTACAGCATCCTCAGCGGGAACGTGGCCGGCCAGTTCTACCTGCAC TCGCTGAGCGGGATCCTGGATGTGATCAACCCCTTGGATTTCGAGGATGTCCAGAAATACTCGCTGAGCATTAAGGCCCAGGATGGGGGC CGGCCCCCGCTCATCAATTCTTCAGGGGTGGTGTCTGTGCAGGTGCTGGATGTCAACGACAACGAGCCTATCTTTGTGAGCAGCCCCTTC CAGGCCACGGTGCTGGAGAATGTGCCCCTGGGCTACCCCGTGGTGCACATTCAGGCGGTGGACGCGGACTCTGGAGAGAACGCCCGGCTG CACTATCGCCTGGTGGACACGGCCTCCACCTTTCTGGGGGGCGGCAGCGCTGGGCCTAAGAATCCTGCCCCCACCCCTGACTTCCCCTTC CAGATCCACAACAGCTCCGGTTGGATCACAGTGTGTGCCGAGCTGGACCGCGAGGAGGTGGAGCACTACAGCTTCGGGGTGGAGGCGGTG GACCACGGCTCGCCCCCCATGAGCTCCTCCACCAGCGTGTCCATCACGGTGCTGGACGTGAATGACAACGACCCGGTGTTCACGCAGCCC ACCTACGAGCTTCGTCTGAATGAGGATGCGGCCGTGGGGAGCAGCGTGCTGACCCTGCAGGCCCGCGACCGTGACGCCAACAGTGTGATT ACCTACCAGCTCACAGGCGGCAACACCCGGAACCGCTTTGCACTCAGCAGCCAGAGAGGGGGCGGCCTCATCACCCTGGCGCTACCTCTG GACTACAAGCAGGAGCAGCAGTACGTGCTGGCGGTGACAGCATCCGACGGCACACGGTCGCACACTGCGCATGTCCTAATCAACGTCACT GATGCCAACACCCACAGGCCTGTCTTTCAGAGCTCCCATTACACAGTGAGTGTCAGTGAGGACAGGCCTGTGGGCACCTCCATTGCTACC CTCAGTGCCAACGATGAGGACACAGGAGAGAATGCCCGCATCACCTACGTGATTCAGGACCCCGTGCCGCAGTTCCGCATTGACCCCGAC AGTGGCACCATGTACACCATGATGGAGCTGGACTATGAGAACCAGGTCGCCTACACGCTGACCATCATGGCCCAGGACAACGGCATCCCG CAGAAATCAGACACCACCACCCTAGAGATCCTCATCCTCGATGCCAATGACAATGCACCCCAGTTCCTGTGGGATTTCTACCAGGGTTCC ATCTTTGAGGATGCTCCACCCTCGACCAGCATCCTCCAGGTCTCTGCCACGGACCGGGACTCAGGTCCCAATGGGCGTCTGCTGTACACC TTCCAGGGTGGGGACGACGGCGATGGGGACTTCTACATCGAGCCCACGTCCGGTGTGATTCGCACCCAGCGCCGGCTGGACCGGGAGAAT GTGGCCGTGTACAACCTTTGGGCTCTGGCTGTGGATCGGGGCAGTCCCACTCCCCTTAGCGCCTCGGTAGAAATCCAGGTGACCATCTTG GACATTAATGACAATGCCCCCATGTTTGAGAAGGACGAACTGGAGCTGTTTGTTGAGGAGAACAACCCAGTGGGGTCGGTGGTGGCAAAG ATTCGTGCTAACGACCCTGATGAAGGCCCTAATGCCCAGATCATGTATCAGATTGTGGAAGGGGACATGCGGCATTTCTTCCAGCTGGAC CTGCTCAACGGGGACCTGCGTGCCATGGTGGAGCTGGACTTTGAGGTCCGGCGGGAGTATGTGCTGGTGGTGCAGGCCACGTCGGCTCCG CTGGTGAGCCGAGCCACGGTGCACATCCTTCTCGTGGACCAGAATGACAACCCGCCTGTGCTGCCCGACTTCCAGATCCTCTTCAACAAC TATGTCACCAACAAGTCCAACAGTTTCCCCACCGGCGTGATCGGCTGCATCCCGGCCCATGACCCCGACGTGTCAGACAGCCTCAACTAC ACCTTCGTGCAGGGCAACGAGCTGCGCCTGTTGCTGCTGGACCCCGCCACGGGCGAACTGCAGCTCAGCCGCGACCTGGACAACAACCGG CCGCTGGAGGCGCTCATGGAGGTGTCTGTGTCTGATGGCATCCACAGCGTCACGGCCTTCTGCACCCTGCGTGTCACCATCATCACGGAC GACATGCTGACCAACAGCATCACTGTCCGCCTGGAGAACATGTCCCAGGAGAAGTTCCTGTCCCCGCTGCTGGCCCTCTTCGTGGAGGGG GTGGCCGCCGTGCTGTCCACCACCAAGGACGACGTCTTCGTCTTCAACGTCCAGAACGACACCGACGTCAGCTCCAACATCCTGAACGTG ACCTTCTCGGCGCTGCTGCCTGGCGGCGTCCGCGGCCAGTTCTTCCCGTCGGAGGACCTGCAGGAGCAGATCTACCTGAATCGGACGCTG CTGACCACCATCTCCACGCAGCGCGTGCTGCCCTTCGACGACAACATCTGCCTGCGCGAGCCCTGCGAGAACTACATGAAGTGCGTGTCC GTTCTGCGATTCGACAGCTCCGCGCCCTTCCTCAGCTCCACCACCGTGCTCTTCCGGCCCATCCACCCCATCAACGGCCTGCGCTGCCGC TGCCCGCCCGGCTTCACCGGCGACTACTGCGAGACGGAGATCGACCTCTGCTACTCCGACCCGTGCGGCGCCAACGGCCGCTGCCGCAGC CGCGAGGGCGGCTACACCTGCGAGTGCTTCGAGGACTTCACTGGAGAGCACTGTGAGGTGGATGCCCGCTCAGGCCGCTGTGCCAACGGG GTGTGCAAGAACGGGGGCACCTGCGTGAACCTGCTCATCGGCGGCTTCCACTGCGTGTGTCCTCCTGGCGAGTATGAGAGGCCCTACTGT GAGGTGACCACCAGGAGCTTCCCGCCCCAGTCCTTCGTCACCTTCCGGGGCCTGAGACAGCGCTTCCACTTCACCATCTCCCTCACGTTT GCCACTCAGGAAAGGAACGGCTTGCTTCTCTACAACGGCCGCTTCAATGAGAAGCACGACTTCATCGCCCTGGAGATCGTGGACGAGCAG GTGCAGCTCACCTTCTCTGCAGGCGAGACAACAACGACCGTGGCACCGAAGGTTCCCAGTGGTGTGAGTGACGGGCGGTGGCACTCTGTG CAGGTGCAGTACTACAACAAGGCAGGCATTCCAGACCCTCCAAACATGTCAGCAGAATTAATCCAATTGAAAGCCAAGGAGCGACACTTT TTGGAGCAATTGTTAGATGGGAAAAAATTGGAAAATTATAAAATTCCAGTACCAATCAATGCTGAACTCAGAAAATATCAGCAGGATGGT GTGAACTGGTTAGCATTTCTTAATAAGTATAAACTTCATGGAATTCTGTGTGATGACATGGGTTTAGGAAAAACTTTACAGTCCATCTGC ATTCTAGCAGGAGATCATTGTCATAGGGCCCAGGAATATGCAAGATCAAAATTAGCAGAATGTATGCCACTTCCTTCCTTAGTGGTTTGT CCGCCAACATTAACAGGCCATTGGGTGGATGAAGTAGGTAAATTTTGCTCTAGAGAATATCTCAACCCGTTGCATTACACTGGACCTCCC ACTGAAAGAATAAGGTTACAGCACCAAGTAAAAAGGCACAATCTAATAGTGGCTTCATATGATGTTGTGAGGAATGACATAGATTTCTTT AGAAATATTAAATTTAACTACTGCATTCTTGATGAAGGCCATGTCATCAAAAATGGAAAAACAAAGTTGTCAAAAGCAGTAAAACAACTG ACTGCTAATTATAGGATTATTCTTTCTGGAACACCAATCCAGAACAACGTTTTGGAGCTGTGGTCATTATTTGATTTCCTCATGCCAGGA TTTTTGGGTACTGAACGCCAGTTTGCTGCTCGATATGGTAAACCTATATTAGCAAGTAGGGATGCTCGAAGCTCCAGTCGAGAGCAAGAA GCAGGTGTTCTTGCTATGGATGCGCTGCACCGCCAAGTACTACCGTTTCTTTTGAGAAGAATGAAAGAAGATGTTTTGCAGGATCTTCCA CCTAAAATTATTCAAGACTATTATTGTACTCTTAGTCCTCTCCAGGTTCAGCTCTATGAAGATTTTGCTAAGTCTCGTGCCAAGTGTGAT GTTGATGAAACAGTTTCTTCAGCTACACTTTCTGAAGAAACTGAAAAACCAAAGCTTAAAGCTACAGGCCACGTATTCCAGGCATTACAG TACTTACGTAAACTGTGCAACCATCCAGCATTAGTCTTAACACCTCAACATCCAGAATTCAAGACCACTGCCGAAAAACTGGCAGTTCAG AATTCTTCTCTACATGATATTCAACATGCCCCTAAGCTCTCAGCTTTGAAACAATTGCTGTTGGACTGCGGTTTGGGAAATGGCAGCACT TCCGAGAGTGGCACAGAGTCTGTTGTGGCCCAGCACAGGATACTGATATTCTGTCAGCTGAAAAGCATGCTTGATATAGTAGAGCATGAT CTCCTCAAACCTCACTTGCCCTCTGTCACTTATTTGAGATTAGATGGCAGCATACCTCCTGGTCAGAGGCATTCCATTGTTTCCCGGTTT AATAATGATCCATCTATAGACGTTCTGTTACTTACCACTCACGTTGGTGGCCTGGGACTTAATTTGACAGGCGCTGACACAGTAGTATTT GTGGAGCATGACTGGAATCCTATGCGAGATCTACAAGCCATGGACCGGGCCCATCGCATTGGGCAGAAACGTGTGGTTAACGTATACCGA TTGATAACCAGAGGAACATTGGAAGAAAAAATAATGGGGTTGCAGAAATTCAAGATGAACATAGCGAATACTGTTATTAGCCAAGAGAAT TCTAGTTTGCAGAGCATGGGGACTGATCAGCTTCTTGATCTGTTTACTCTTGATAAGGATGGCAAAGCAGAAAAAGCTGACACCTCTACT TCTGGGAAAGCAAGTATGAAATCAATTCTTGAAAACCTGAGTGATCTTTGGGATCAAGAGCAGTATGATTCAGAGTACAGCCTGGAAAAT TTTATGCATTCTCTCAAGTAACTATCAAATATTGTAAATGCAATTGCTGCTAGTTCAGTTACATTTCTGCTGATATTCAGCAAATTTCTA AGTTTATGGTGAACTTTTAACTCAATGTGTAAATAGATCTGGAGAATATTCAGCATAATGCTGGCTCTTGTTTCACTGGGGGAAACAAGT TATTCTCAAATATTTGCACTGTATTAAATTTAAATGTTAATGTGAAATGGTTTAAATTTTACATGCTGAAAAGCTGCAGAGCAGAGGAAC CAAACCAGGTTTATTTGTGCTACCAGGAGGTGTTCTTAATGGGAACAGGCCTCCTATCTTTAGTCACTTTGTACAGGATGATATCCATGA GTACTTTAGTGTTTGTATATGTTTTTTCTTTTAAATAGAACTTGTTTTTATAATTGATTTAAAATAGGGCTTTTTTTCTATAAAAGCCTT AATGAGCCATAATTTTAAGAATATAAGAATAATGACTATGATATTGGTCAGTCTTAGAACATGAAAATGTCAGACAATGAATTTAATCGG GCCCTTGGTGGAAACATTTATATATTTAATGCAGATGCATAGTGTTTTTCAAATCTTTAACATCATTTTTTCAACTTTTAAGATATAATA TCAACTATTCTAAATACAGGTAATGGTATATTTAAGGCTACCATAACATCCTATTTAGAGGGTGGTTTTTTTTAATTTATCTAATGGCTA CGATATAGCCAGATTCAAATAACATATGTACTCAATAGGTTTCAATCATATATATAATATATTTTTTGTAACTATGAACATACACAGTTT TCCAGAAATAGATTTTACACTGTTTAGAATTCCAACACCTACAGTGGAAAGACTGTTTATTGTAGTAATGCATGGCTGAAGCATAAAAGG GAGAGAGAATTTCTTTATATACACAAACATACATGAATATGCATATCTATATGCATAGATGTACCTATCCTGCACCCAAAAAGGTGCCTG GTATTGGCACTAAAATCATGTAGTTATACTGGGCAGCAATAATAATTGGGCATGAGGCTGATTACTCAATGGTGAGGGTAGGTATATGTA GATAGATGTGCATGTATGTGTTTGTATATATAATTTTATAAGTTTCACGCATAAATTAATACATGCCTGTGTGCATACATATGTATGGGA TATATTTGTATATATATGTACAGGTAATAAACATCCCCAAGGTTTGTGGCTGGCCATACACATAGGCATCAGTTTAACAACCATCAGACC TCAGCTGTACAATAACAGGTGTTTTGTTTAATTAAAATTATACTGTTCTGCAGCATTTAGACATTTGTCACATTTCATTAGCTTTGACAA >15640_15640_1_CELSR1-BTAF1_CELSR1_chr22_46829290_ENST00000262738_BTAF1_chr10_93767880_ENST00000265990_length(amino acids)=2166AA_BP=1537 MAPPPPPVLPVLLLLAAAAALPAMGLRAAAWEPRVPGGTRAFALRPGCTYAVGAACTPRAPRELLDVGRDGRLAGRRRVSGAGRPLPLQV RLVARSAPTALSRRLRARTHLPGCGARARLCGTGARLCGALCFPVPGGCAAAQHSALAAPTTLPACRCPPRPRPRCPGRPICLPPGGSVR LRLLCALRRAAGAVRVGLALEAATAGTPSASPSPSPPLPPNLPEARAGPARRARRGTSGRGSLKFPMPNYQVALFENEPAGTLILQLHAH YTIEGEEERVSYYMEGLFDERSRGYFRIDSATGAVSTDSVLDRETKETHVLRVKAVDYSTPPRSATTYITVLVKDTNDHSPVFEQSEYRE RVRENLEVGYEVLTIRASDRDSPINANLRYRVLGGAWDVFQLNESSGVVSTRAVLDREEAAEYQLLVEANDQGRNPGPLSATATVYIEVE DENDNYPQFSEQNYVVQVPEDVGLNTAVLRVQATDRDQGQNAAIHYSILSGNVAGQFYLHSLSGILDVINPLDFEDVQKYSLSIKAQDGG RPPLINSSGVVSVQVLDVNDNEPIFVSSPFQATVLENVPLGYPVVHIQAVDADSGENARLHYRLVDTASTFLGGGSAGPKNPAPTPDFPF QIHNSSGWITVCAELDREEVEHYSFGVEAVDHGSPPMSSSTSVSITVLDVNDNDPVFTQPTYELRLNEDAAVGSSVLTLQARDRDANSVI TYQLTGGNTRNRFALSSQRGGGLITLALPLDYKQEQQYVLAVTASDGTRSHTAHVLINVTDANTHRPVFQSSHYTVSVSEDRPVGTSIAT LSANDEDTGENARITYVIQDPVPQFRIDPDSGTMYTMMELDYENQVAYTLTIMAQDNGIPQKSDTTTLEILILDANDNAPQFLWDFYQGS IFEDAPPSTSILQVSATDRDSGPNGRLLYTFQGGDDGDGDFYIEPTSGVIRTQRRLDRENVAVYNLWALAVDRGSPTPLSASVEIQVTIL DINDNAPMFEKDELELFVEENNPVGSVVAKIRANDPDEGPNAQIMYQIVEGDMRHFFQLDLLNGDLRAMVELDFEVRREYVLVVQATSAP LVSRATVHILLVDQNDNPPVLPDFQILFNNYVTNKSNSFPTGVIGCIPAHDPDVSDSLNYTFVQGNELRLLLLDPATGELQLSRDLDNNR PLEALMEVSVSDGIHSVTAFCTLRVTIITDDMLTNSITVRLENMSQEKFLSPLLALFVEGVAAVLSTTKDDVFVFNVQNDTDVSSNILNV TFSALLPGGVRGQFFPSEDLQEQIYLNRTLLTTISTQRVLPFDDNICLREPCENYMKCVSVLRFDSSAPFLSSTTVLFRPIHPINGLRCR CPPGFTGDYCETEIDLCYSDPCGANGRCRSREGGYTCECFEDFTGEHCEVDARSGRCANGVCKNGGTCVNLLIGGFHCVCPPGEYERPYC EVTTRSFPPQSFVTFRGLRQRFHFTISLTFATQERNGLLLYNGRFNEKHDFIALEIVDEQVQLTFSAGETTTTVAPKVPSGVSDGRWHSV QVQYYNKAGIPDPPNMSAELIQLKAKERHFLEQLLDGKKLENYKIPVPINAELRKYQQDGVNWLAFLNKYKLHGILCDDMGLGKTLQSIC ILAGDHCHRAQEYARSKLAECMPLPSLVVCPPTLTGHWVDEVGKFCSREYLNPLHYTGPPTERIRLQHQVKRHNLIVASYDVVRNDIDFF RNIKFNYCILDEGHVIKNGKTKLSKAVKQLTANYRIILSGTPIQNNVLELWSLFDFLMPGFLGTERQFAARYGKPILASRDARSSSREQE AGVLAMDALHRQVLPFLLRRMKEDVLQDLPPKIIQDYYCTLSPLQVQLYEDFAKSRAKCDVDETVSSATLSEETEKPKLKATGHVFQALQ YLRKLCNHPALVLTPQHPEFKTTAEKLAVQNSSLHDIQHAPKLSALKQLLLDCGLGNGSTSESGTESVVAQHRILIFCQLKSMLDIVEHD LLKPHLPSVTYLRLDGSIPPGQRHSIVSRFNNDPSIDVLLLTTHVGGLGLNLTGADTVVFVEHDWNPMRDLQAMDRAHRIGQKRVVNVYR LITRGTLEEKIMGLQKFKMNIANTVISQENSSLQSMGTDQLLDLFTLDKDGKAEKADTSTSGKASMKSILENLSDLWDQEQYDSEYSLEN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CELSR1-BTAF1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CELSR1-BTAF1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CELSR1-BTAF1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
