
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CEP70-GYG1 (FusionGDB2 ID:15881) |
Fusion Gene Summary for CEP70-GYG1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CEP70-GYG1 | Fusion gene ID: 15881 | Hgene | Tgene | Gene symbol | CEP70 | GYG1 | Gene ID | 80321 | 2992 |
| Gene name | centrosomal protein 70 | glycogenin 1 | |
| Synonyms | BITE | GSD15|GYG | |
| Cytomap | 3q22.3 | 3q24 | |
| Type of gene | protein-coding | protein-coding | |
| Description | centrosomal protein of 70 kDacentrosomal protein 70kDap10-binding proteintesticular tissue protein Li 36 | glycogenin-1GN-1glycogenin glucosyltransferase | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q8NHQ1 | P46976 | |
| Ensembl transtripts involved in fusion gene | ENST00000264982, ENST00000464035, ENST00000481834, ENST00000484888, ENST00000542237, ENST00000478673, ENST00000489254, | ENST00000479119, ENST00000484197, ENST00000296048, ENST00000345003, ENST00000483267, | |
| Fusion gene scores | * DoF score | 11 X 13 X 6=858 | 9 X 7 X 3=189 |
| # samples | 14 | 11 | |
| ** MAII score | log2(14/858*10)=-2.61555082055458 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/189*10)=-0.780882710696413 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CEP70 [Title/Abstract] AND GYG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CEP70(138289876)-GYG1(148741840), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CEP70-GYG1 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CEP70 (5'-gene) Fusion gene breakpoints across CEP70 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
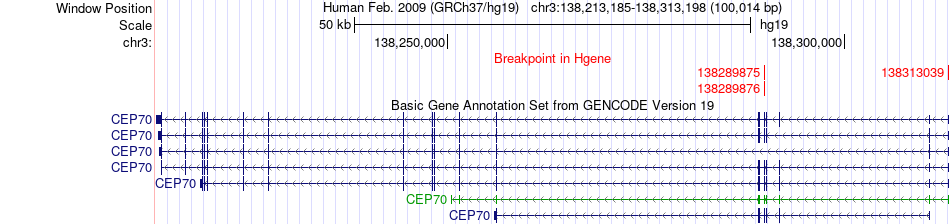 |
 Fusion gene breakpoints across GYG1 (3'-gene) Fusion gene breakpoints across GYG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
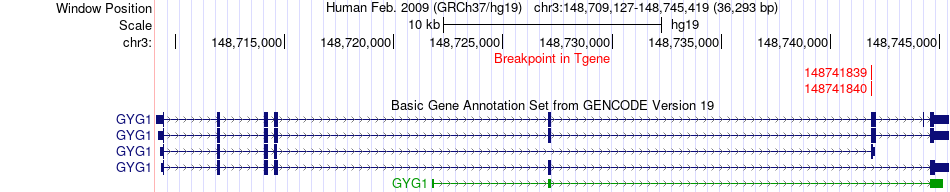 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM-01A | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM-01A | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
Top |
Fusion Gene ORF analysis for CEP70-GYG1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000264982 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000264982 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000264982 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000264982 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000464035 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000464035 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000464035 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000464035 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000481834 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000481834 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000481834 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000481834 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000484888 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000484888 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000484888 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000484888 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000542237 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000542237 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5CDS-intron | ENST00000542237 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5CDS-intron | ENST00000542237 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000264982 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000264982 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000264982 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000478673 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000481834 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000481834 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000481834 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000484888 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000484888 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000484888 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000489254 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000489254 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000489254 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000542237 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000542237 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-3CDS | ENST00000542237 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000264982 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000264982 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000478673 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5UTR-intron | ENST00000478673 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000478673 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000478673 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| 5UTR-intron | ENST00000478673 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000478673 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000481834 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000481834 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000484888 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000484888 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000489254 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000489254 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000542237 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| 5UTR-intron | ENST00000542237 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000264982 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000264982 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000464035 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000464035 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000464035 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000464035 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000481834 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000481834 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000484888 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000484888 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000484888 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000484888 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| Frame-shift | ENST00000542237 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| Frame-shift | ENST00000542237 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000264982 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000264982 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000264982 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000264982 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000464035 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000464035 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000481834 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000481834 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000481834 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000481834 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000484888 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000484888 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000542237 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000542237 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| In-frame | ENST00000542237 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| In-frame | ENST00000542237 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| intron-3CDS | ENST00000464035 | ENST00000296048 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| intron-3CDS | ENST00000464035 | ENST00000345003 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| intron-3CDS | ENST00000464035 | ENST00000483267 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| intron-3CDS | ENST00000489254 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| intron-3CDS | ENST00000489254 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| intron-3CDS | ENST00000489254 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| intron-3CDS | ENST00000489254 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| intron-3CDS | ENST00000489254 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| intron-3CDS | ENST00000489254 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| intron-intron | ENST00000464035 | ENST00000479119 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| intron-intron | ENST00000464035 | ENST00000484197 | CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + |
| intron-intron | ENST00000489254 | ENST00000479119 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| intron-intron | ENST00000489254 | ENST00000479119 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
| intron-intron | ENST00000489254 | ENST00000484197 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + |
| intron-intron | ENST00000489254 | ENST00000484197 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264982 | CEP70 | chr3 | 138289876 | - | ENST00000345003 | GYG1 | chr3 | 148741840 | + | 1695 | 551 | 267 | 995 | 242 |
| ENST00000264982 | CEP70 | chr3 | 138289876 | - | ENST00000296048 | GYG1 | chr3 | 148741840 | + | 1644 | 551 | 267 | 944 | 225 |
| ENST00000542237 | CEP70 | chr3 | 138289876 | - | ENST00000345003 | GYG1 | chr3 | 148741840 | + | 1513 | 369 | 145 | 813 | 222 |
| ENST00000542237 | CEP70 | chr3 | 138289876 | - | ENST00000296048 | GYG1 | chr3 | 148741840 | + | 1462 | 369 | 145 | 762 | 205 |
| ENST00000484888 | CEP70 | chr3 | 138289876 | - | ENST00000483267 | GYG1 | chr3 | 148741840 | + | 674 | 471 | 148 | 672 | 175 |
| ENST00000481834 | CEP70 | chr3 | 138289876 | - | ENST00000345003 | GYG1 | chr3 | 148741840 | + | 1587 | 443 | 159 | 887 | 242 |
| ENST00000481834 | CEP70 | chr3 | 138289876 | - | ENST00000296048 | GYG1 | chr3 | 148741840 | + | 1536 | 443 | 159 | 836 | 225 |
| ENST00000464035 | CEP70 | chr3 | 138289876 | - | ENST00000483267 | GYG1 | chr3 | 148741840 | + | 647 | 444 | 121 | 645 | 175 |
| ENST00000264982 | CEP70 | chr3 | 138289875 | - | ENST00000345003 | GYG1 | chr3 | 148741839 | + | 1695 | 551 | 267 | 995 | 242 |
| ENST00000264982 | CEP70 | chr3 | 138289875 | - | ENST00000296048 | GYG1 | chr3 | 148741839 | + | 1644 | 551 | 267 | 944 | 225 |
| ENST00000542237 | CEP70 | chr3 | 138289875 | - | ENST00000345003 | GYG1 | chr3 | 148741839 | + | 1513 | 369 | 145 | 813 | 222 |
| ENST00000542237 | CEP70 | chr3 | 138289875 | - | ENST00000296048 | GYG1 | chr3 | 148741839 | + | 1462 | 369 | 145 | 762 | 205 |
| ENST00000484888 | CEP70 | chr3 | 138289875 | - | ENST00000483267 | GYG1 | chr3 | 148741839 | + | 674 | 471 | 148 | 672 | 175 |
| ENST00000481834 | CEP70 | chr3 | 138289875 | - | ENST00000345003 | GYG1 | chr3 | 148741839 | + | 1587 | 443 | 159 | 887 | 242 |
| ENST00000481834 | CEP70 | chr3 | 138289875 | - | ENST00000296048 | GYG1 | chr3 | 148741839 | + | 1536 | 443 | 159 | 836 | 225 |
| ENST00000464035 | CEP70 | chr3 | 138289875 | - | ENST00000483267 | GYG1 | chr3 | 148741839 | + | 647 | 444 | 121 | 645 | 175 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264982 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.00072881 | 0.9992712 |
| ENST00000264982 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.000501637 | 0.99949837 |
| ENST00000542237 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.000938075 | 0.9990619 |
| ENST00000542237 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.001135748 | 0.9988643 |
| ENST00000484888 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.61706734 | 0.38293263 |
| ENST00000481834 | ENST00000345003 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.000722638 | 0.9992774 |
| ENST00000481834 | ENST00000296048 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.000536181 | 0.99946386 |
| ENST00000464035 | ENST00000483267 | CEP70 | chr3 | 138289876 | - | GYG1 | chr3 | 148741840 | + | 0.5994379 | 0.4005621 |
| ENST00000264982 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.00072881 | 0.9992712 |
| ENST00000264982 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000501637 | 0.99949837 |
| ENST00000542237 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000938075 | 0.9990619 |
| ENST00000542237 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.001135748 | 0.9988643 |
| ENST00000484888 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.61706734 | 0.38293263 |
| ENST00000481834 | ENST00000345003 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000722638 | 0.9992774 |
| ENST00000481834 | ENST00000296048 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000536181 | 0.99946386 |
| ENST00000464035 | ENST00000483267 | CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.5994379 | 0.4005621 |
Top |
Fusion Genomic Features for CEP70-GYG1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000911707 | 0.9990883 |
| CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000911707 | 0.9990883 |
| CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + | 0.000571804 | 0.9994282 |
| CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000911707 | 0.9990883 |
| CEP70 | chr3 | 138289875 | - | GYG1 | chr3 | 148741839 | + | 0.000911707 | 0.9990883 |
| CEP70 | chr3 | 138313039 | - | GYG1 | chr3 | 148741839 | + | 0.000571804 | 0.9994282 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
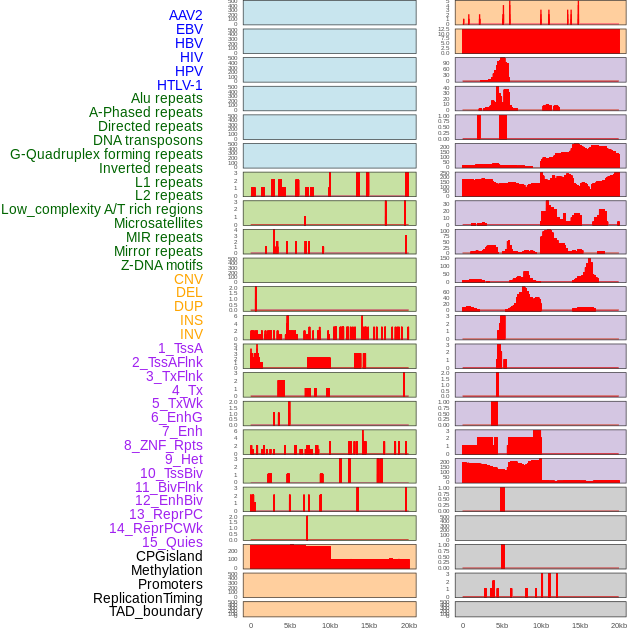 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
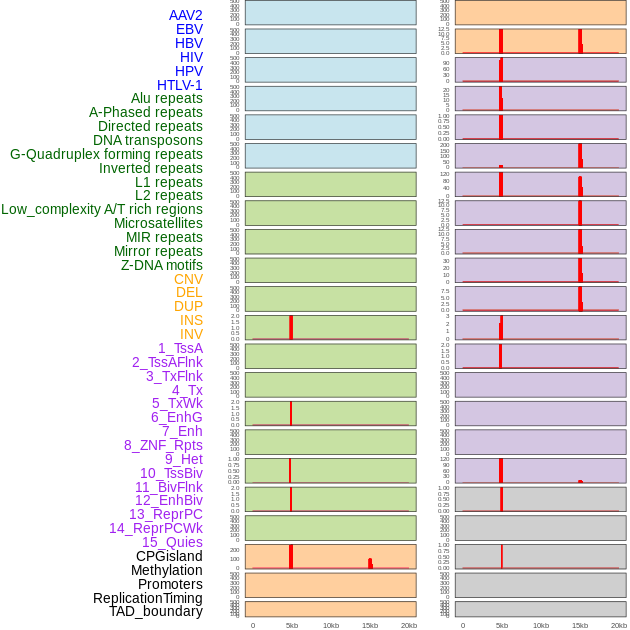 |
Top |
Fusion Protein Features for CEP70-GYG1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:138289876/chr3:148741840) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CEP70 | GYG1 |
| FUNCTION: Plays a role in the organization of both preexisting and nascent microtubules in interphase cells. During mitosis, required for the organization and orientation of the mitotic spindle. | FUNCTION: Self-glucosylates, via an inter-subunit mechanism, to form an oligosaccharide primer that serves as substrate for glycogen synthase. {ECO:0000269|PubMed:22160680, ECO:0000269|PubMed:30356213}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000296048 | 4 | 7 | 212_218 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000345003 | 4 | 8 | 212_218 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000484197 | 0 | 6 | 102_104 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000484197 | 0 | 6 | 133_135 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000484197 | 0 | 6 | 160_164 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000484197 | 0 | 6 | 212_218 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000484197 | 0 | 6 | 9_15 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000296048 | 4 | 7 | 212_218 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000345003 | 4 | 8 | 212_218 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000484197 | 0 | 6 | 102_104 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000484197 | 0 | 6 | 133_135 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000484197 | 0 | 6 | 160_164 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000484197 | 0 | 6 | 212_218 | 0 | 280.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000484197 | 0 | 6 | 9_15 | 0 | 280.0 | Region | Substrate binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000264982 | - | 5 | 18 | 254_326 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000264982 | - | 5 | 18 | 75_179 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000464035 | - | 4 | 6 | 254_326 | 94 | 213.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000464035 | - | 4 | 6 | 75_179 | 94 | 213.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000481834 | - | 5 | 16 | 254_326 | 94 | 555.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000481834 | - | 5 | 16 | 75_179 | 94 | 555.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000484888 | - | 5 | 18 | 254_326 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000484888 | - | 5 | 18 | 75_179 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000542237 | - | 4 | 17 | 254_326 | 74 | 578.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000542237 | - | 4 | 17 | 75_179 | 74 | 578.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000264982 | - | 5 | 18 | 254_326 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000264982 | - | 5 | 18 | 75_179 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000464035 | - | 4 | 6 | 254_326 | 94 | 213.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000464035 | - | 4 | 6 | 75_179 | 94 | 213.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000481834 | - | 5 | 16 | 254_326 | 94 | 555.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000481834 | - | 5 | 16 | 75_179 | 94 | 555.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000484888 | - | 5 | 18 | 254_326 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000484888 | - | 5 | 18 | 75_179 | 94 | 598.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000542237 | - | 4 | 17 | 254_326 | 74 | 578.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000542237 | - | 4 | 17 | 75_179 | 74 | 578.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000264982 | - | 5 | 18 | 483_516 | 94 | 598.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000464035 | - | 4 | 6 | 483_516 | 94 | 213.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000481834 | - | 5 | 16 | 483_516 | 94 | 555.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000484888 | - | 5 | 18 | 483_516 | 94 | 598.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289875 | chr3:148741839 | ENST00000542237 | - | 4 | 17 | 483_516 | 74 | 578.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000264982 | - | 5 | 18 | 483_516 | 94 | 598.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000464035 | - | 4 | 6 | 483_516 | 94 | 213.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000481834 | - | 5 | 16 | 483_516 | 94 | 555.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000484888 | - | 5 | 18 | 483_516 | 94 | 598.0 | Repeat | Note=TPR |
| Hgene | CEP70 | chr3:138289876 | chr3:148741840 | ENST00000542237 | - | 4 | 17 | 483_516 | 74 | 578.0 | Repeat | Note=TPR |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000296048 | 4 | 7 | 102_104 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000296048 | 4 | 7 | 133_135 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000296048 | 4 | 7 | 160_164 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000296048 | 4 | 7 | 9_15 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000345003 | 4 | 8 | 102_104 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000345003 | 4 | 8 | 133_135 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000345003 | 4 | 8 | 160_164 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000345003 | 4 | 8 | 9_15 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000296048 | 4 | 7 | 102_104 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000296048 | 4 | 7 | 133_135 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000296048 | 4 | 7 | 160_164 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000296048 | 4 | 7 | 9_15 | 202 | 334.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000345003 | 4 | 8 | 102_104 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000345003 | 4 | 8 | 133_135 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000345003 | 4 | 8 | 160_164 | 202 | 351.0 | Region | Substrate binding | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000345003 | 4 | 8 | 9_15 | 202 | 351.0 | Region | Substrate binding |
Top |
Fusion Gene Sequence for CEP70-GYG1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >15881_15881_1_CEP70-GYG1_CEP70_chr3_138289875_ENST00000264982_GYG1_chr3_148741839_ENST00000296048_length(transcript)=1644nt_BP=551nt ATTCGCAGGCCAAAGAGGGGCTGGCCCTCGACGCGCTCGGGCTCGATTGGCCGCCCTCCCCCGGCGGTGGTAACGGTCGCGGAGGCGGGG CCTGACGTAGCTGATCCCAGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACA TGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATG TTTCCGGTAGCCCCTAAACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGC ATAAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAG TCATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAAT CAACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAA AGTGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCT CTGCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGG GAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGAT TCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGT TTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTT TTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTG TTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTG CAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGA CCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTG ATTCATAAACATATCAACCAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAA GGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTTCTCTGATGGTTACCTGCAGTGGCACCCTGTACA >15881_15881_1_CEP70-GYG1_CEP70_chr3_138289875_ENST00000264982_GYG1_chr3_148741839_ENST00000296048_length(amino acids)=225AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVEDVSGAISHLSL -------------------------------------------------------------- >15881_15881_2_CEP70-GYG1_CEP70_chr3_138289875_ENST00000264982_GYG1_chr3_148741839_ENST00000345003_length(transcript)=1695nt_BP=551nt ATTCGCAGGCCAAAGAGGGGCTGGCCCTCGACGCGCTCGGGCTCGATTGGCCGCCCTCCCCCGGCGGTGGTAACGGTCGCGGAGGCGGGG CCTGACGTAGCTGATCCCAGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACA TGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATG TTTCCGGTAGCCCCTAAACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGC ATAAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAG TCATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAAT CAACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAA AGTGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCT CTGCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGCTTTCAGACTTGGTCTATACACTGGCTTTCTCTTGTGGC TTCTGTAGAAAGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAA GAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTC CAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTC TGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGC AGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAA AGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCA GACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTG AGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAACCAATAGCATTAACCCATTTTA TTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATT >15881_15881_2_CEP70-GYG1_CEP70_chr3_138289875_ENST00000264982_GYG1_chr3_148741839_ENST00000345003_length(amino acids)=242AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSC -------------------------------------------------------------- >15881_15881_3_CEP70-GYG1_CEP70_chr3_138289875_ENST00000464035_GYG1_chr3_148741839_ENST00000483267_length(transcript)=647nt_BP=444nt ATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATA TGCTATGACTGAGGCAACTCAAATTTTTTAATTGCCAATTTCTGTTTCTTTTGATTCTACAAAAGTAACTATGTTTCCGGTAGCCCCTAA ACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGCATAAATGTGCTATTGAT GATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTCATCACAAAGGATGAG ACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCAACAGCTTAGGTTTGG TGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAGTGTCAAAAGTGAGGC CCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCTGCTTCAACAATTTGG >15881_15881_3_CEP70-GYG1_CEP70_chr3_138289875_ENST00000464035_GYG1_chr3_148741839_ENST00000483267_length(amino acids)=175AA_BP=107 MPISVSFDSTKVTMFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEET -------------------------------------------------------------- >15881_15881_4_CEP70-GYG1_CEP70_chr3_138289875_ENST00000481834_GYG1_chr3_148741839_ENST00000296048_length(transcript)=1536nt_BP=443nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATGTTTCCGGTAGCCCCTAAA CCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGCATAAATGTGCTATTGATG ATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTCATCACAAAGGATGAGA CAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCAACAGCTTAGGTTTGGT GCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAGTGTCAAAAGTGAGGCC CATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCTGCTTCAACAATTTGGC CTTGTCAAAGACACCTGCTCATATGTAAATGTGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCA CAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAG AGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCT AGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGA ACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTT TTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAAT CAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGC CCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAAC CAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGC TTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTTCTCTGATGGTTACCTGCAGTGGCACCCTGTACAAAAAATAAAAGACTTATT >15881_15881_4_CEP70-GYG1_CEP70_chr3_138289875_ENST00000481834_GYG1_chr3_148741839_ENST00000296048_length(amino acids)=225AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVEDVSGAISHLSL -------------------------------------------------------------- >15881_15881_5_CEP70-GYG1_CEP70_chr3_138289875_ENST00000481834_GYG1_chr3_148741839_ENST00000345003_length(transcript)=1587nt_BP=443nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATGTTTCCGGTAGCCCCTAAA CCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGCATAAATGTGCTATTGATG ATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTCATCACAAAGGATGAGA CAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCAACAGCTTAGGTTTGGT GCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAGTGTCAAAAGTGAGGCC CATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCTGCTTCAACAATTTGGC CTTGTCAAAGACACCTGCTCATATGTAAATGTGCTTTCAGACTTGGTCTATACACTGGCTTTCTCTTGTGGCTTCTGTAGAAAGGAAGAT GTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGG GAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATT TTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGG TTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGG ACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGA TATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCT TGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCA GGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAACCAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTC TGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTT >15881_15881_5_CEP70-GYG1_CEP70_chr3_138289875_ENST00000481834_GYG1_chr3_148741839_ENST00000345003_length(amino acids)=242AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSC -------------------------------------------------------------- >15881_15881_6_CEP70-GYG1_CEP70_chr3_138289875_ENST00000484888_GYG1_chr3_148741839_ENST00000483267_length(transcript)=674nt_BP=471nt CTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAAT GACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGGCAACTCAAATTTTTTAATTGCCAATTTCTGTTTCTTTTGATTCTACAAA AGTAACTATGTTTCCGGTAGCCCCTAAACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGA ATGGGAAAGCATAAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTT TGACAAACAGTCATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTAT AGAAACTAATCAACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCC CAAAACAAAAAGTGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAA >15881_15881_6_CEP70-GYG1_CEP70_chr3_138289875_ENST00000484888_GYG1_chr3_148741839_ENST00000483267_length(amino acids)=175AA_BP=107 MPISVSFDSTKVTMFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEET -------------------------------------------------------------- >15881_15881_7_CEP70-GYG1_CEP70_chr3_138289875_ENST00000542237_GYG1_chr3_148741839_ENST00000296048_length(transcript)=1462nt_BP=369nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGCAGGAAGAAGCAGAATGGGAAAGCAT AAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTC ATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCA ACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAG TGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCT GCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGA GATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTC CTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTT CTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTT AGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTT TCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCA GAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACC TGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGAT TCATAAACATATCAACCAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGG CAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTTCTCTGATGGTTACCTGCAGTGGCACCCTGTACAAA >15881_15881_7_CEP70-GYG1_CEP70_chr3_138289875_ENST00000542237_GYG1_chr3_148741839_ENST00000296048_length(amino acids)=205AA_BP=74 MTEQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIETNQQLRFGASAKVVHFLGRVK PWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVEDVSGAISHLSLGEIPAMAQPFVSSEERKERW -------------------------------------------------------------- >15881_15881_8_CEP70-GYG1_CEP70_chr3_138289875_ENST00000542237_GYG1_chr3_148741839_ENST00000345003_length(transcript)=1513nt_BP=369nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGCAGGAAGAAGCAGAATGGGAAAGCAT AAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTC ATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCA ACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAG TGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCT GCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGCTTTCAGACTTGGTCTATACACTGGCTTTCTCTTGTGGCTT CTGTAGAAAGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGA ACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCA GTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTG TTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAG CAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAG TAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGA CATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAG TGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAACCAATAGCATTAACCCATTTTATT TCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTG >15881_15881_8_CEP70-GYG1_CEP70_chr3_138289875_ENST00000542237_GYG1_chr3_148741839_ENST00000345003_length(amino acids)=222AA_BP=74 MTEQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIETNQQLRFGASAKVVHFLGRVK PWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSCGFCRKEDVSGAISHLSLGEI -------------------------------------------------------------- >15881_15881_9_CEP70-GYG1_CEP70_chr3_138289876_ENST00000264982_GYG1_chr3_148741840_ENST00000296048_length(transcript)=1644nt_BP=551nt ATTCGCAGGCCAAAGAGGGGCTGGCCCTCGACGCGCTCGGGCTCGATTGGCCGCCCTCCCCCGGCGGTGGTAACGGTCGCGGAGGCGGGG CCTGACGTAGCTGATCCCAGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACA TGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATG TTTCCGGTAGCCCCTAAACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGC ATAAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAG TCATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAAT CAACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAA AGTGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCT CTGCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGG GAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGAT TCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGT TTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTT TTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTG TTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTG CAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGA CCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTG ATTCATAAACATATCAACCAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAA GGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTTCTCTGATGGTTACCTGCAGTGGCACCCTGTACA >15881_15881_9_CEP70-GYG1_CEP70_chr3_138289876_ENST00000264982_GYG1_chr3_148741840_ENST00000296048_length(amino acids)=225AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVEDVSGAISHLSL -------------------------------------------------------------- >15881_15881_10_CEP70-GYG1_CEP70_chr3_138289876_ENST00000264982_GYG1_chr3_148741840_ENST00000345003_length(transcript)=1695nt_BP=551nt ATTCGCAGGCCAAAGAGGGGCTGGCCCTCGACGCGCTCGGGCTCGATTGGCCGCCCTCCCCCGGCGGTGGTAACGGTCGCGGAGGCGGGG CCTGACGTAGCTGATCCCAGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACA TGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATG TTTCCGGTAGCCCCTAAACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGC ATAAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAG TCATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAAT CAACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAA AGTGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCT CTGCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGCTTTCAGACTTGGTCTATACACTGGCTTTCTCTTGTGGC TTCTGTAGAAAGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAA GAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTC CAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTC TGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGC AGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAA AGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCA GACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTG AGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAACCAATAGCATTAACCCATTTTA TTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATT >15881_15881_10_CEP70-GYG1_CEP70_chr3_138289876_ENST00000264982_GYG1_chr3_148741840_ENST00000345003_length(amino acids)=242AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSC -------------------------------------------------------------- >15881_15881_11_CEP70-GYG1_CEP70_chr3_138289876_ENST00000464035_GYG1_chr3_148741840_ENST00000483267_length(transcript)=647nt_BP=444nt ATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATA TGCTATGACTGAGGCAACTCAAATTTTTTAATTGCCAATTTCTGTTTCTTTTGATTCTACAAAAGTAACTATGTTTCCGGTAGCCCCTAA ACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGCATAAATGTGCTATTGAT GATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTCATCACAAAGGATGAG ACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCAACAGCTTAGGTTTGG TGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAGTGTCAAAAGTGAGGC CCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCTGCTTCAACAATTTGG >15881_15881_11_CEP70-GYG1_CEP70_chr3_138289876_ENST00000464035_GYG1_chr3_148741840_ENST00000483267_length(amino acids)=175AA_BP=107 MPISVSFDSTKVTMFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEET -------------------------------------------------------------- >15881_15881_12_CEP70-GYG1_CEP70_chr3_138289876_ENST00000481834_GYG1_chr3_148741840_ENST00000296048_length(transcript)=1536nt_BP=443nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATGTTTCCGGTAGCCCCTAAA CCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGCATAAATGTGCTATTGATG ATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTCATCACAAAGGATGAGA CAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCAACAGCTTAGGTTTGGT GCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAGTGTCAAAAGTGAGGCC CATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCTGCTTCAACAATTTGGC CTTGTCAAAGACACCTGCTCATATGTAAATGTGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCA CAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAG AGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCT AGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGA ACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTT TTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAAT CAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGC CCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAAC CAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGC TTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTTCTCTGATGGTTACCTGCAGTGGCACCCTGTACAAAAAATAAAAGACTTATT >15881_15881_12_CEP70-GYG1_CEP70_chr3_138289876_ENST00000481834_GYG1_chr3_148741840_ENST00000296048_length(amino acids)=225AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVEDVSGAISHLSL -------------------------------------------------------------- >15881_15881_13_CEP70-GYG1_CEP70_chr3_138289876_ENST00000481834_GYG1_chr3_148741840_ENST00000345003_length(transcript)=1587nt_BP=443nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGTAACTATGTTTCCGGTAGCCCCTAAA CCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGAATGGGAAAGCATAAATGTGCTATTGATG ATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTCATCACAAAGGATGAGA CAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCAACAGCTTAGGTTTGGT GCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAGTGTCAAAAGTGAGGCC CATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCTGCTTCAACAATTTGGC CTTGTCAAAGACACCTGCTCATATGTAAATGTGCTTTCAGACTTGGTCTATACACTGGCTTTCTCTTGTGGCTTCTGTAGAAAGGAAGAT GTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGG GAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATT TTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGG TTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGG ACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGA TATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCT TGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCA GGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAACCAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTC TGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTT >15881_15881_13_CEP70-GYG1_CEP70_chr3_138289876_ENST00000481834_GYG1_chr3_148741840_ENST00000345003_length(amino acids)=242AA_BP=94 MFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIET NQQLRFGASAKVVHFLGRVKPWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSC -------------------------------------------------------------- >15881_15881_14_CEP70-GYG1_CEP70_chr3_138289876_ENST00000484888_GYG1_chr3_148741840_ENST00000483267_length(transcript)=674nt_BP=471nt CTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGATCATTTCATTGAGATCAACCTGAAT GACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGGCAACTCAAATTTTTTAATTGCCAATTTCTGTTTCTTTTGATTCTACAAA AGTAACTATGTTTCCGGTAGCCCCTAAACCCCAGGATTCCAGTCAACCATCAGACAGACTCATGACTGAAAAACAGCAGGAAGAAGCAGA ATGGGAAAGCATAAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTT TGACAAACAGTCATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTAT AGAAACTAATCAACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCC CAAAACAAAAAGTGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAA >15881_15881_14_CEP70-GYG1_CEP70_chr3_138289876_ENST00000484888_GYG1_chr3_148741840_ENST00000483267_length(amino acids)=175AA_BP=107 MPISVSFDSTKVTMFPVAPKPQDSSQPSDRLMTEKQQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEET -------------------------------------------------------------- >15881_15881_15_CEP70-GYG1_CEP70_chr3_138289876_ENST00000542237_GYG1_chr3_148741840_ENST00000296048_length(transcript)=1462nt_BP=369nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGCAGGAAGAAGCAGAATGGGAAAGCAT AAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTC ATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCA ACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAG TGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCT GCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGA GATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGAACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTC CTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCAGTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTT CTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTGTTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTT AGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAGCAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTT TCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAGTAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCA GAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGACATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACC TGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAGTGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGAT TCATAAACATATCAACCAATAGCATTAACCCATTTTATTTCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGG CAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTGAAAATAGCAGTGTGTTCTCTGATGGTTACCTGCAGTGGCACCCTGTACAAA >15881_15881_15_CEP70-GYG1_CEP70_chr3_138289876_ENST00000542237_GYG1_chr3_148741840_ENST00000296048_length(amino acids)=205AA_BP=74 MTEQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIETNQQLRFGASAKVVHFLGRVK PWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVEDVSGAISHLSLGEIPAMAQPFVSSEERKERW -------------------------------------------------------------- >15881_15881_16_CEP70-GYG1_CEP70_chr3_138289876_ENST00000542237_GYG1_chr3_148741840_ENST00000345003_length(transcript)=1513nt_BP=369nt AGCCTAACCTGGCCCGGCTGGCTGCTGGTGGGAGGCCGGCGACTGAGTAAGATGAAGATCCAGTTTCAGACATGAGACTCGTTGGGTGAT CATTTCATTGAGATCAACCTGAATGACCAGGTGTAAAGTGCAAGAGTAATATGCTATGACTGAGCAGGAAGAAGCAGAATGGGAAAGCAT AAATGTGCTATTGATGATGCATGGCTTAAAACCTTTGTCTCTAGTCAAAAGAACAGATCTCAAAGATCTCATCATTTTTGACAAACAGTC ATCACAAAGGATGAGACAGAATTTGAAATTGTTGGTGGAAGAAACATCATGTCAACAGAACATGATACAGGAGCTTATAGAAACTAATCA ACAGCTTAGGTTTGGTGCAAGTGCCAAAGTTGTGCATTTCCTGGGACGAGTCAAACCATGGAATTATACTTATGATCCCAAAACAAAAAG TGTCAAAAGTGAGGCCCATGATCCCAACATGACTCATCCAGAGTTTCTCATCCTGTGGTGGAACATCTTTACCACCAACGTTTTACCTCT GCTTCAACAATTTGGCCTTGTCAAAGACACCTGCTCATATGTAAATGTGCTTTCAGACTTGGTCTATACACTGGCTTTCTCTTGTGGCTT CTGTAGAAAGGAAGATGTCTCAGGAGCCATATCACATCTGTCCCTTGGGGAGATCCCAGCTATGGCACAGCCGTTTGTATCCTCGGAAGA ACGGAAGGAACGATGGGAACAGGGCCAGGCTGATTATATGGGAGCAGATTCCTTTGACAACATCAAGAGGAAACTTGACACTTACCTCCA GTAGAAACACTGCATTTTTCTGTGAACACATCCACTTCACAAGCCTTGTTTCTGATACTTAGTATCTAGAGCTGGGTTGAGAAAAGTCTG TTACAGTTGCTAGAGGTTTTCATTAAAACTTATCAGATGAGAGGCTTTTTTAGGATAAGAGGTGAGAACTGGGCAAAAGTTGTGAAGCAG CAATTCTGTTATATGGACAGTGTTCTGCTTTTTAATCCTATTTAGCTTGTTTCAGAAATTCTCACTTTTGTTGACTGCCAACATACAAAG TAAGGGAAACTCAAGATATTAAGATGGCTGTATCAGTTCTTAAAATCTGCAGAGCCTGGTTCAAAATCAGTCACTCCCTTCAGAAGCAGA CATGGCATCTGTTCCTTGCTTGCTTGTTGGTTGTGTACCTTTCACGAGACCTGAATTTTAGAATTGCCCAGTGCTGCCAGAGTGAGTGAG TGTAATTCTCCTTTCAGGTAAAGATAGGCTATCTCAACACTGCTGAGTGATTCATAAACATATCAACCAATAGCATTAACCCATTTTATT TCCTGTCCTTAGTGTCTGAAGATGCTCACCAGTTTTCTGTGTACAGTAAGGCAGCATGCTAAAATGCTTTTGTTCAGTTCTGTATATTTG >15881_15881_16_CEP70-GYG1_CEP70_chr3_138289876_ENST00000542237_GYG1_chr3_148741840_ENST00000345003_length(amino acids)=222AA_BP=74 MTEQEEAEWESINVLLMMHGLKPLSLVKRTDLKDLIIFDKQSSQRMRQNLKLLVEETSCQQNMIQELIETNQQLRFGASAKVVHFLGRVK PWNYTYDPKTKSVKSEAHDPNMTHPEFLILWWNIFTTNVLPLLQQFGLVKDTCSYVNVLSDLVYTLAFSCGFCRKEDVSGAISHLSLGEI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CEP70-GYG1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000296048 | 4 | 7 | 301_333 | 202.66666666666666 | 334.0 | GYS1 | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000345003 | 4 | 8 | 301_333 | 202.66666666666666 | 351.0 | GYS1 | |
| Tgene | GYG1 | chr3:138289875 | chr3:148741839 | ENST00000484197 | 0 | 6 | 301_333 | 0 | 280.0 | GYS1 | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000296048 | 4 | 7 | 301_333 | 202.66666666666666 | 334.0 | GYS1 | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000345003 | 4 | 8 | 301_333 | 202.66666666666666 | 351.0 | GYS1 | |
| Tgene | GYG1 | chr3:138289876 | chr3:148741840 | ENST00000484197 | 0 | 6 | 301_333 | 0 | 280.0 | GYS1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CEP70-GYG1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CEP70-GYG1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
