
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHM-KLHL4 (FusionGDB2 ID:16519) |
Fusion Gene Summary for CHM-KLHL4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHM-KLHL4 | Fusion gene ID: 16519 | Hgene | Tgene | Gene symbol | CHM | KLHL4 | Gene ID | 4359 | 56062 |
| Gene name | myelin protein zero | kelch like family member 4 | |
| Synonyms | CHM|CHN2|CMT1|CMT1B|CMT2I|CMT2J|CMT4E|CMTDI3|CMTDID|DSS|HMSNIB|MPP|P0 | DKELCHL|KHL4 | |
| Cytomap | 1q23.3 | Xq21.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | myelin protein P0Charcot-Marie-Tooth neuropathy 1Bmyelin peripheral protein | kelch-like protein 4 | |
| Modification date | 20200328 | 20200313 | |
| UniProtAcc | P24386 | Q9P2K6 | |
| Ensembl transtripts involved in fusion gene | ENST00000467744, ENST00000357749, ENST00000358786, ENST00000537751, | ENST00000373114, ENST00000373119, | |
| Fusion gene scores | * DoF score | 7 X 8 X 4=224 | 1 X 1 X 1=1 |
| # samples | 8 | 1 | |
| ** MAII score | log2(8/224*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: CHM [Title/Abstract] AND KLHL4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CHM(85282495)-KLHL4(86868880), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CHM (5'-gene) Fusion gene breakpoints across CHM (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
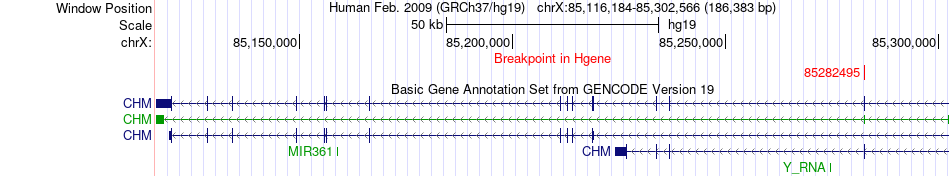 |
 Fusion gene breakpoints across KLHL4 (3'-gene) Fusion gene breakpoints across KLHL4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
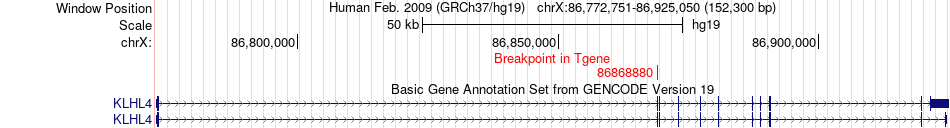 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CR-7367-01A | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
Top |
Fusion Gene ORF analysis for CHM-KLHL4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000467744 | ENST00000373114 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| 5UTR-3CDS | ENST00000467744 | ENST00000373119 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| In-frame | ENST00000357749 | ENST00000373114 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| In-frame | ENST00000357749 | ENST00000373119 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| In-frame | ENST00000358786 | ENST00000373114 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| In-frame | ENST00000358786 | ENST00000373119 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| intron-3CDS | ENST00000537751 | ENST00000373114 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
| intron-3CDS | ENST00000537751 | ENST00000373119 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000357749 | CHM | chrX | 85282495 | - | ENST00000373119 | KLHL4 | chrX | 86868880 | + | 5397 | 146 | 30 | 1880 | 616 |
| ENST00000357749 | CHM | chrX | 85282495 | - | ENST00000373114 | KLHL4 | chrX | 86868880 | + | 2089 | 146 | 30 | 1886 | 618 |
| ENST00000358786 | CHM | chrX | 85282495 | - | ENST00000373119 | KLHL4 | chrX | 86868880 | + | 5374 | 123 | 7 | 1857 | 616 |
| ENST00000358786 | CHM | chrX | 85282495 | - | ENST00000373114 | KLHL4 | chrX | 86868880 | + | 2066 | 123 | 7 | 1863 | 618 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000357749 | ENST00000373119 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + | 0.000175587 | 0.99982446 |
| ENST00000357749 | ENST00000373114 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + | 0.000937753 | 0.99906224 |
| ENST00000358786 | ENST00000373119 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + | 0.000179052 | 0.999821 |
| ENST00000358786 | ENST00000373114 | CHM | chrX | 85282495 | - | KLHL4 | chrX | 86868880 | + | 0.001030633 | 0.99896944 |
Top |
Fusion Genomic Features for CHM-KLHL4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CHM | chrX | 85282494 | - | KLHL4 | chrX | 86868879 | + | 2.08E-06 | 0.999998 |
| CHM | chrX | 85282494 | - | KLHL4 | chrX | 86868879 | + | 2.08E-06 | 0.999998 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for CHM-KLHL4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:85282495/chrX:86868880) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHM | KLHL4 |
| FUNCTION: Substrate-binding subunit of the Rab geranylgeranyltransferase (GGTase) complex. Binds unprenylated Rab proteins and presents the substrate peptide to the catalytic component B composed of RABGGTA and RABGGTB, and remains bound to it after the geranylgeranyl transfer reaction. The component A is thought to be regenerated by transferring its prenylated Rab back to the donor membrane. Besides, a pre-formed complex consisting of CHM and the Rab GGTase dimer (RGGT or component B) can bind to and prenylate Rab proteins; this alternative pathway is proposed to be the predominant pathway for Rab protein geranylgeranylation. {ECO:0000269|PubMed:18532927, ECO:0000269|PubMed:7957092}. | FUNCTION: Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex required for mitotic progression and cytokinesis. The BCR(KLHL42) E3 ubiquitin ligase complex mediates the ubiquitination and subsequent degradation of KATNA1. Involved in microtubule dynamics throughout mitosis. {ECO:0000269|PubMed:19261606}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 182_249 | 140 | 721.0 | Domain | BTB | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 182_249 | 140 | 719.0 | Domain | BTB | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 430_476 | 140 | 721.0 | Repeat | Note=Kelch 1 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 477_523 | 140 | 721.0 | Repeat | Note=Kelch 2 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 525_570 | 140 | 721.0 | Repeat | Note=Kelch 3 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 571_617 | 140 | 721.0 | Repeat | Note=Kelch 4 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 619_670 | 140 | 721.0 | Repeat | Note=Kelch 5 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373114 | 0 | 11 | 671_717 | 140 | 721.0 | Repeat | Note=Kelch 6 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 430_476 | 140 | 719.0 | Repeat | Note=Kelch 1 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 477_523 | 140 | 719.0 | Repeat | Note=Kelch 2 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 525_570 | 140 | 719.0 | Repeat | Note=Kelch 3 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 571_617 | 140 | 719.0 | Repeat | Note=Kelch 4 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 619_670 | 140 | 719.0 | Repeat | Note=Kelch 5 | |
| Tgene | KLHL4 | chrX:85282495 | chrX:86868880 | ENST00000373119 | 0 | 11 | 671_717 | 140 | 719.0 | Repeat | Note=Kelch 6 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for CHM-KLHL4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16519_16519_1_CHM-KLHL4_CHM_chrX_85282495_ENST00000357749_KLHL4_chrX_86868880_ENST00000373114_length(transcript)=2089nt_BP=146nt TAATAGTCACATGACACGTTTCCCGTCAAGATGGCGGATACTCTCCCTTCGGAGTTTGATGTGATCGTAATAGGGACGGGTTTGCCTGAA TCCATCATTGCAGCTGCATGTTCAAGAAGTGGCCGGAGAGTTCTGCATGTTGATTCATTAGATACACAACACTCTGAAGACATGAATGCC ACCAGATCTGAAGAGCAGTTCCATGTTATAAACCACGCAGAGCAAACTCTTCGTAAAATGGAGAACTACTTGAAAGAGAAACAACTATGT GATGTGCTACTGATTGCAGGACACCTCCGCATCCCAGCCCATAGGTTGGTTCTCAGCGCAGTGTCTGATTATTTTGCTGCAATGTTTACT AATGATGTGCTTGAAGCCAAACAAGAAGAGGTCAGGATGGAAGGAGTAGATCCAAATGCACTAAATTCCTTGGTGCAGTATGCTTACACA GGAGTCCTGCAATTGAAAGAAGATACCATTGAAAGTTTGCTGGCTGCAGCTTGTCTTCTGCAGCTGACTCAGGTCATTGATGTTTGCTCC AATTTTCTCATAAAGCAGCTCCATCCTTCAAACTGCTTAGGGATTCGATCATTTGGAGATGCCCAAGGCTGTACAGAACTTCTGAACGTG GCACACAAATACACTATGGAACACTTCATTGAGGTAATAAAAAACCAAGAATTCCTCCTGCTTCCAGCTAATGAAATTTCAAAACTTCTG TGCAGTGATGACATTAATGTGCCTGATGAAGAGACCATTTTTCATGCTCTAATGCAGTGGGTGGGGCATGATGTGCAGAATAGGCAAGGA GAACTGGGGATGCTGCTTTCTTACATCAGACTGCCATTACTCCCACCACAGTTACTGGCAGATCTTGAAACCAGTTCCATGTTTACTGGT GATCTTGAGTGTCAGAAGCTCCTGATGGAAGCTATGAAGTATCATCTTTTGCCTGAGAGAAGATCCATGATGCAAAGCCCTCGGACAAAG CCTAGAAAATCAACTGTGGGGGCACTTTATGCTGTAGGAGGCATGGATGCTATGAAAGGTACTACTACTATTGAAAAATATGACCTCAGG ACCAACAGTTGGCTACATATTGGCACCATGAATGGCCGTAGGCTTCAATTTGGAGTCGCAGTTATTGATAATAAGCTCTATGTCGTGGGA GGAAGAGACGGTTTAAAAACTTTGAATACAGTGGAATGTTTTAATCCAGTTGGCAAAATCTGGACTGTGATGCCTCCCATGTCAACACAT CGGCACGGCTTAGGTGTAGCCACTCTTGAAGGACCAATGTATGCTGTAGGTGGTCATGATGGATGGAGCTATCTAAATACTGTAGAAAGA TGGGACCCTGAGGGACGACAGTGGAATTACGTAGCCAGTATGTCAACTCCTAGAAGCACAGTTGGTGTTGTTGCATTAAACAACAAATTA TATGCTATTGGTGGACGTGATGGAAGTTCCTGCCTCAAATCAATGGAATACTTTGACCCACACACTAACAAGTGGAGTTTGTGTGCTCCA ATGTCCAAAAGACGTGGAGGTGTGGGAGTTGCCACATACAATGGATTCTTATATGTTGTAGGGGGGCATGATGCCCCTGCTTCCAACCAT TGCTCCAGGCTTTCTGACTGTGTGGAACGGTATGATCCAAAAGGTGATTCATGGTCAACTGTGGCACCTCTGAGTGTTCCTCGAGATGCT GTTGCTGTGTGCCCTCTTGGAGACAAACTCTACGTGGTTGGAGGATATGACGGACATACTTATTTGAACACAGTTGAGTCATATGATGCA CAGAGAAATGAATGGAAAGAGAGCATGCAAGAACTTCTACAAAACTTCTATACCACACAGAAGCTGAAAGAGACTCTGGGGCACTAAATT CAGATCCTTTCTCCACACAGCAATGACATTTATCAATCTGACTCAGGTGTGGCTGTTGCCCCTTTTTGATATGGAGGGAACACTGAAAGA CAGTTTCCTTCTGGAGTTCCAAATGCAATATAGAAAAAAAAACCCTGTGACTCTGGATTTTATTTTAAGTTCTCTAACAAGACTCCTACC >16519_16519_1_CHM-KLHL4_CHM_chrX_85282495_ENST00000357749_KLHL4_chrX_86868880_ENST00000373114_length(amino acids)=618AA_BP=39 MADTLPSEFDVIVIGTGLPESIIAAACSRSGRRVLHVDSLDTQHSEDMNATRSEEQFHVINHAEQTLRKMENYLKEKQLCDVLLIAGHLR IPAHRLVLSAVSDYFAAMFTNDVLEAKQEEVRMEGVDPNALNSLVQYAYTGVLQLKEDTIESLLAAACLLQLTQVIDVCSNFLIKQLHPS NCLGIRSFGDAQGCTELLNVAHKYTMEHFIEVIKNQEFLLLPANEISKLLCSDDINVPDEETIFHALMQWVGHDVQNRQGELGMLLSYIR LPLLPPQLLADLETSSMFTGDLECQKLLMEAMKYHLLPERRSMMQSPRTKPRKSTVGALYAVGGMDAMKGTTTIEKYDLRTNSWLHIGTM NGRRLQFGVAVIDNKLYVVGGRDGLKTLNTVECFNPVGKIWTVMPPMSTHRHGLGVATLEGPMYAVGGHDGWSYLNTVERWDPEGRQWNY VASMSTPRSTVGVVALNNKLYAIGGRDGSSCLKSMEYFDPHTNKWSLCAPMSKRRGGVGVATYNGFLYVVGGHDAPASNHCSRLSDCVER -------------------------------------------------------------- >16519_16519_2_CHM-KLHL4_CHM_chrX_85282495_ENST00000357749_KLHL4_chrX_86868880_ENST00000373119_length(transcript)=5397nt_BP=146nt TAATAGTCACATGACACGTTTCCCGTCAAGATGGCGGATACTCTCCCTTCGGAGTTTGATGTGATCGTAATAGGGACGGGTTTGCCTGAA TCCATCATTGCAGCTGCATGTTCAAGAAGTGGCCGGAGAGTTCTGCATGTTGATTCATTAGATACACAACACTCTGAAGACATGAATGCC ACCAGATCTGAAGAGCAGTTCCATGTTATAAACCACGCAGAGCAAACTCTTCGTAAAATGGAGAACTACTTGAAAGAGAAACAACTATGT GATGTGCTACTGATTGCAGGACACCTCCGCATCCCAGCCCATAGGTTGGTTCTCAGCGCAGTGTCTGATTATTTTGCTGCAATGTTTACT AATGATGTGCTTGAAGCCAAACAAGAAGAGGTCAGGATGGAAGGAGTAGATCCAAATGCACTAAATTCCTTGGTGCAGTATGCTTACACA GGAGTCCTGCAATTGAAAGAAGATACCATTGAAAGTTTGCTGGCTGCAGCTTGTCTTCTGCAGCTGACTCAGGTCATTGATGTTTGCTCC AATTTTCTCATAAAGCAGCTCCATCCTTCAAACTGCTTAGGGATTCGATCATTTGGAGATGCCCAAGGCTGTACAGAACTTCTGAACGTG GCACACAAATACACTATGGAACACTTCATTGAGGTAATAAAAAACCAAGAATTCCTCCTGCTTCCAGCTAATGAAATTTCAAAACTTCTG TGCAGTGATGACATTAATGTGCCTGATGAAGAGACCATTTTTCATGCTCTAATGCAGTGGGTGGGGCATGATGTGCAGAATAGGCAAGGA GAACTGGGGATGCTGCTTTCTTACATCAGACTGCCATTACTCCCACCACAGTTACTGGCAGATCTTGAAACCAGTTCCATGTTTACTGGT GATCTTGAGTGTCAGAAGCTCCTGATGGAAGCTATGAAGTATCATCTTTTGCCTGAGAGAAGATCCATGATGCAAAGCCCTCGGACAAAG CCTAGAAAATCAACTGTGGGGGCACTTTATGCTGTAGGAGGCATGGATGCTATGAAAGGTACTACTACTATTGAAAAATATGACCTCAGG ACCAACAGTTGGCTACATATTGGCACCATGAATGGCCGTAGGCTTCAATTTGGAGTCGCAGTTATTGATAATAAGCTCTATGTCGTGGGA GGAAGAGACGGTTTAAAAACTTTGAATACAGTGGAATGTTTTAATCCAGTTGGCAAAATCTGGACTGTGATGCCTCCCATGTCAACACAT CGGCACGGCTTAGGTGTAGCCACTCTTGAAGGACCAATGTATGCTGTAGGTGGTCATGATGGATGGAGCTATCTAAATACTGTAGAAAGA TGGGACCCTGAGGGACGACAGTGGAATTACGTAGCCAGTATGTCAACTCCTAGAAGCACAGTTGGTGTTGTTGCATTAAACAACAAATTA TATGCTATTGGTGGACGTGATGGAAGTTCCTGCCTCAAATCAATGGAATACTTTGACCCACACACTAACAAGTGGAGTTTGTGTGCTCCA ATGTCCAAAAGACGTGGAGGTGTGGGAGTTGCCACATACAATGGATTCTTATATGTTGTAGGGGGGCATGATGCCCCTGCTTCCAACCAT TGCTCCAGGCTTTCTGACTGTGTGGAACGGTATGATCCAAAAGGTGATTCATGGTCAACTGTGGCACCTCTGAGTGTTCCTCGAGATGCT GTTGCTGTGTGCCCTCTTGGAGACAAACTCTACGTGGTTGGAGGATATGACGGACATACTTATTTGAACACAGTTGAGTCATATGATGCA CAGAGAAATGAATGGAAAGAGGAAGTTCCTGTTAACATTGGAAGAGCTGGTGCATGTGTTGTAGTGGTGAAGCTACCCTAAAGCTATCTA TCTTTATCAAATGGAATGAAACTAGATAATTTCAAGAAACTGAGTAGGACAAAGGGAGAAAGAAATACATGTTCTTTTTCCTGCAATTAA TAATCAGACTGGAAAATTGTTGTATCATTTTAATTTGTAGTTACAATTGCTTTCATTCGTGAAGCCGAAACGTTTTTAAACATGAATTAC ATATGAATTATTAAGCATATGTGCTTTCGCAGCTGATAATATAAAAGGAAATCCCACAGTCTAGATATAGCCCCATTACTACAAAATGCT AAAATATTTAATGAAAATTGATGGTGGCCACAGTGTGCAGGTTATAAAAGCATTAATACATTTCAAGGTAAGAGCCTTAAAAGTTAAAAA CATTTTCAGTTTTTTTTTAAAAAACGTACTCTTATTATCTGGAACATAGAAATATAAAAGGTAACATCTAAAGCTTAGAATAGTGTGATT TTTAGTAAGCCATTATTCTCCTATTCAAATAATATCCCAAAGAGCTAAACAATTCCTTACATTTACCAAGAGGAAAGCTTTTACTGTGTT GAAGCTAAAAAAATAATGGCTCTTTGACAAAACTTGTTATGTTGATCGCGGTATGTCAAAATTTTTACAGGTTTGCTCATCTGCCAGAGC ACACATATAAATTTGGTATTTCTTAACATATTATCTTGTTAGATTTGTTACCAGTAAAATATTACTGTAATTTCATATACACAGTCTATA CAATGAAATAATGAATATTTATCATATTGATACAAACTGTGACCTCAGCTTCAGAGTGTCAGGGCCTCACTTGTATAGAATGTAATGTTC TCCTCAAACATTTATGTTAACTCTATAAACAAATATCGTTAAGTTAAACAAGTTTTCAAAAACAAAACAATTTTTAAAGTACCTTAAAAT TGAGGATGTTACTCAGTGTTAACACATGGGAACACCAAAATATTCAATAAGCCTGGTCAATTCTATAGTTATCTTTTTTGTACCAACACA TGCTTTTCTGTTACTGTTATATTATCCAGTAGAAAATGTTAGGATATGTGTGCTATATAAAAAAAAAAAAAGACTTGTTAAGTTTTAAAA TAACAAAAATGGCTAGTTGAATAGTATTTTATGTGTAATTCTTCCATTTATTCTGTTTAATTATACAACTAAGATGAAATATTGAAAAAC CCTTTGTGAAAGTAACTTTTCAAGTAAATGCACAACTTTAGAATTTCTACAAATAAGTTCTTTTAAACAGTCTTTTTATTGTGGATTGTG AAATCAAAATCTGGAGAAATGCTTATAAAATATACTACTAGCTTTTAAGTTTTAAGAAAGAAGAACGTAAGTTGTACAAAGATATTTGTA CTTTGACAAACTGAATTTAAATAAACTTTATTTCCTCTCACTATAGTGTGGCTTTAGAATATATCAAGTACAACCTGAATGGACTTTTTG ATTTTACATTTCCTATATCACTAAAATACTTAAACACAAAAATAAAATTCCACTCCTTTATTTCTTCTTACACCTGCCATTGTCTCTAAT TCTAAAGAGAGAGCTATAGATTTCTGCAGTTCTAGAGGTGAAGATAGAGACATAGAGAGGCTGTGAAACACACATACAGCCCTGTTAGGT CTTTTGAATTCACACCTTATTGAAATCAGTGTAGAAGTAGAGTTACCTAAACCTTTATCGCCAATGCACAGCTTGGCCTGTTAAGTTAAA TGTGGGTGTCAACCAGTGGAAAAGATCAAAGGACCTTTTGCAGCCTAGCTTGCCAATCTTCCAGTAACAACGAGGTGCTACCAATAGGCA GACTGTGTCTAGAGTCATGCATGGACAAGAGGAAAGTGAAAGTTGAATACTCTCTTCTCAATGGCATAGCTGGACTTGATTAAGTATTGA CTCATAGAAGAGACATGTATGTTTCCCGGGGCTACCCCTTCATAGCTGCATTTAAAAATGGTGTTCAGGCCACTATGGCTGCTGTGGTTT GAATATTTTTTTCCCTCGAAAACTCATGTTGAAACTTAACTCCTAATGTAGCAGTACTGAGAGGCAGGGCCCTTAAGAGGTGATTGGGCC ATGAGGTCTGTGCCCTCCTGAATGGATTAATCCATTCATGGATTAATGGATTATTATGAGTGGGGGACTGGTGACTTTATATGAAAAGGA AGGAAGAGTGACCTAAGCTAGCATGCTCAGGGTGCACCGTATGTCCCCTGGTCATGTGATGCTATGTGTTACCTTGGGACTTACCTGACA GCTCTTACCAACAAGAAGGCCATCCCAGATATGGCCTCTCAACCTCAGACTTCTCAGCCTGCCTAACTGTAAGAAATTAATTCTTTTTCT TTATAAATTACCCAGGTTCTGTTTTAAGCCTCAGAAAATGAACTAAGACAATGCCCTAGATGAGCTGCCAAAATCTGAGCAGATATGAGC CATCAGAATAGAGACCTGCAGAGGCCCAGGGCACTCAAACATAACTGTGCTTGTCTCAGTCAAACTTAATTGGGAAAAGGAGGAGTAAGA GCAGGCCCTTTCTGACATGCTTTAGCAGAGATAACTTATCAGGGCTAGTTTAAACAAATGTGTATAGGAAAGGATGTTATTTATATATTC TTACAAGAGTGTAAGGGCTCACACATTTACTGTACTCAGATCTGAAAGACAATGTTGCTTCTTGATTTTTTTCTATAATCACTATGACCG TGTTCACGATTCCCTACTCTGCAACTCTCAGCATGCATCATGATGATTCTCTAACACTGTCTGTCACCTTCCTGTATTTCCACAGAGCAT GCAAGAACTTCTACAAAACTTCTATACCACACAGAAGCTGAAAGAGACTCTGGGGCACTAAATTCAGATCCTTTCTCCACACAGCAATGA CATTTATCAATCTGACTCAGGTGTGGCTGTTGCCCCTTTTTGATATGGAGGGAACACTGAAAGACAGTTTCCTTCTGGAGTTCCAAATGC AATATAGAAAAAAAAACCCTGTGACTCTGGATTTTATTTTAAGTTCTCTAACAAGACTCCTACCTTGAGATCTTAGTCTAGGTAAAGAAA AAAAAAAAAAGGTCATGAAACACAAATGTCCTCAACTGTCATCATTCTATGTTTACTTTCTCCCAATCACATCACCTGAGGTAGAAGAGA TTTTTCAGTTGCTATTTCTCTTCGTAATGAATTTTCTTTACATCCTGTCCACACTGCTGGAGATACTAAGTCCCAACCTAAATGATGGAA GGAGCATCTCCCCTCTTGTCTGTGGAAGATATCACATGAACTACCTAATTTTCAAGCATGGATCTCCATATTTCAAAGGAAAGTTTTAAA AATGTTTTTAAAACTTCAAATATCTCACCACAGTGAATTATGGGTAAGGTGATAATATGTTTAATTAGCTTGATTTAATCATCCCACACT >16519_16519_2_CHM-KLHL4_CHM_chrX_85282495_ENST00000357749_KLHL4_chrX_86868880_ENST00000373119_length(amino acids)=616AA_BP=39 MADTLPSEFDVIVIGTGLPESIIAAACSRSGRRVLHVDSLDTQHSEDMNATRSEEQFHVINHAEQTLRKMENYLKEKQLCDVLLIAGHLR IPAHRLVLSAVSDYFAAMFTNDVLEAKQEEVRMEGVDPNALNSLVQYAYTGVLQLKEDTIESLLAAACLLQLTQVIDVCSNFLIKQLHPS NCLGIRSFGDAQGCTELLNVAHKYTMEHFIEVIKNQEFLLLPANEISKLLCSDDINVPDEETIFHALMQWVGHDVQNRQGELGMLLSYIR LPLLPPQLLADLETSSMFTGDLECQKLLMEAMKYHLLPERRSMMQSPRTKPRKSTVGALYAVGGMDAMKGTTTIEKYDLRTNSWLHIGTM NGRRLQFGVAVIDNKLYVVGGRDGLKTLNTVECFNPVGKIWTVMPPMSTHRHGLGVATLEGPMYAVGGHDGWSYLNTVERWDPEGRQWNY VASMSTPRSTVGVVALNNKLYAIGGRDGSSCLKSMEYFDPHTNKWSLCAPMSKRRGGVGVATYNGFLYVVGGHDAPASNHCSRLSDCVER -------------------------------------------------------------- >16519_16519_3_CHM-KLHL4_CHM_chrX_85282495_ENST00000358786_KLHL4_chrX_86868880_ENST00000373114_length(transcript)=2066nt_BP=123nt CGTCAAGATGGCGGATACTCTCCCTTCGGAGTTTGATGTGATCGTAATAGGGACGGGTTTGCCTGAATCCATCATTGCAGCTGCATGTTC AAGAAGTGGCCGGAGAGTTCTGCATGTTGATTCATTAGATACACAACACTCTGAAGACATGAATGCCACCAGATCTGAAGAGCAGTTCCA TGTTATAAACCACGCAGAGCAAACTCTTCGTAAAATGGAGAACTACTTGAAAGAGAAACAACTATGTGATGTGCTACTGATTGCAGGACA CCTCCGCATCCCAGCCCATAGGTTGGTTCTCAGCGCAGTGTCTGATTATTTTGCTGCAATGTTTACTAATGATGTGCTTGAAGCCAAACA AGAAGAGGTCAGGATGGAAGGAGTAGATCCAAATGCACTAAATTCCTTGGTGCAGTATGCTTACACAGGAGTCCTGCAATTGAAAGAAGA TACCATTGAAAGTTTGCTGGCTGCAGCTTGTCTTCTGCAGCTGACTCAGGTCATTGATGTTTGCTCCAATTTTCTCATAAAGCAGCTCCA TCCTTCAAACTGCTTAGGGATTCGATCATTTGGAGATGCCCAAGGCTGTACAGAACTTCTGAACGTGGCACACAAATACACTATGGAACA CTTCATTGAGGTAATAAAAAACCAAGAATTCCTCCTGCTTCCAGCTAATGAAATTTCAAAACTTCTGTGCAGTGATGACATTAATGTGCC TGATGAAGAGACCATTTTTCATGCTCTAATGCAGTGGGTGGGGCATGATGTGCAGAATAGGCAAGGAGAACTGGGGATGCTGCTTTCTTA CATCAGACTGCCATTACTCCCACCACAGTTACTGGCAGATCTTGAAACCAGTTCCATGTTTACTGGTGATCTTGAGTGTCAGAAGCTCCT GATGGAAGCTATGAAGTATCATCTTTTGCCTGAGAGAAGATCCATGATGCAAAGCCCTCGGACAAAGCCTAGAAAATCAACTGTGGGGGC ACTTTATGCTGTAGGAGGCATGGATGCTATGAAAGGTACTACTACTATTGAAAAATATGACCTCAGGACCAACAGTTGGCTACATATTGG CACCATGAATGGCCGTAGGCTTCAATTTGGAGTCGCAGTTATTGATAATAAGCTCTATGTCGTGGGAGGAAGAGACGGTTTAAAAACTTT GAATACAGTGGAATGTTTTAATCCAGTTGGCAAAATCTGGACTGTGATGCCTCCCATGTCAACACATCGGCACGGCTTAGGTGTAGCCAC TCTTGAAGGACCAATGTATGCTGTAGGTGGTCATGATGGATGGAGCTATCTAAATACTGTAGAAAGATGGGACCCTGAGGGACGACAGTG GAATTACGTAGCCAGTATGTCAACTCCTAGAAGCACAGTTGGTGTTGTTGCATTAAACAACAAATTATATGCTATTGGTGGACGTGATGG AAGTTCCTGCCTCAAATCAATGGAATACTTTGACCCACACACTAACAAGTGGAGTTTGTGTGCTCCAATGTCCAAAAGACGTGGAGGTGT GGGAGTTGCCACATACAATGGATTCTTATATGTTGTAGGGGGGCATGATGCCCCTGCTTCCAACCATTGCTCCAGGCTTTCTGACTGTGT GGAACGGTATGATCCAAAAGGTGATTCATGGTCAACTGTGGCACCTCTGAGTGTTCCTCGAGATGCTGTTGCTGTGTGCCCTCTTGGAGA CAAACTCTACGTGGTTGGAGGATATGACGGACATACTTATTTGAACACAGTTGAGTCATATGATGCACAGAGAAATGAATGGAAAGAGAG CATGCAAGAACTTCTACAAAACTTCTATACCACACAGAAGCTGAAAGAGACTCTGGGGCACTAAATTCAGATCCTTTCTCCACACAGCAA TGACATTTATCAATCTGACTCAGGTGTGGCTGTTGCCCCTTTTTGATATGGAGGGAACACTGAAAGACAGTTTCCTTCTGGAGTTCCAAA >16519_16519_3_CHM-KLHL4_CHM_chrX_85282495_ENST00000358786_KLHL4_chrX_86868880_ENST00000373114_length(amino acids)=618AA_BP=39 MADTLPSEFDVIVIGTGLPESIIAAACSRSGRRVLHVDSLDTQHSEDMNATRSEEQFHVINHAEQTLRKMENYLKEKQLCDVLLIAGHLR IPAHRLVLSAVSDYFAAMFTNDVLEAKQEEVRMEGVDPNALNSLVQYAYTGVLQLKEDTIESLLAAACLLQLTQVIDVCSNFLIKQLHPS NCLGIRSFGDAQGCTELLNVAHKYTMEHFIEVIKNQEFLLLPANEISKLLCSDDINVPDEETIFHALMQWVGHDVQNRQGELGMLLSYIR LPLLPPQLLADLETSSMFTGDLECQKLLMEAMKYHLLPERRSMMQSPRTKPRKSTVGALYAVGGMDAMKGTTTIEKYDLRTNSWLHIGTM NGRRLQFGVAVIDNKLYVVGGRDGLKTLNTVECFNPVGKIWTVMPPMSTHRHGLGVATLEGPMYAVGGHDGWSYLNTVERWDPEGRQWNY VASMSTPRSTVGVVALNNKLYAIGGRDGSSCLKSMEYFDPHTNKWSLCAPMSKRRGGVGVATYNGFLYVVGGHDAPASNHCSRLSDCVER -------------------------------------------------------------- >16519_16519_4_CHM-KLHL4_CHM_chrX_85282495_ENST00000358786_KLHL4_chrX_86868880_ENST00000373119_length(transcript)=5374nt_BP=123nt CGTCAAGATGGCGGATACTCTCCCTTCGGAGTTTGATGTGATCGTAATAGGGACGGGTTTGCCTGAATCCATCATTGCAGCTGCATGTTC AAGAAGTGGCCGGAGAGTTCTGCATGTTGATTCATTAGATACACAACACTCTGAAGACATGAATGCCACCAGATCTGAAGAGCAGTTCCA TGTTATAAACCACGCAGAGCAAACTCTTCGTAAAATGGAGAACTACTTGAAAGAGAAACAACTATGTGATGTGCTACTGATTGCAGGACA CCTCCGCATCCCAGCCCATAGGTTGGTTCTCAGCGCAGTGTCTGATTATTTTGCTGCAATGTTTACTAATGATGTGCTTGAAGCCAAACA AGAAGAGGTCAGGATGGAAGGAGTAGATCCAAATGCACTAAATTCCTTGGTGCAGTATGCTTACACAGGAGTCCTGCAATTGAAAGAAGA TACCATTGAAAGTTTGCTGGCTGCAGCTTGTCTTCTGCAGCTGACTCAGGTCATTGATGTTTGCTCCAATTTTCTCATAAAGCAGCTCCA TCCTTCAAACTGCTTAGGGATTCGATCATTTGGAGATGCCCAAGGCTGTACAGAACTTCTGAACGTGGCACACAAATACACTATGGAACA CTTCATTGAGGTAATAAAAAACCAAGAATTCCTCCTGCTTCCAGCTAATGAAATTTCAAAACTTCTGTGCAGTGATGACATTAATGTGCC TGATGAAGAGACCATTTTTCATGCTCTAATGCAGTGGGTGGGGCATGATGTGCAGAATAGGCAAGGAGAACTGGGGATGCTGCTTTCTTA CATCAGACTGCCATTACTCCCACCACAGTTACTGGCAGATCTTGAAACCAGTTCCATGTTTACTGGTGATCTTGAGTGTCAGAAGCTCCT GATGGAAGCTATGAAGTATCATCTTTTGCCTGAGAGAAGATCCATGATGCAAAGCCCTCGGACAAAGCCTAGAAAATCAACTGTGGGGGC ACTTTATGCTGTAGGAGGCATGGATGCTATGAAAGGTACTACTACTATTGAAAAATATGACCTCAGGACCAACAGTTGGCTACATATTGG CACCATGAATGGCCGTAGGCTTCAATTTGGAGTCGCAGTTATTGATAATAAGCTCTATGTCGTGGGAGGAAGAGACGGTTTAAAAACTTT GAATACAGTGGAATGTTTTAATCCAGTTGGCAAAATCTGGACTGTGATGCCTCCCATGTCAACACATCGGCACGGCTTAGGTGTAGCCAC TCTTGAAGGACCAATGTATGCTGTAGGTGGTCATGATGGATGGAGCTATCTAAATACTGTAGAAAGATGGGACCCTGAGGGACGACAGTG GAATTACGTAGCCAGTATGTCAACTCCTAGAAGCACAGTTGGTGTTGTTGCATTAAACAACAAATTATATGCTATTGGTGGACGTGATGG AAGTTCCTGCCTCAAATCAATGGAATACTTTGACCCACACACTAACAAGTGGAGTTTGTGTGCTCCAATGTCCAAAAGACGTGGAGGTGT GGGAGTTGCCACATACAATGGATTCTTATATGTTGTAGGGGGGCATGATGCCCCTGCTTCCAACCATTGCTCCAGGCTTTCTGACTGTGT GGAACGGTATGATCCAAAAGGTGATTCATGGTCAACTGTGGCACCTCTGAGTGTTCCTCGAGATGCTGTTGCTGTGTGCCCTCTTGGAGA CAAACTCTACGTGGTTGGAGGATATGACGGACATACTTATTTGAACACAGTTGAGTCATATGATGCACAGAGAAATGAATGGAAAGAGGA AGTTCCTGTTAACATTGGAAGAGCTGGTGCATGTGTTGTAGTGGTGAAGCTACCCTAAAGCTATCTATCTTTATCAAATGGAATGAAACT AGATAATTTCAAGAAACTGAGTAGGACAAAGGGAGAAAGAAATACATGTTCTTTTTCCTGCAATTAATAATCAGACTGGAAAATTGTTGT ATCATTTTAATTTGTAGTTACAATTGCTTTCATTCGTGAAGCCGAAACGTTTTTAAACATGAATTACATATGAATTATTAAGCATATGTG CTTTCGCAGCTGATAATATAAAAGGAAATCCCACAGTCTAGATATAGCCCCATTACTACAAAATGCTAAAATATTTAATGAAAATTGATG GTGGCCACAGTGTGCAGGTTATAAAAGCATTAATACATTTCAAGGTAAGAGCCTTAAAAGTTAAAAACATTTTCAGTTTTTTTTTAAAAA ACGTACTCTTATTATCTGGAACATAGAAATATAAAAGGTAACATCTAAAGCTTAGAATAGTGTGATTTTTAGTAAGCCATTATTCTCCTA TTCAAATAATATCCCAAAGAGCTAAACAATTCCTTACATTTACCAAGAGGAAAGCTTTTACTGTGTTGAAGCTAAAAAAATAATGGCTCT TTGACAAAACTTGTTATGTTGATCGCGGTATGTCAAAATTTTTACAGGTTTGCTCATCTGCCAGAGCACACATATAAATTTGGTATTTCT TAACATATTATCTTGTTAGATTTGTTACCAGTAAAATATTACTGTAATTTCATATACACAGTCTATACAATGAAATAATGAATATTTATC ATATTGATACAAACTGTGACCTCAGCTTCAGAGTGTCAGGGCCTCACTTGTATAGAATGTAATGTTCTCCTCAAACATTTATGTTAACTC TATAAACAAATATCGTTAAGTTAAACAAGTTTTCAAAAACAAAACAATTTTTAAAGTACCTTAAAATTGAGGATGTTACTCAGTGTTAAC ACATGGGAACACCAAAATATTCAATAAGCCTGGTCAATTCTATAGTTATCTTTTTTGTACCAACACATGCTTTTCTGTTACTGTTATATT ATCCAGTAGAAAATGTTAGGATATGTGTGCTATATAAAAAAAAAAAAAGACTTGTTAAGTTTTAAAATAACAAAAATGGCTAGTTGAATA GTATTTTATGTGTAATTCTTCCATTTATTCTGTTTAATTATACAACTAAGATGAAATATTGAAAAACCCTTTGTGAAAGTAACTTTTCAA GTAAATGCACAACTTTAGAATTTCTACAAATAAGTTCTTTTAAACAGTCTTTTTATTGTGGATTGTGAAATCAAAATCTGGAGAAATGCT TATAAAATATACTACTAGCTTTTAAGTTTTAAGAAAGAAGAACGTAAGTTGTACAAAGATATTTGTACTTTGACAAACTGAATTTAAATA AACTTTATTTCCTCTCACTATAGTGTGGCTTTAGAATATATCAAGTACAACCTGAATGGACTTTTTGATTTTACATTTCCTATATCACTA AAATACTTAAACACAAAAATAAAATTCCACTCCTTTATTTCTTCTTACACCTGCCATTGTCTCTAATTCTAAAGAGAGAGCTATAGATTT CTGCAGTTCTAGAGGTGAAGATAGAGACATAGAGAGGCTGTGAAACACACATACAGCCCTGTTAGGTCTTTTGAATTCACACCTTATTGA AATCAGTGTAGAAGTAGAGTTACCTAAACCTTTATCGCCAATGCACAGCTTGGCCTGTTAAGTTAAATGTGGGTGTCAACCAGTGGAAAA GATCAAAGGACCTTTTGCAGCCTAGCTTGCCAATCTTCCAGTAACAACGAGGTGCTACCAATAGGCAGACTGTGTCTAGAGTCATGCATG GACAAGAGGAAAGTGAAAGTTGAATACTCTCTTCTCAATGGCATAGCTGGACTTGATTAAGTATTGACTCATAGAAGAGACATGTATGTT TCCCGGGGCTACCCCTTCATAGCTGCATTTAAAAATGGTGTTCAGGCCACTATGGCTGCTGTGGTTTGAATATTTTTTTCCCTCGAAAAC TCATGTTGAAACTTAACTCCTAATGTAGCAGTACTGAGAGGCAGGGCCCTTAAGAGGTGATTGGGCCATGAGGTCTGTGCCCTCCTGAAT GGATTAATCCATTCATGGATTAATGGATTATTATGAGTGGGGGACTGGTGACTTTATATGAAAAGGAAGGAAGAGTGACCTAAGCTAGCA TGCTCAGGGTGCACCGTATGTCCCCTGGTCATGTGATGCTATGTGTTACCTTGGGACTTACCTGACAGCTCTTACCAACAAGAAGGCCAT CCCAGATATGGCCTCTCAACCTCAGACTTCTCAGCCTGCCTAACTGTAAGAAATTAATTCTTTTTCTTTATAAATTACCCAGGTTCTGTT TTAAGCCTCAGAAAATGAACTAAGACAATGCCCTAGATGAGCTGCCAAAATCTGAGCAGATATGAGCCATCAGAATAGAGACCTGCAGAG GCCCAGGGCACTCAAACATAACTGTGCTTGTCTCAGTCAAACTTAATTGGGAAAAGGAGGAGTAAGAGCAGGCCCTTTCTGACATGCTTT AGCAGAGATAACTTATCAGGGCTAGTTTAAACAAATGTGTATAGGAAAGGATGTTATTTATATATTCTTACAAGAGTGTAAGGGCTCACA CATTTACTGTACTCAGATCTGAAAGACAATGTTGCTTCTTGATTTTTTTCTATAATCACTATGACCGTGTTCACGATTCCCTACTCTGCA ACTCTCAGCATGCATCATGATGATTCTCTAACACTGTCTGTCACCTTCCTGTATTTCCACAGAGCATGCAAGAACTTCTACAAAACTTCT ATACCACACAGAAGCTGAAAGAGACTCTGGGGCACTAAATTCAGATCCTTTCTCCACACAGCAATGACATTTATCAATCTGACTCAGGTG TGGCTGTTGCCCCTTTTTGATATGGAGGGAACACTGAAAGACAGTTTCCTTCTGGAGTTCCAAATGCAATATAGAAAAAAAAACCCTGTG ACTCTGGATTTTATTTTAAGTTCTCTAACAAGACTCCTACCTTGAGATCTTAGTCTAGGTAAAGAAAAAAAAAAAAAGGTCATGAAACAC AAATGTCCTCAACTGTCATCATTCTATGTTTACTTTCTCCCAATCACATCACCTGAGGTAGAAGAGATTTTTCAGTTGCTATTTCTCTTC GTAATGAATTTTCTTTACATCCTGTCCACACTGCTGGAGATACTAAGTCCCAACCTAAATGATGGAAGGAGCATCTCCCCTCTTGTCTGT GGAAGATATCACATGAACTACCTAATTTTCAAGCATGGATCTCCATATTTCAAAGGAAAGTTTTAAAAATGTTTTTAAAACTTCAAATAT CTCACCACAGTGAATTATGGGTAAGGTGATAATATGTTTAATTAGCTTGATTTAATCATCCCACACTGTATACATATGCCAAAATATCAT >16519_16519_4_CHM-KLHL4_CHM_chrX_85282495_ENST00000358786_KLHL4_chrX_86868880_ENST00000373119_length(amino acids)=616AA_BP=39 MADTLPSEFDVIVIGTGLPESIIAAACSRSGRRVLHVDSLDTQHSEDMNATRSEEQFHVINHAEQTLRKMENYLKEKQLCDVLLIAGHLR IPAHRLVLSAVSDYFAAMFTNDVLEAKQEEVRMEGVDPNALNSLVQYAYTGVLQLKEDTIESLLAAACLLQLTQVIDVCSNFLIKQLHPS NCLGIRSFGDAQGCTELLNVAHKYTMEHFIEVIKNQEFLLLPANEISKLLCSDDINVPDEETIFHALMQWVGHDVQNRQGELGMLLSYIR LPLLPPQLLADLETSSMFTGDLECQKLLMEAMKYHLLPERRSMMQSPRTKPRKSTVGALYAVGGMDAMKGTTTIEKYDLRTNSWLHIGTM NGRRLQFGVAVIDNKLYVVGGRDGLKTLNTVECFNPVGKIWTVMPPMSTHRHGLGVATLEGPMYAVGGHDGWSYLNTVERWDPEGRQWNY VASMSTPRSTVGVVALNNKLYAIGGRDGSSCLKSMEYFDPHTNKWSLCAPMSKRRGGVGVATYNGFLYVVGGHDAPASNHCSRLSDCVER -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHM-KLHL4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHM-KLHL4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHM-KLHL4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
