
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHD7-ASPH (FusionGDB2 ID:16372) |
Fusion Gene Summary for CHD7-ASPH |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHD7-ASPH | Fusion gene ID: 16372 | Hgene | Tgene | Gene symbol | CHD7 | ASPH | Gene ID | 55636 | 444 |
| Gene name | chromodomain helicase DNA binding protein 7 | aspartate beta-hydroxylase | |
| Synonyms | CRG|HH5|IS3|KAL5 | AAH|BAH|CASQ2BP1|FDLAB|HAAH|JCTN|junctin | |
| Cytomap | 8q12.2 | 8q12.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | chromodomain-helicase-DNA-binding protein 7ATP-dependent helicase CHD7CHARGE associationchromodomain helicase DNA binding protein 7 isoform CRA_e | aspartyl/asparaginyl beta-hydroxylaseA beta H-J-JASP beta-hydroxylasecardiac junctinhumbugjunctatepeptide-aspartate beta-dioxygenase | |
| Modification date | 20200328 | 20200315 | |
| UniProtAcc | Q9P2D1 | Q6ICH7 | |
| Ensembl transtripts involved in fusion gene | ENST00000423902, ENST00000524602, ENST00000525508, ENST00000529472, | ENST00000356457, ENST00000379449, ENST00000389204, ENST00000445642, ENST00000517661, ENST00000517847, ENST00000517856, ENST00000517903, ENST00000518068, ENST00000522603, ENST00000522835, ENST00000522919, ENST00000523897, ENST00000379454, ENST00000541428, | |
| Fusion gene scores | * DoF score | 12 X 8 X 8=768 | 17 X 17 X 5=1445 |
| # samples | 17 | 19 | |
| ** MAII score | log2(17/768*10)=-2.17557156458345 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/1445*10)=-2.92699816905709 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CHD7 [Title/Abstract] AND ASPH [Title/Abstract] AND fusion [Title/Abstract] | ||
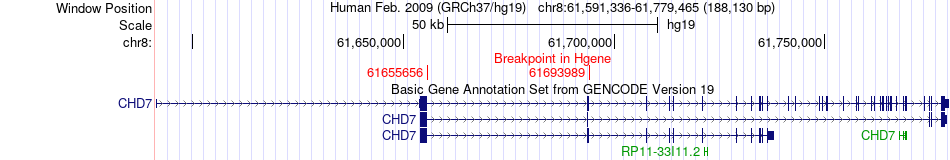
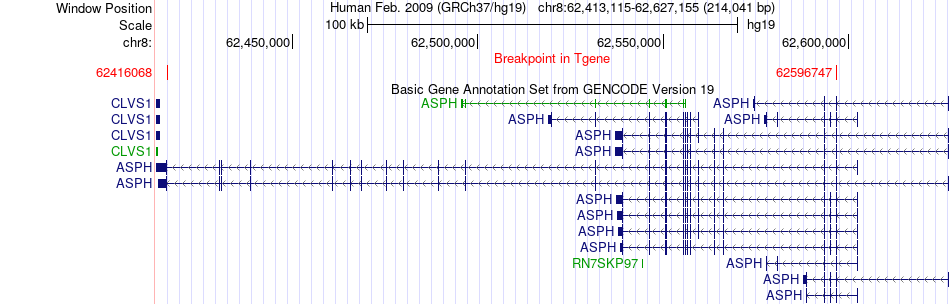
| Most frequent breakpoint | CHD7(61655656)-ASPH(62596747), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CHD7-ASPH seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. CHD7-ASPH seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ASPH | GO:0045862 | positive regulation of proteolysis | 18387192 |
| Tgene | ASPH | GO:0097202 | activation of cysteine-type endopeptidase activity | 18387192 |
 Fusion gene breakpoints across CHD7 (5'-gene) Fusion gene breakpoints across CHD7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across ASPH (3'-gene) Fusion gene breakpoints across ASPH (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A7-A2KD-01A | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| ChimerDB4 | BRCA | TCGA-AR-A1AU-01A | CHD7 | chr8 | 61655656 | - | ASPH | chr8 | 62596747 | - |
| ChimerDB4 | BRCA | TCGA-AR-A1AU-01A | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
Top |
Fusion Gene ORF analysis for CHD7-ASPH |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000423902 | ENST00000356457 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000356457 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000379449 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000379449 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000389204 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000389204 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000445642 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000445642 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517661 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517661 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517847 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517847 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517856 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517856 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517903 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000517903 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000518068 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000518068 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000522603 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000522603 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000522835 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000522835 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000522919 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000522919 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000423902 | ENST00000523897 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000423902 | ENST00000523897 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000356457 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000379449 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000389204 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000445642 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000517661 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000517847 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000517856 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000517903 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000518068 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000522603 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000522835 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000522919 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000524602 | ENST00000523897 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000356457 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000356457 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000379449 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000379449 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000389204 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000389204 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000445642 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000445642 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517661 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517661 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517847 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517847 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517856 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517856 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517903 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000517903 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000518068 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000518068 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000522603 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000522603 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000522835 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000522835 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000522919 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000522919 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| 5CDS-intron | ENST00000525508 | ENST00000523897 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| 5CDS-intron | ENST00000525508 | ENST00000523897 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| Frame-shift | ENST00000423902 | ENST00000379454 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| Frame-shift | ENST00000423902 | ENST00000541428 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| Frame-shift | ENST00000423902 | ENST00000541428 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| Frame-shift | ENST00000524602 | ENST00000379454 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| Frame-shift | ENST00000524602 | ENST00000541428 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| Frame-shift | ENST00000525508 | ENST00000379454 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| Frame-shift | ENST00000525508 | ENST00000541428 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| Frame-shift | ENST00000525508 | ENST00000541428 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| In-frame | ENST00000423902 | ENST00000379454 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| In-frame | ENST00000525508 | ENST00000379454 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-3CDS | ENST00000524602 | ENST00000379454 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-3CDS | ENST00000524602 | ENST00000541428 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-3CDS | ENST00000529472 | ENST00000379454 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-3CDS | ENST00000529472 | ENST00000379454 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-3CDS | ENST00000529472 | ENST00000541428 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-3CDS | ENST00000529472 | ENST00000541428 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000524602 | ENST00000356457 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000379449 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000389204 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000445642 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000517661 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000517847 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000517856 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000517903 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000518068 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000522603 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000522835 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000522919 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000524602 | ENST00000523897 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000356457 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000356457 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000379449 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000379449 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000389204 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000389204 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000445642 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000445642 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000517661 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000517661 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000517847 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000517847 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000517856 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000517856 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000517903 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000517903 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000518068 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000518068 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000522603 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000522603 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000522835 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000522835 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000522919 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000522919 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
| intron-intron | ENST00000529472 | ENST00000523897 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - |
| intron-intron | ENST00000529472 | ENST00000523897 | CHD7 | chr8 | 61655656 | + | ASPH | chr8 | 62596747 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000423902 | CHD7 | chr8 | 61693989 | + | ENST00000379454 | ASPH | chr8 | 62416068 | - | 4856 | 2575 | 446 | 2725 | 759 |
| ENST00000525508 | CHD7 | chr8 | 61693989 | + | ENST00000379454 | ASPH | chr8 | 62416068 | - | 4377 | 2096 | 0 | 2246 | 748 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000423902 | ENST00000379454 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - | 0.002884837 | 0.99711514 |
| ENST00000525508 | ENST00000379454 | CHD7 | chr8 | 61693989 | + | ASPH | chr8 | 62416068 | - | 0.002561413 | 0.99743855 |
Top |
Fusion Genomic Features for CHD7-ASPH |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
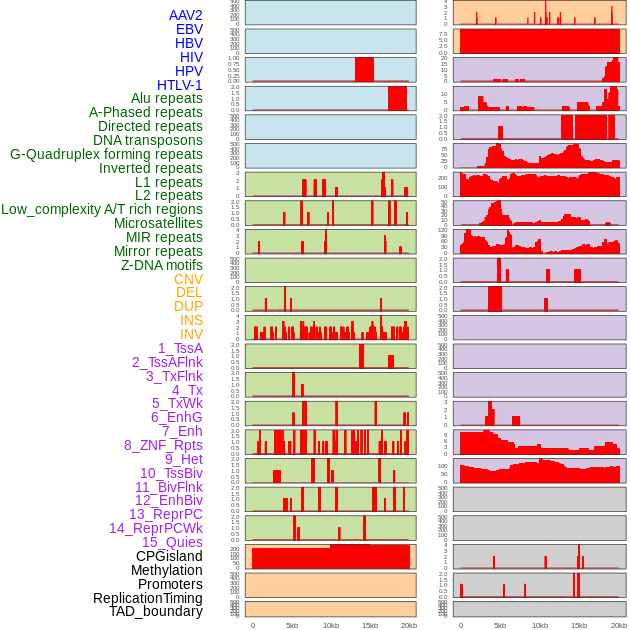
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CHD7-ASPH |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:61655656/chr8:62596747) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHD7 | ASPH |
| FUNCTION: Probable transcription regulator. Maybe involved in the in 45S precursor rRNA production. {ECO:0000269|PubMed:22646239}. | FUNCTION: May function as 2-oxoglutarate-dependent dioxygenase. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 151_222 | 698 | 2998.0 | Compositional bias | Note=Gln-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 383_568 | 698 | 2998.0 | Compositional bias | Note=Pro-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 151_222 | 698 | 1139.0 | Compositional bias | Note=Gln-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 383_568 | 698 | 1139.0 | Compositional bias | Note=Pro-rich |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 90_102 | 0 | 314.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 90_102 | 0 | 204.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 90_102 | 0 | 226.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 90_102 | 0 | 651.6666666666666 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 90_102 | 0 | 300.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 90_102 | 0 | 299.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 90_102 | 0 | 271.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 90_102 | 0 | 211.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 90_102 | 0 | 257.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 111_312 | 0 | 314.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 323_332 | 0 | 314.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 9_29 | 0 | 314.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 111_312 | 0 | 204.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 323_332 | 0 | 204.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 9_29 | 0 | 204.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 111_312 | 0 | 226.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 323_332 | 0 | 226.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 9_29 | 0 | 226.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 111_312 | 0 | 651.6666666666666 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 323_332 | 0 | 651.6666666666666 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 9_29 | 0 | 651.6666666666666 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 111_312 | 0 | 300.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 323_332 | 0 | 300.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 9_29 | 0 | 300.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 111_312 | 0 | 299.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 323_332 | 0 | 299.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 9_29 | 0 | 299.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 111_312 | 0 | 271.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 323_332 | 0 | 271.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 9_29 | 0 | 271.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 111_312 | 0 | 211.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 323_332 | 0 | 211.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 9_29 | 0 | 211.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 111_312 | 0 | 257.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 323_332 | 0 | 257.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 9_29 | 0 | 257.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 688_690 | 0 | 314.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 688_690 | 0 | 204.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 688_690 | 0 | 226.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 688_690 | 0 | 651.6666666666666 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 688_690 | 0 | 300.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 688_690 | 0 | 299.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 688_690 | 0 | 271.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 688_690 | 0 | 211.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 688_690 | 0 | 257.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 688_690 | 679 | 730.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 341_374 | 0 | 314.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 454_487 | 0 | 314.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 489_521 | 0 | 314.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 525_557 | 0 | 314.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 341_374 | 0 | 204.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 454_487 | 0 | 204.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 489_521 | 0 | 204.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 525_557 | 0 | 204.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 341_374 | 0 | 226.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 454_487 | 0 | 226.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 489_521 | 0 | 226.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 525_557 | 0 | 226.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 341_374 | 0 | 651.6666666666666 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 454_487 | 0 | 651.6666666666666 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 489_521 | 0 | 651.6666666666666 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 525_557 | 0 | 651.6666666666666 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 341_374 | 0 | 300.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 454_487 | 0 | 300.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 489_521 | 0 | 300.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 525_557 | 0 | 300.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 341_374 | 0 | 299.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 454_487 | 0 | 299.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 489_521 | 0 | 299.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 525_557 | 0 | 299.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 341_374 | 0 | 271.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 454_487 | 0 | 271.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 489_521 | 0 | 271.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 525_557 | 0 | 271.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 341_374 | 0 | 211.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 454_487 | 0 | 211.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 489_521 | 0 | 211.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 525_557 | 0 | 211.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 341_374 | 0 | 257.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 454_487 | 0 | 257.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 489_521 | 0 | 257.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 525_557 | 0 | 257.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 1_53 | 0 | 314.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 75_758 | 0 | 314.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 1_53 | 0 | 204.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 75_758 | 0 | 204.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 1_53 | 0 | 226.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 75_758 | 0 | 226.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 1_53 | 0 | 651.6666666666666 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 75_758 | 0 | 651.6666666666666 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 1_53 | 0 | 300.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 75_758 | 0 | 300.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 1_53 | 0 | 299.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 75_758 | 0 | 299.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 1_53 | 0 | 271.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 75_758 | 0 | 271.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 1_53 | 0 | 211.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 75_758 | 0 | 211.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 1_53 | 0 | 257.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 75_758 | 0 | 257.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000356457 | 0 | 14 | 54_74 | 0 | 314.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379449 | 0 | 5 | 54_74 | 0 | 204.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000389204 | 0 | 6 | 54_74 | 0 | 226.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000445642 | 0 | 16 | 54_74 | 0 | 651.6666666666666 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517847 | 0 | 15 | 54_74 | 0 | 300.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000517903 | 0 | 15 | 54_74 | 0 | 299.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000518068 | 0 | 13 | 54_74 | 0 | 271.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522603 | 0 | 5 | 54_74 | 0 | 211.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000522835 | 0 | 14 | 54_74 | 0 | 257.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 2401_2431 | 698 | 2998.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 2401_2431 | 0 | 949.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 2401_2431 | 698 | 1139.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 1939_1945 | 698 | 2998.0 | Compositional bias | Note=Poly-Arg |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 2165_2258 | 698 | 2998.0 | Compositional bias | Note=Glu-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 2398_2405 | 698 | 2998.0 | Compositional bias | Note=Poly-Arg |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 2726_2736 | 698 | 2998.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 597_718 | 698 | 2998.0 | Compositional bias | Note=Lys-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 151_222 | 0 | 949.0 | Compositional bias | Note=Gln-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 1939_1945 | 0 | 949.0 | Compositional bias | Note=Poly-Arg |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 2165_2258 | 0 | 949.0 | Compositional bias | Note=Glu-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 2398_2405 | 0 | 949.0 | Compositional bias | Note=Poly-Arg |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 2726_2736 | 0 | 949.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 383_568 | 0 | 949.0 | Compositional bias | Note=Pro-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 597_718 | 0 | 949.0 | Compositional bias | Note=Lys-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 1939_1945 | 698 | 1139.0 | Compositional bias | Note=Poly-Arg |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 2165_2258 | 698 | 1139.0 | Compositional bias | Note=Glu-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 2398_2405 | 698 | 1139.0 | Compositional bias | Note=Poly-Arg |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 2726_2736 | 698 | 1139.0 | Compositional bias | Note=Poly-Ala |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 597_718 | 698 | 1139.0 | Compositional bias | Note=Lys-rich |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 1294_1464 | 698 | 2998.0 | Domain | Helicase C-terminal |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 800_867 | 698 | 2998.0 | Domain | Chromo 1 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 882_947 | 698 | 2998.0 | Domain | Chromo 2 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 980_1154 | 698 | 2998.0 | Domain | Helicase ATP-binding |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 1294_1464 | 0 | 949.0 | Domain | Helicase C-terminal |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 800_867 | 0 | 949.0 | Domain | Chromo 1 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 882_947 | 0 | 949.0 | Domain | Chromo 2 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 980_1154 | 0 | 949.0 | Domain | Helicase ATP-binding |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 1294_1464 | 698 | 1139.0 | Domain | Helicase C-terminal |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 800_867 | 698 | 1139.0 | Domain | Chromo 1 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 882_947 | 698 | 1139.0 | Domain | Chromo 2 |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 980_1154 | 698 | 1139.0 | Domain | Helicase ATP-binding |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 1105_1108 | 698 | 2998.0 | Motif | Note=DEAH box |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 1105_1108 | 0 | 949.0 | Motif | Note=DEAH box |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 1105_1108 | 698 | 1139.0 | Motif | Note=DEAH box |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000423902 | + | 3 | 38 | 993_1000 | 698 | 2998.0 | Nucleotide binding | ATP |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000524602 | + | 1 | 5 | 993_1000 | 0 | 949.0 | Nucleotide binding | ATP |
| Hgene | CHD7 | chr8:61693989 | chr8:62416068 | ENST00000525508 | + | 2 | 12 | 993_1000 | 698 | 1139.0 | Nucleotide binding | ATP |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 90_102 | 708 | 759.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 90_102 | 679 | 730.0 | Calcium binding | Ontology_term=ECO:0000305 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 111_312 | 708 | 759.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 323_332 | 708 | 759.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 9_29 | 708 | 759.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 111_312 | 679 | 730.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 323_332 | 679 | 730.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 9_29 | 679 | 730.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 688_690 | 708 | 759.0 | Region | 2-oxoglutarate binding | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 341_374 | 708 | 759.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 454_487 | 708 | 759.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 489_521 | 708 | 759.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 525_557 | 708 | 759.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 341_374 | 679 | 730.0 | Repeat | Note=TPR 1 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 454_487 | 679 | 730.0 | Repeat | Note=TPR 2 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 489_521 | 679 | 730.0 | Repeat | Note=TPR 3 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 525_557 | 679 | 730.0 | Repeat | Note=TPR 4 | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 1_53 | 708 | 759.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 75_758 | 708 | 759.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 1_53 | 679 | 730.0 | Topological domain | Cytoplasmic | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 75_758 | 679 | 730.0 | Topological domain | Lumenal | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000379454 | 23 | 25 | 54_74 | 708 | 759.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | ASPH | chr8:61693989 | chr8:62416068 | ENST00000541428 | 23 | 25 | 54_74 | 679 | 730.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for CHD7-ASPH |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16372_16372_1_CHD7-ASPH_CHD7_chr8_61693989_ENST00000423902_ASPH_chr8_62416068_ENST00000379454_length(transcript)=4856nt_BP=2575nt CGGCGGCGGCGGCAGCGGCGGCGGCGGCGGCGGCGCGGGGGTTGAGTCGTGGTGGTGCGGACGCGCTCGTGCTCGGGAACTATCGGATTA AACTTGAATCGAGTGAAATTACACAAAGGAGCGCCGCGGAGGAGGCGGCCCGGGGACCCGGACACCCTGAAACTCACCAGAGACCCGTTC GCCCCCGGCCAACTCCGTGCCCGTGGATTCAGCCCCCTGGCCGCAGCTGCCGAGCCAACTCCGGAGCCCGCTCTGCGTTTTGTTTTCCCC TCGGCACTAGGCAGCGGAGGAGCCCGACCGACCCGGACCTATATCCAGACTTTGCCTGACACTGCAGGGTCCAAGAGAATTAAAGAAATA TGGAATGACATGAAGAAGATTAGTTAAGGATTATAGGCTTTGAGGGCAAACACCTCAGTGAAGTGAAGCACAGGCAAGCTCCTGAGCTGT GGTTTGGAGGAGCCGTGTGTTGGAAGAAGATGGCAGATCCAGGAATGATGAGTCTTTTTGGCGAGGATGGGAATATTTTCAGTGAAGGTC TTGAAGGCCTCGGAGAATGTGGTTACCCGGAAAATCCAGTAAATCCTATGGGTCAGCAAATGCCAATAGACCAAGGCTTTGCCTCTTTAC AGCCATCCCTTCATCATCCTTCAACTAATCAAAATCAAACAAAGCTGACACATTTTGATCACTATAATCAGTATGAACAACAAAAGATGC ATCTGATGGATCAGCCGAACAGAATGATGAGCAACACCCCTGGGAACGGACTCGCGTCTCCGCACTCGCAGTATCACACCCCTCCCGTTC CTCAGGTGCCCCATGGTGGCAGTGGTGGCGGTCAGATGGGTGTCTACCCTGGCATGCAGAATGAGAGGCATGGGCAATCCTTTGTGGACA GCAGCTCCATGTGGGGCCCCAGGGCTGTTCAGGTACCAGACCAGATACGAGCCCCCTACCAGCAGCAGCAGCCACAGCCGCAGCCACCGC AGCCGGCTCCGTCGGGGCCCCCTGCACAGGGCCACCCTCAGCACATGCAGCAGATGGGCAGCTATATGGCACGTGGGGATTTTTCCATGC AGCAGCATGGTCAGCCACAGCAGAGGATGAGCCAGTTTTCCCAAGGCCAAGAGGGCCTCAATCAGGGAAATCCTTTTATTGCCACCTCAG GACCTGGCCACTTGTCCCACGTGCCCCAGCAGAGTCCCAGCATGGCACCTTCCTTGCGTCACTCGGTGCAGCAGTTCCATCACCACCCCT CTACTGCTCTCCATGGAGAATCCGTTGCCCACAGTCCCAGATTCTCCCCGAATCCTCCCCAACAAGGGGCTGTTAGGCCGCAAACCCTTA ACTTTAGTTCTCGGAGCCAGACAGTCCCCTCTCCTACTATAAACAACTCAGGGCAGTATTCTCGATATCCTTACAGTAACCTAAATCAGG GATTAGTTAACAATACAGGGATGAATCAAAATTTAGGCCTTACAAATAATACTCCAATGAATCAGTCCGTACCAAGATACCCCAATGCTG TAGGATTCCCATCAAACAGTGGTCAAGGACTAATGCACCAGCAGCCCATCCACCCCAGTGGCTCACTTAACCAAATGAACACACAAACTA TGCATCCTTCACAGCCTCAGGGAACTTATGCCTCTCCACCTCCCATGTCACCCATGAAAGCAATGAGTAATCCAGCAGGCACTCCTCCTC CACAAGTCAGGCCGGGAAGTGCTGGGATACCAATGGAAGTTGGCAGTTATCCAAATATGCCCCATCCTCAGCCATCTCACCAGCCCCCTG GTGCCATGGGAATCGGACAGAGGAATATGGGCCCCAGAAACATGCAGCAGTCTCGTCCATTTATAGGCATGTCCTCGGCACCAAGGGAAT TGACTGGGCACATGAGGCCAAATGGTTGTCCTGGTGTTGGCCTTGGAGACCCACAAGCAATCCAGGAACGACTGATACCTGGCCAACAAC ATCCTGGTCAACAGCCATCTTTTCAGCAGTTGCCAACCTGTCCTCCACTGCAGCCTCACCCGGGCTTGCACCACCAGTCTTCACCTCCAC ACCCTCATCACCAGCCTTGGGCACAGCTCCACCCATCACCCCAGAACACCCCGCAGAAAGTGCCTGTGCATCAGCATTCCCCGTCGGAGC CCTTTCTAGAGAAACCAGTGCCGGATATGACTCAGGTTAGTGGACCGAATGCTCAGCTAGTGAAGAGTGATGATTACCTGCCATCAATAG AACAGCAGCCACAACAAAAGAAGAAGAAAAAGAAAAACAACCACATTGTAGCAGAGGATCCCAGTAAAGGTTTTGGTAAAGATGACTTCC CTGGTGGGGTAGATAACCAAGAACTAAATAGGAACTCACTGGATGGGTCCCAAGAAGAAAAAAAGAAAAAGAAAAGGTCAAAGGCAAAAA AAGACCCGAAGGAACCGAAAGAACCCAAGGAGAAAAAAGAGCCCAAGGAACCCAAGACCCCGAAAGCCCCTAAGATTCCCAAAGAGCCAA AGGAAAAGAAAGCAAAAACTGCCACGCCAAAACCCAAATCCAGCAAAAAGTCAAGGACCTGGGAGGAAGGCAAGGTGCTCATCTTTGATG ACTCCTTTGAGCACGAGGTATGGCAGGATGCCTCATCTTTCCGGCTGATATTCATCGTGGATGTGTGGCATCCGGAACTGACACCACAGC AGAGACGCAGCCTTCCAGCAATTTAGCATGAATTCATGCAAGCTTGGGAAACTCTGGAGAGAGGCTGCCTTTCTGGTTCCATCTCCTTGG GTGTGAGGATAGAATTTCGAACACCAAGAGTCAATTCCCTTGACTTGCAGCCCGAGTAATTCAAAGCCTCCTCCTAGGGTCAGAAGACAC TAAAGGGAATATTTGCCTCGCTGCAATTCATTTAGGAAACACCCTGCTGTGTGTCATCTCATGACAGCACTGGTCTTCTGCCAGTATTTA AGGTGAACATTTGATAGCTTCTACCTTACCAGCCAAAGATATTTTTTCCACATAGAATAGGTCTAATTCAATGTATAATGAGAACATATG TAGAAACTGTGAATGGATTGCTTTAGTTTGTAATTTTTCTATGCAGTTATATTTTTCTAGTGTAGCTAGACTATTTTGTCATCATGTACC ACTACATTTTTGTTTATTTTAATGACAAGCTGTATAAATGCTTTACTTCTAGCTATTTAATGGTAGCATTACTGGGGAACTCAGACTTCC CTCTTTTAATTCTTCTTAGTAAAAGATACTCATGAAAAAAGCAGTTTTATTTTCCTAACAAAAAAGAAAGAGCTCATTATGTCAGTGTCT ATGAACTGTACCCATCCCAACTCTCAAATCGTTTGGTTTTTTTTATCTTGATTGAGATCCTCTTCTCACTATGCTAGTGGTGGAGATATT GACAAAATCCTATTTCTTTCAAAGAGGAACTTTTCACACCGAAAAAAGAGCATGGAATTATTTTATATTGTTATAAAAATCCCAGATGCA AATTTTTTTAATGCCAATTATTAGAGCTTCTGGGGAAAAAGTATAGTTCACGGAAATAAAACTATGTTCTTTCAGGGTTGGGTGGATAGG TGGCTGCTAGGGTGTCTGGCTCCTGGCGGCTTTGCCATCCATGAGGCAAGGGCTGGGAACACAGTGTCTTTGCCTATGGTAGATCCATGT GAATGTCAGGAAGCCAGCTCTTCAGTCTTGGAGATGATTTCTGCTACAATTCTGTAGAAAGATTAAGGATGGCAGAGTAAAAGGTTACCA AGAATGCCAGGATGTTTTTCTTGGGCGTAGGAGGTCCAGATTACTTTCCTTTTTGATGAAAGAGTTTGGAAGACTGTCCCATCTCTCTGG CTTGAGAAATCTCTGCCATTTTAAACATCACTGTGAAATAGCAATTATTATCATCTGTATTTAGTTTTAACATTACCCACAACATAGAAA TAATAGGTAAAAATCGTCTTGCCTACTCATTCCAAAGATGATCAAGTCATTAATCTAGCAAAGTATTCATGTATCAGATTTTGTATATTT TGAATCAAAGCTAACTAGGAATGTTAGATATAAGAATGTAATGATATTCATGCACTGAATTCTAAGCCAATATGAACAAAAATGCTGCAT GAATGGCACATATAGGTCACCAAAGTTCATTCACAGGTAGAAAAAACTTGTGCTTTCTTTTCCATCTAAAAACAAAAGGAGACTTTCTTT ATCTCATTTAAAGAACAGCTCTTTGAAATTGAAATTGACCCTTTTTGCTTGACCTTAAGGAGATTAGCTTCCAGTAGATGAGTTTGCAAA ATACTTTTCCTGTTCTTTTGTTTTGCTGGTATTGAAAACATCCCACTAAATCAGATGAAGAGGCATGGGAGGAAAAATATCCAAATTAAT TACTAAAATCGAGAAGAGAAGGCAAACTCTTGAAAAGTAAAAAGGTGTTTGTGACCTTCAGTATTTATTGAACAGAGGAAATAACTGACA AGGGCAATACAATTCAATGTTCATGTAGTAACATTCATGTCACTTGTTGAATTTGGTTCTCATATGTATATTGCATACACATAAATTCAA ACTATAAGTCGTCATTTTTGAGCCATCATCTTACATTCATGTAATGAAATTATGGAAGAGAGTAAAAACTAGCTCTTAACTTAGTAAATA TAATATGGTATTTAAAATCAGGTCACTACAGTAAGGTTCTAAGTATTGCCAATTGAAAAGCTAGAAATGGTATTACTGTTGCAAAGTGTT >16372_16372_1_CHD7-ASPH_CHD7_chr8_61693989_ENST00000423902_ASPH_chr8_62416068_ENST00000379454_length(amino acids)=759AA_BP=704 MWFGGAVCWKKMADPGMMSLFGEDGNIFSEGLEGLGECGYPENPVNPMGQQMPIDQGFASLQPSLHHPSTNQNQTKLTHFDHYNQYEQQK MHLMDQPNRMMSNTPGNGLASPHSQYHTPPVPQVPHGGSGGGQMGVYPGMQNERHGQSFVDSSSMWGPRAVQVPDQIRAPYQQQQPQPQP PQPAPSGPPAQGHPQHMQQMGSYMARGDFSMQQHGQPQQRMSQFSQGQEGLNQGNPFIATSGPGHLSHVPQQSPSMAPSLRHSVQQFHHH PSTALHGESVAHSPRFSPNPPQQGAVRPQTLNFSSRSQTVPSPTINNSGQYSRYPYSNLNQGLVNNTGMNQNLGLTNNTPMNQSVPRYPN AVGFPSNSGQGLMHQQPIHPSGSLNQMNTQTMHPSQPQGTYASPPPMSPMKAMSNPAGTPPPQVRPGSAGIPMEVGSYPNMPHPQPSHQP PGAMGIGQRNMGPRNMQQSRPFIGMSSAPRELTGHMRPNGCPGVGLGDPQAIQERLIPGQQHPGQQPSFQQLPTCPPLQPHPGLHHQSSP PHPHHQPWAQLHPSPQNTPQKVPVHQHSPSEPFLEKPVPDMTQVSGPNAQLVKSDDYLPSIEQQPQQKKKKKKNNHIVAEDPSKGFGKDD FPGGVDNQELNRNSLDGSQEEKKKKKRSKAKKDPKEPKEPKEKKEPKEPKTPKAPKIPKEPKEKKAKTATPKPKSSKKSRTWEEGKVLIF -------------------------------------------------------------- >16372_16372_2_CHD7-ASPH_CHD7_chr8_61693989_ENST00000525508_ASPH_chr8_62416068_ENST00000379454_length(transcript)=4377nt_BP=2096nt ATGGCAGATCCAGGAATGATGAGTCTTTTTGGCGAGGATGGGAATATTTTCAGTGAAGGTCTTGAAGGCCTCGGAGAATGTGGTTACCCG GAAAATCCAGTAAATCCTATGGGTCAGCAAATGCCAATAGACCAAGGCTTTGCCTCTTTACAGCCATCCCTTCATCATCCTTCAACTAAT CAAAATCAAACAAAGCTGACACATTTTGATCACTATAATCAGTATGAACAACAAAAGATGCATCTGATGGATCAGCCGAACAGAATGATG AGCAACACCCCTGGGAACGGACTCGCGTCTCCGCACTCGCAGTATCACACCCCTCCCGTTCCTCAGGTGCCCCATGGTGGCAGTGGTGGC GGTCAGATGGGTGTCTACCCTGGCATGCAGAATGAGAGGCATGGGCAATCCTTTGTGGACAGCAGCTCCATGTGGGGCCCCAGGGCTGTT CAGGTACCAGACCAGATACGAGCCCCCTACCAGCAGCAGCAGCCACAGCCGCAGCCACCGCAGCCGGCTCCGTCGGGGCCCCCTGCACAG GGCCACCCTCAGCACATGCAGCAGATGGGCAGCTATATGGCACGTGGGGATTTTTCCATGCAGCAGCATGGTCAGCCACAGCAGAGGATG AGCCAGTTTTCCCAAGGCCAAGAGGGCCTCAATCAGGGAAATCCTTTTATTGCCACCTCAGGACCTGGCCACTTGTCCCACGTGCCCCAG CAGAGTCCCAGCATGGCACCTTCCTTGCGTCACTCGGTGCAGCAGTTCCATCACCACCCCTCTACTGCTCTCCATGGAGAATCCGTTGCC CACAGTCCCAGATTCTCCCCGAATCCTCCCCAACAAGGGGCTGTTAGGCCGCAAACCCTTAACTTTAGTTCTCGGAGCCAGACAGTCCCC TCTCCTACTATAAACAACTCAGGGCAGTATTCTCGATATCCTTACAGTAACCTAAATCAGGGATTAGTTAACAATACAGGGATGAATCAA AATTTAGGCCTTACAAATAATACTCCAATGAATCAGTCCGTACCAAGATACCCCAATGCTGTAGGATTCCCATCAAACAGTGGTCAAGGA CTAATGCACCAGCAGCCCATCCACCCCAGTGGCTCACTTAACCAAATGAACACACAAACTATGCATCCTTCACAGCCTCAGGGAACTTAT GCCTCTCCACCTCCCATGTCACCCATGAAAGCAATGAGTAATCCAGCAGGCACTCCTCCTCCACAAGTCAGGCCGGGAAGTGCTGGGATA CCAATGGAAGTTGGCAGTTATCCAAATATGCCCCATCCTCAGCCATCTCACCAGCCCCCTGGTGCCATGGGAATCGGACAGAGGAATATG GGCCCCAGAAACATGCAGCAGTCTCGTCCATTTATAGGCATGTCCTCGGCACCAAGGGAATTGACTGGGCACATGAGGCCAAATGGTTGT CCTGGTGTTGGCCTTGGAGACCCACAAGCAATCCAGGAACGACTGATACCTGGCCAACAACATCCTGGTCAACAGCCATCTTTTCAGCAG TTGCCAACCTGTCCTCCACTGCAGCCTCACCCGGGCTTGCACCACCAGTCTTCACCTCCACACCCTCATCACCAGCCTTGGGCACAGCTC CACCCATCACCCCAGAACACCCCGCAGAAAGTGCCTGTGCATCAGCATTCCCCGTCGGAGCCCTTTCTAGAGAAACCAGTGCCGGATATG ACTCAGGTTAGTGGACCGAATGCTCAGCTAGTGAAGAGTGATGATTACCTGCCATCAATAGAACAGCAGCCACAACAAAAGAAGAAGAAA AAGAAAAACAACCACATTGTAGCAGAGGATCCCAGTAAAGGTTTTGGTAAAGATGACTTCCCTGGTGGGGTAGATAACCAAGAACTAAAT AGGAACTCACTGGATGGGTCCCAAGAAGAAAAAAAGAAAAAGAAAAGGTCAAAGGCAAAAAAAGACCCGAAGGAACCGAAAGAACCCAAG GAGAAAAAAGAGCCCAAGGAACCCAAGACCCCGAAAGCCCCTAAGATTCCCAAAGAGCCAAAGGAAAAGAAAGCAAAAACTGCCACGCCA AAACCCAAATCCAGCAAAAAGTCAAGGACCTGGGAGGAAGGCAAGGTGCTCATCTTTGATGACTCCTTTGAGCACGAGGTATGGCAGGAT GCCTCATCTTTCCGGCTGATATTCATCGTGGATGTGTGGCATCCGGAACTGACACCACAGCAGAGACGCAGCCTTCCAGCAATTTAGCAT GAATTCATGCAAGCTTGGGAAACTCTGGAGAGAGGCTGCCTTTCTGGTTCCATCTCCTTGGGTGTGAGGATAGAATTTCGAACACCAAGA GTCAATTCCCTTGACTTGCAGCCCGAGTAATTCAAAGCCTCCTCCTAGGGTCAGAAGACACTAAAGGGAATATTTGCCTCGCTGCAATTC ATTTAGGAAACACCCTGCTGTGTGTCATCTCATGACAGCACTGGTCTTCTGCCAGTATTTAAGGTGAACATTTGATAGCTTCTACCTTAC CAGCCAAAGATATTTTTTCCACATAGAATAGGTCTAATTCAATGTATAATGAGAACATATGTAGAAACTGTGAATGGATTGCTTTAGTTT GTAATTTTTCTATGCAGTTATATTTTTCTAGTGTAGCTAGACTATTTTGTCATCATGTACCACTACATTTTTGTTTATTTTAATGACAAG CTGTATAAATGCTTTACTTCTAGCTATTTAATGGTAGCATTACTGGGGAACTCAGACTTCCCTCTTTTAATTCTTCTTAGTAAAAGATAC TCATGAAAAAAGCAGTTTTATTTTCCTAACAAAAAAGAAAGAGCTCATTATGTCAGTGTCTATGAACTGTACCCATCCCAACTCTCAAAT CGTTTGGTTTTTTTTATCTTGATTGAGATCCTCTTCTCACTATGCTAGTGGTGGAGATATTGACAAAATCCTATTTCTTTCAAAGAGGAA CTTTTCACACCGAAAAAAGAGCATGGAATTATTTTATATTGTTATAAAAATCCCAGATGCAAATTTTTTTAATGCCAATTATTAGAGCTT CTGGGGAAAAAGTATAGTTCACGGAAATAAAACTATGTTCTTTCAGGGTTGGGTGGATAGGTGGCTGCTAGGGTGTCTGGCTCCTGGCGG CTTTGCCATCCATGAGGCAAGGGCTGGGAACACAGTGTCTTTGCCTATGGTAGATCCATGTGAATGTCAGGAAGCCAGCTCTTCAGTCTT GGAGATGATTTCTGCTACAATTCTGTAGAAAGATTAAGGATGGCAGAGTAAAAGGTTACCAAGAATGCCAGGATGTTTTTCTTGGGCGTA GGAGGTCCAGATTACTTTCCTTTTTGATGAAAGAGTTTGGAAGACTGTCCCATCTCTCTGGCTTGAGAAATCTCTGCCATTTTAAACATC ACTGTGAAATAGCAATTATTATCATCTGTATTTAGTTTTAACATTACCCACAACATAGAAATAATAGGTAAAAATCGTCTTGCCTACTCA TTCCAAAGATGATCAAGTCATTAATCTAGCAAAGTATTCATGTATCAGATTTTGTATATTTTGAATCAAAGCTAACTAGGAATGTTAGAT ATAAGAATGTAATGATATTCATGCACTGAATTCTAAGCCAATATGAACAAAAATGCTGCATGAATGGCACATATAGGTCACCAAAGTTCA TTCACAGGTAGAAAAAACTTGTGCTTTCTTTTCCATCTAAAAACAAAAGGAGACTTTCTTTATCTCATTTAAAGAACAGCTCTTTGAAAT TGAAATTGACCCTTTTTGCTTGACCTTAAGGAGATTAGCTTCCAGTAGATGAGTTTGCAAAATACTTTTCCTGTTCTTTTGTTTTGCTGG TATTGAAAACATCCCACTAAATCAGATGAAGAGGCATGGGAGGAAAAATATCCAAATTAATTACTAAAATCGAGAAGAGAAGGCAAACTC TTGAAAAGTAAAAAGGTGTTTGTGACCTTCAGTATTTATTGAACAGAGGAAATAACTGACAAGGGCAATACAATTCAATGTTCATGTAGT AACATTCATGTCACTTGTTGAATTTGGTTCTCATATGTATATTGCATACACATAAATTCAAACTATAAGTCGTCATTTTTGAGCCATCAT CTTACATTCATGTAATGAAATTATGGAAGAGAGTAAAAACTAGCTCTTAACTTAGTAAATATAATATGGTATTTAAAATCAGGTCACTAC AGTAAGGTTCTAAGTATTGCCAATTGAAAAGCTAGAAATGGTATTACTGTTGCAAAGTGTTGTCAATAATTGACTCCAATAGCATTGTAA >16372_16372_2_CHD7-ASPH_CHD7_chr8_61693989_ENST00000525508_ASPH_chr8_62416068_ENST00000379454_length(amino acids)=748AA_BP=693 MADPGMMSLFGEDGNIFSEGLEGLGECGYPENPVNPMGQQMPIDQGFASLQPSLHHPSTNQNQTKLTHFDHYNQYEQQKMHLMDQPNRMM SNTPGNGLASPHSQYHTPPVPQVPHGGSGGGQMGVYPGMQNERHGQSFVDSSSMWGPRAVQVPDQIRAPYQQQQPQPQPPQPAPSGPPAQ GHPQHMQQMGSYMARGDFSMQQHGQPQQRMSQFSQGQEGLNQGNPFIATSGPGHLSHVPQQSPSMAPSLRHSVQQFHHHPSTALHGESVA HSPRFSPNPPQQGAVRPQTLNFSSRSQTVPSPTINNSGQYSRYPYSNLNQGLVNNTGMNQNLGLTNNTPMNQSVPRYPNAVGFPSNSGQG LMHQQPIHPSGSLNQMNTQTMHPSQPQGTYASPPPMSPMKAMSNPAGTPPPQVRPGSAGIPMEVGSYPNMPHPQPSHQPPGAMGIGQRNM GPRNMQQSRPFIGMSSAPRELTGHMRPNGCPGVGLGDPQAIQERLIPGQQHPGQQPSFQQLPTCPPLQPHPGLHHQSSPPHPHHQPWAQL HPSPQNTPQKVPVHQHSPSEPFLEKPVPDMTQVSGPNAQLVKSDDYLPSIEQQPQQKKKKKKNNHIVAEDPSKGFGKDDFPGGVDNQELN RNSLDGSQEEKKKKKRSKAKKDPKEPKEPKEKKEPKEPKTPKAPKIPKEPKEKKAKTATPKPKSSKKSRTWEEGKVLIFDDSFEHEVWQD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHD7-ASPH |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHD7-ASPH |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHD7-ASPH |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
