
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHMP3-EPB41L5 (FusionGDB2 ID:16537) |
Fusion Gene Summary for CHMP3-EPB41L5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHMP3-EPB41L5 | Fusion gene ID: 16537 | Hgene | Tgene | Gene symbol | CHMP3 | EPB41L5 | Gene ID | 100526767 | 57669 |
| Gene name | RNF103-CHMP3 readthrough | erythrocyte membrane protein band 4.1 like 5 | |
| Synonyms | CGI149|CHMP3|NEDF|RNF103-VPS24|VPS24|hVps24 | BE37|LULU|LULU1|YMO1|YRT | |
| Cytomap | 2p11.2 | 2q14.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | RNF103-CHMP3 proteinCharged multivesicular body protein 3Chromatin-modifying protein 3Neuroendocrine differentiation factorRNF103-CHMP3 read-throughRNF103-VPS24 readthroughVacuolar protein sorting-associated protein 24 | band 4.1-like protein 5yurt homolog | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9Y3E7 | Q9HCM4 | |
| Ensembl transtripts involved in fusion gene | ENST00000263856, ENST00000409727, ENST00000409225, ENST00000439940, ENST00000494623, | ENST00000331393, ENST00000443124, ENST00000452780, ENST00000488691, ENST00000263713, ENST00000443902, | |
| Fusion gene scores | * DoF score | 10 X 6 X 6=360 | 9 X 9 X 4=324 |
| # samples | 11 | 9 | |
| ** MAII score | log2(11/360*10)=-1.71049338280502 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/324*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CHMP3 [Title/Abstract] AND EPB41L5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CHMP3(86790426)-EPB41L5(120900578), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CHMP3 (5'-gene) Fusion gene breakpoints across CHMP3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
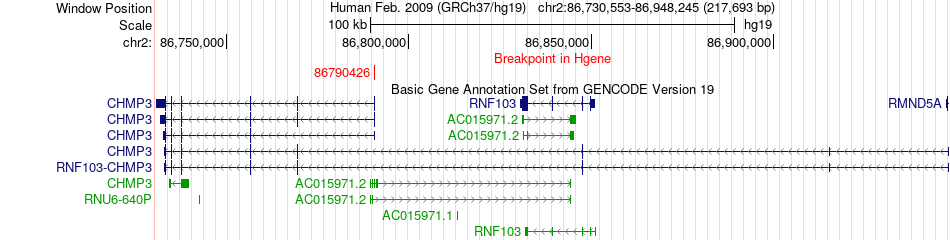 |
 Fusion gene breakpoints across EPB41L5 (3'-gene) Fusion gene breakpoints across EPB41L5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
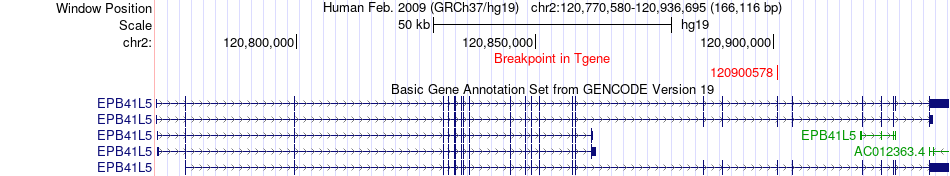 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-4267 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
Top |
Fusion Gene ORF analysis for CHMP3-EPB41L5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000263856 | ENST00000331393 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000263856 | ENST00000443124 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000263856 | ENST00000452780 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000263856 | ENST00000488691 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000409727 | ENST00000331393 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000409727 | ENST00000443124 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000409727 | ENST00000452780 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5CDS-intron | ENST00000409727 | ENST00000488691 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5UTR-3CDS | ENST00000409225 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5UTR-3CDS | ENST00000409225 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5UTR-intron | ENST00000409225 | ENST00000331393 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5UTR-intron | ENST00000409225 | ENST00000443124 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5UTR-intron | ENST00000409225 | ENST00000452780 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| 5UTR-intron | ENST00000409225 | ENST00000488691 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| In-frame | ENST00000263856 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| In-frame | ENST00000263856 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| In-frame | ENST00000409727 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| In-frame | ENST00000409727 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-3CDS | ENST00000439940 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-3CDS | ENST00000439940 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-3CDS | ENST00000494623 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-3CDS | ENST00000494623 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000439940 | ENST00000331393 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000439940 | ENST00000443124 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000439940 | ENST00000452780 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000439940 | ENST00000488691 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000494623 | ENST00000331393 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000494623 | ENST00000443124 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000494623 | ENST00000452780 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
| intron-intron | ENST00000494623 | ENST00000488691 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000263856 | CHMP3 | chr2 | 86790426 | - | ENST00000263713 | EPB41L5 | chr2 | 120900578 | + | 4989 | 174 | 129 | 776 | 215 |
| ENST00000263856 | CHMP3 | chr2 | 86790426 | - | ENST00000443902 | EPB41L5 | chr2 | 120900578 | + | 1600 | 174 | 129 | 638 | 169 |
| ENST00000409727 | CHMP3 | chr2 | 86790426 | - | ENST00000263713 | EPB41L5 | chr2 | 120900578 | + | 4886 | 71 | 26 | 673 | 215 |
| ENST00000409727 | CHMP3 | chr2 | 86790426 | - | ENST00000443902 | EPB41L5 | chr2 | 120900578 | + | 1497 | 71 | 26 | 535 | 169 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000263856 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + | 0.002035973 | 0.997964 |
| ENST00000263856 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + | 0.004303587 | 0.9956964 |
| ENST00000409727 | ENST00000263713 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + | 0.002030049 | 0.9979699 |
| ENST00000409727 | ENST00000443902 | CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + | 0.004669717 | 0.99533033 |
Top |
Fusion Genomic Features for CHMP3-EPB41L5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + | 0.005589369 | 0.9944106 |
| CHMP3 | chr2 | 86790426 | - | EPB41L5 | chr2 | 120900578 | + | 0.005589369 | 0.9944106 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
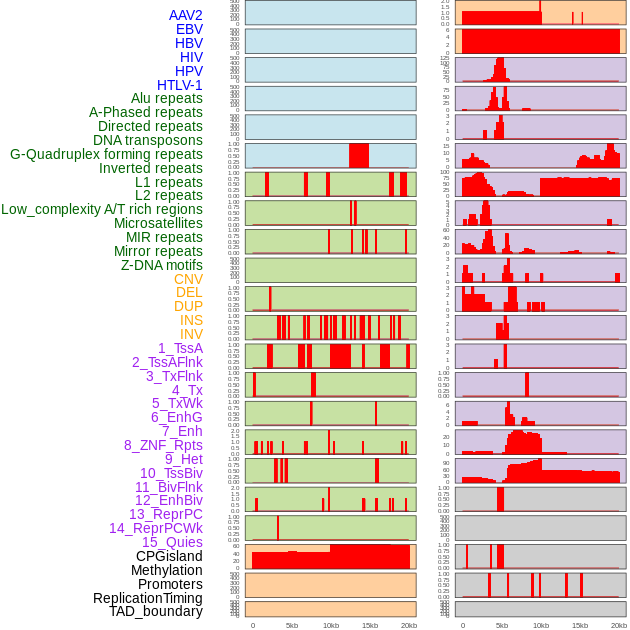 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
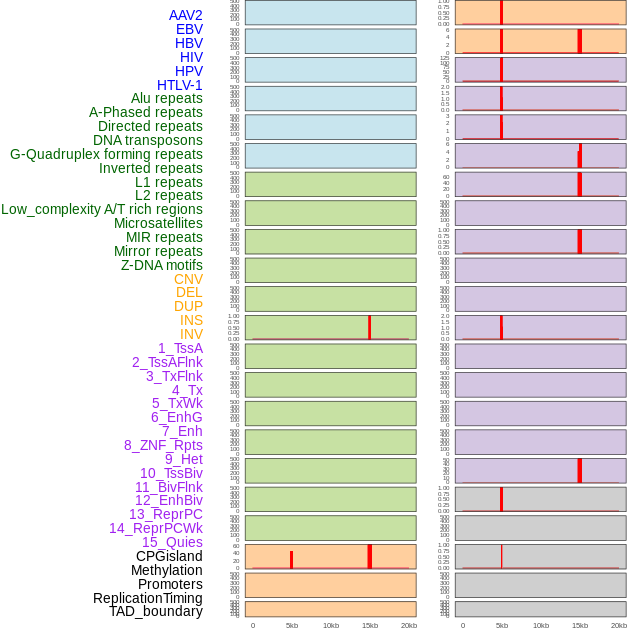 |
Top |
Fusion Protein Features for CHMP3-EPB41L5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:86790426/chr2:120900578) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHMP3 | EPB41L5 |
| FUNCTION: Probable core component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and mostly are delivered to lysosomes enabling degradation of membrane proteins, such as stimulated growth factor receptors, lysosomal enzymes and lipids. The MVB pathway appears to require the sequential function of ESCRT-O, -I,-II and -III complexes. ESCRT-III proteins mostly dissociate from the invaginating membrane before the ILV is released. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis and the budding of enveloped viruses (HIV-1 and other lentiviruses). ESCRT-III proteins are believed to mediate the necessary vesicle extrusion and/or membrane fission activities, possibly in conjunction with the AAA ATPase VPS4. Selectively binds to phosphatidylinositol 3,5-bisphosphate PtdIns(3,5)P2 and PtdIns(3,4)P2 in preference to other phosphoinositides tested. Involved in late stages of cytokinesis. Plays a role in endosomal sorting/trafficking of EGF receptor. Isoform 2 prevents stress-mediated cell death and accumulation of reactive oxygen species when expressed in yeast cells. {ECO:0000269|PubMed:14505570, ECO:0000269|PubMed:15707591, ECO:0000269|PubMed:16740483, ECO:0000269|PubMed:17331679, ECO:0000269|PubMed:18076377}. | FUNCTION: Plays a role in the formation and organization of tight junctions during the establishment of polarity in epithelial cells. {ECO:0000269|PubMed:17920587}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EPB41L5 | chr2:86790426 | chr2:120900578 | ENST00000331393 | 0 | 17 | 43_327 | 0 | 506.0 | Domain | FERM | |
| Tgene | EPB41L5 | chr2:86790426 | chr2:120900578 | ENST00000443124 | 0 | 17 | 43_327 | 0 | 506.0 | Domain | FERM |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000263856 | - | 1 | 6 | 141_222 | 15 | 223.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000263856 | - | 1 | 6 | 22_54 | 15 | 223.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409225 | - | 1 | 5 | 141_222 | 0 | 157.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409225 | - | 1 | 5 | 22_54 | 0 | 157.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409727 | - | 1 | 6 | 141_222 | 15 | 183.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409727 | - | 1 | 6 | 22_54 | 15 | 183.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000439940 | - | 1 | 8 | 141_222 | 0 | 252.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000439940 | - | 1 | 8 | 22_54 | 0 | 252.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000263856 | - | 1 | 6 | 201_211 | 15 | 223.0 | Motif | Note=MIT-interacting motif |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409225 | - | 1 | 5 | 201_211 | 0 | 157.0 | Motif | Note=MIT-interacting motif |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409727 | - | 1 | 6 | 201_211 | 15 | 183.0 | Motif | Note=MIT-interacting motif |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000439940 | - | 1 | 8 | 201_211 | 0 | 252.0 | Motif | Note=MIT-interacting motif |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000263856 | - | 1 | 6 | 59_64 | 15 | 223.0 | Region | Note=Important for autoinhibitory function |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409225 | - | 1 | 5 | 59_64 | 0 | 157.0 | Region | Note=Important for autoinhibitory function |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409727 | - | 1 | 6 | 59_64 | 15 | 183.0 | Region | Note=Important for autoinhibitory function |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000439940 | - | 1 | 8 | 59_64 | 0 | 252.0 | Region | Note=Important for autoinhibitory function |
| Tgene | EPB41L5 | chr2:86790426 | chr2:120900578 | ENST00000263713 | 17 | 25 | 43_327 | 533 | 734.0 | Domain | FERM | |
| Tgene | EPB41L5 | chr2:86790426 | chr2:120900578 | ENST00000443902 | 17 | 24 | 43_327 | 533 | 688.0 | Domain | FERM | |
| Tgene | EPB41L5 | chr2:86790426 | chr2:120900578 | ENST00000452780 | 16 | 24 | 43_327 | 533 | 733.0 | Domain | FERM |
Top |
Fusion Gene Sequence for CHMP3-EPB41L5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16537_16537_1_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000263856_EPB41L5_chr2_120900578_ENST00000263713_length(transcript)=4989nt_BP=174nt TGCCGCCGCTCCCTACTCTGGGGGTTGGGACTACCTCCTTTTCCGCGGGCCCCGCCCAGGCGGCTGCCCGTGACCTGCCTGGGCGCGGGG AACTGAAAGCCGGAAGGGGCAAGACGGGTTCAGTTCGTCATGGGGCTGTTTGGAAAGACCCAGGAGAAGCCGCCCAAAGAACTGGAGGAA GTGGTGAAGTTGACTGAGAAATGCCTTAATAATGTCATTGAGAGCCCAGGATTGAATGTCATGAGAGTTCCTCCTGACTTCAAGAGTAAC ATTTTGAAGGCTCAAGTAGAAGCAGTGCATAAGGTTACAAAAGAAGATAGCTTATTAAGTCATAAAAATGCCAATGTTCAGGATGCTGCC ACAAACAGTGCTGTGTTAAATGAGAATAATGTGCCCCTCCCCAAAGAGTCTCTTGAGACTCTGATGCTTATCACACCTGCCGACAGTGGT TCTGTTCTAAAGGAAGCTACAGATGAATTGGATGCCTTGCTTGCATCTCTAACTGAGAATCTAATTGATCACACAGTTGCACCTCAGGTG TCTTCCACATCCATGATCACACCCCGGTGGATTGTTCCGCAGAGTGGTGCCATGTCTAATGGACTTGCGGGATGTGAAATGCTTTTGACA GGGAAGGAGGGACATGGTAATAAAGATGGAATCTCACTGATCTCTCCCCCAGCGCCATTCTTGGTAGATGCTGTGACCAGCTCTGGTCCC ATTTTGGCAGAAGAAGCTGTCCTGAAGCAGAAGTGTTTACTGACCACTGAGCTCTGAGGGCCTGTAGCTGGAATACGCATCTCTCCAGCA TTCCGTCCTGGGATCCGTTTCAGCTAGAATATGTTGGATTCAGGAGCTTGTCCATTATTTGTAGGTAAAAAAAGCTGCACGTAGATTTGA CTTCAACTCCGTAAAAAAGACAGCTGTATTTTCCGTCCAACTGGAATTGTTGAATCACACTGCATAGCTGCCCAAAAGAGAGTGTTTGGT CTTGAACTTTCTATACTTTTATAAATGTTACAAATTCCCGAAAGAAGGGAATTTCTTTTTCTGGGGTTTCCTTCAAACTCTTGGCTCCAC CTAGCGGTTCTATTTGTTCATAACAACTTCATAACAAGCCTGCCTCTGGTAGTCAACAGCCTTTTGAAAGCTATTTCCATCTAGTATCAG GGTGAGAGCATCCTTGATCTGGCTGCCTGTTAGAGAAATTGCACTTTTCCTGACTTACCTAGAAATCAAGAATTTAGGAAATTAATGTGG ACACTATAAAGGCAGACTTAGGGCCAACTTTTTTTTTTTTTTACAATTATTACAACACTAAAGAGAAGTTTAGAATATAGAGAGTTTTTA AATGTCTCCCATTCTTTTGATTTCTTACTGTACTGGCTATCTTAATATTTCAAGTTTACATCAAGATAAACCCTGAGAAGAACTACGGAG AAATCAAATAAAATCCTGTCATATTTTTTTCACCCTGCCTTTCCACAGGAAGCACTCACAGGCACCACACACGTATCATGTAACTTATCA GTGGGGTGGGTTACTGTTGAAGAGACCCTGGGGCATTTACCTCAGGCATCTGCACTCCTCCGAGCCCGGTGGAGAATGCAGGCTGCTGTA GTCTCAGGTAATGAAGGCACAGCACAGCAGTACTCCACATTGTTTCCTATTTGGACATAGACTTCATTTCCTTTCAGTATAAGCTGAATA AATTTAGAGCTTTCAAACTGGAAAAAAAAATGAAACAAAACAAATACACCAAGACCAAAATAGGCAATAGGAACAGGGGTGAAGGGATGT TGTTTCTTAAATACCTACCATGTATGAAGCTATACACAGCATATACCGAAAGAACCTGCATTGCACTAGGAATTCTGTGTTAGTTTAAAA GAGATCTCTAAAACTTCCCCATCCCTTTGGGCCTGTACAAAGAAATTCTGGATGTTAAAATAATATATACTCATCACAGAAAAATAAAGT ATAGCAATGTCCATCTGTAATTCTAATACCCAGAAATAACGCTATTTAGCTTTATAATTTTCGAATGAACAAGGTAAACCTCGGTTGCCA TGGGGAAGAAGGATGATGTGGAAACCATATTGGTAAAGTTGTTAATCCCTCGTTATGGAGAACTGATCTTAAGCTATACCTCCTGGAATT TGCTTTCTAGTTTTCTGTCCTGCAATATGTATATAATTAAGCACTAATTTGTACTGCTTAGCATAAAAGAACATCCAGTCTTAGATCCTT AAAACTTCATGGATTGGACTTTCCTGGGCTCCTTATAACATAATCGTGTGTCCAGGCAAACGCACACTAGTGTCTGACTGGAAAGCTCAG GAATTTTAATCTTGCACTGTTTCCCAGGGAGCTGTAGTGATTGGAACCCACGTTTGCACAAAACATTTTTGCAGAAGGAAAGTCAACACT TCTTGCTGGCTGCCTCCCCTTAGCCATTATGCTAAAAACAGCTTCTGAGTTTCACTGGTGGGGCTCTTGCCAGTTCTTAATTATAGGACA TATTTTCTCAAAGCTGAAGGTGACACCTAGAACCAGGGGCTTGACCCAGGACATGATGGAATGAGCATCAAATTTTCAGTGTCTTGGCAA CCGTAGATGTCCTACAGGGTTACCGTTGTGCTGCTCACCACAGAGCAGCTGAGGCATTATGCCTTGGAAGACCTAAATCTCCCATCCAGT TCAGGAGGTGACAACATCCTTATTTTAAACTTCCTAAAATTAGGAATTAGGTAGTTGGACATAGTCTGTGACCTTTATGTCGTTGGATAC CTGTATTCTTGACAGTTAGAATATTGGTAGGGACTTTGTTAAAATTCACTTGAATTTCAAGCTCAGAGGAAACTTTGTCTCATGCCCTGA CATGAAGTGGCAAACACGGAAGTTCATACTTGAATGCTGAATTGGCCCCGACAGATTAAATGCGTGTTGGGGATTGGTTTCCTGTCATAG CTGCTGCTGCTGCCATGCGCAGAGCTGCTGTAACAGCTCTTCCTGTTCTGCTCCCCTGAGAACAGTGTGGTGGGGAGAGGCAGGGCTGAG GTGGTCTACGAATGTGGCAGGTAGGGAAGGGGAGATGTCTGTCTCTTGAGAAGAGAGAGGCATGTGTGCCGGCATCCTTGATGGGTTCAA AGAGAAAGGTTGGAGATGATAGTGGGTGAGAAGCAGGCTGGTGAGACTGGGCTGAGGTTGGAGAAGGGGCAGCCGGGGGCACTGCTGAGG GTTTGCCGTGCACGCCTCGGACGGAGCACGGTGGGGTGGCGGGGAGCAGATAGTGCTGCCTCCCTGCGGGCAGACAGGAGAGGAACCTCA ACTCAGTCCATTTCATAGCCCTGATAGGGGAAGTGGGAGTTGACAGGATGGTTTAAAATAAGACGTGAAGGTTTCAGTTACCTGCTCTAG ACTTTGCCTGAGAACTTGTAAATTAATCAGTGAGACCTAATTTGTGACATGTCAGTAGCATCATCTTTTGACACACAGGAGGTCATGGTC ATTTCATTCCTACTCTTCAGGAGACACTGCTGAACAGAGGAATGATTCTGTTCCTTGTGTGCTTACTTCCTTAACATTTATACATTGTTT TAAGAAAAAACTTTTAAAAATATTTCTTATAGTCTCCTAACATTTGTCTCTAGCCTTTGCCTTTGTACAATCACAGATATCCTATGGAGA TTTAAGGATGAAAGCCCTGAGTTGTTCTTGGGTTCTTGGATCTGGACTACTTGTTATCTTATGCTTCTCACTTCTGGCTAAAACTTGCAC CTCTTCTTCTCTTAGCTAAGCCCCAAAATGAAGATTTCCTTCAGAAGTCTTGTTAGCAGAATTATTTATCAGTCACAGAGAGAAAAATCT GCTATTTTTCTAAGTAAGAGTCTCGAGAAGCAGAGTTTTTGTCTTGTCATTGAGAGGAGTCAGCAGTCTTGTTCTGTAAAGGACCAGAGA TGGTAAATACTGTCCCACTCAGCTCTGCTGGCGCAGTACAGCAGCAGCAGCCCCAGCACAGCTGTGTTCCTGCGGAGTCCCCTTTACAAA GCCGCTGACCCCTGATGTGAAACTTTGTAGAGCAGCAGAGTGGCTGCGTGAAACGGGAGGCTGGCAGGTCCTCAGATAGGTTCTGCAGTG TTACCTGTCACTTGGAGGCAGCCAACACTTCTGGACATTGCATCCTTATTCACACATGTGGCAGCTGAACGAGGTGCTGTTGTGGGTGTC TCAGCTCTGAGGGTCTTTGTGAGCTCCCACTGTTGTGGGTGTCTCAGCTCTGAGGGTCTTTGTGAGCTCCCACTGTTGTGGGTGTCTCAG CTCTGAGGGTCTTTGTGAGTTCCCACCAACTTTTAATTATTCATGCCCTTGACCATGTGGTTGCTTGGAGACCTGGGGTCTTCTGCAGAC TGAAGAAGACACATTTCTAGATTATTTGTCCTTTTTATCCTCTCAAAAATTTAAACACTGTACCTCTTCAGTGGTCAGAAGAAAGTTGGA AACTTTTCCTACATATTAGGCGTTAGTATGAGGACATTTGTTTGAATTATAGAAATTTGCCCTGAGCTGAACTGGGTTGTGTTAACACAT TGGTAGAGCTATGATTCCTTCCCAGTTCTAAGAGATACGATCTGTAAGTCCCTATGTCACCACATTGCTTGAGATGATCATTCAGTTACT TGTCAGGATTTCTCCTCTTCAGAGAGATTTTTTTTTATAGCACAGATTCCTTTGCCCCTTTTATCTCCTTATCTGGATATGATAAGTGGT TATGAGGGTCTCACTAACTATTTTGTGTTTACCTTTTATATGTGTAAAACTTTGCAGTAGCATTTAAAGTGTAATTTATTTTTCTATCAA GTGCACTATTCATTTAGTGTGTTCCAGTTTTATATGACTTGTATTAGAAACACTGCACTGAGTTGTTTGTACACTGAAATGAGAACTCTA >16537_16537_1_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000263856_EPB41L5_chr2_120900578_ENST00000263713_length(amino acids)=215AA_BP=15 MGLFGKTQEKPPKELEEVVKLTEKCLNNVIESPGLNVMRVPPDFKSNILKAQVEAVHKVTKEDSLLSHKNANVQDAATNSAVLNENNVPL PKESLETLMLITPADSGSVLKEATDELDALLASLTENLIDHTVAPQVSSTSMITPRWIVPQSGAMSNGLAGCEMLLTGKEGHGNKDGISL -------------------------------------------------------------- >16537_16537_2_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000263856_EPB41L5_chr2_120900578_ENST00000443902_length(transcript)=1600nt_BP=174nt TGCCGCCGCTCCCTACTCTGGGGGTTGGGACTACCTCCTTTTCCGCGGGCCCCGCCCAGGCGGCTGCCCGTGACCTGCCTGGGCGCGGGG AACTGAAAGCCGGAAGGGGCAAGACGGGTTCAGTTCGTCATGGGGCTGTTTGGAAAGACCCAGGAGAAGCCGCCCAAAGAACTGGAGGAA GTGGTGAAGTTGACTGAGAAATGCCTTAATAATGTCATTGAGAGCCCAGGATTGAATGTCATGAGAGTTCCTCCTGACTTCAAGAGTAAC ATTTTGAAGGCTCAAGTAGAAGCAGTGCATAAGGTTACAAAAGAAGATAGCTTATTAAGTCATAAAAATGCCAATGTTCAGGATGCTGCC ACAAACAGTGCTGTGTTAAATGAGAATAATGTGCCCCTCCCCAAAGAGTCTCTTGAGACTCTGATGCTTATCACACCTGCCGACAGTGGT TCTGTTCTAAAGGAAGCTACAGATGAATTGGATGCCTTGCTTGCATCTCTAACTGAGAATCTAATTGATCACACAGTTGCACCTCAGGTG TCTTCCACATCCATGATCACACCCCGGTGGATTGTTCCGCTCTGGTCCCATTTTGGCAGAAGAAGCTGTCCTGAAGCAGAAGTGTTTACT GACCACTGAGCTCTGAGGGCCTGTAGCTGGAATACGCATCTCTCCAGCATTCCGTCCTGGGATCCGTTTCAGCTAGAATATGTTGGATTC AGGAGCTTGTCCATTATTTGTAGGTAAAAAAAGCTGCACGTAGATTTGACTTCAACTCCGTAAAAAAGACAGCTGTATTTTCCGTCCAAC TGGAATTGTTGAATCACACTGCATAGCTGCCCAAAAGAGAGTGTTTGGTCTTGAACTTTCTATACTTTTATAAATGTTACAAATTCCCGA AAGAAGGGAATTTCTTTTTCTGGGGTTTCCTTCAAACTCTTGGCTCCACCTAGCGGTTCTATTTGTTCATAACAACTTCATAACAAGCCT GCCTCTGGTAGTCAACAGCCTTTTGAAAGCTATTTCCATCTAGTATCAGGGTGAGAGCATCCTTGATCTGGCTGCCTGTTAGAGAAATTG CACTTTTCCTGACTTACCTAGAAATCAAGAATTTAGGAAATTAATGTGGACACTATAAAGGCAGACTTAGGGCCAACTTTTTTTTTTTTT TACAATTATTACAACACTAAAGAGAAGTTTAGAATATAGAGAGTTTTTAAATGTCTCCCATTCTTTTGATTTCTTACTGTACTGGCTATC TTAATATTTCAAGTTTACATCAAGATAAACCCTGAGAAGAACTACGGAGAAATCAAATAAAATCCTGTCATATTTTTTTCACCCTGCCTT TCCACAGGAAGCACTCACAGGCACCACACACGTATCATGTAACTTATCAGTGGGGTGGGTTACTGTTGAAGAGACCCTGGGGCATTTACC TCAGGCATCTGCACTCCTCCGAGCCCGGTGGAGAATGCAGGCTGCTGTAGTCTCAGGTAATGAAGGCACAGCACAGCAGTACTCCACATT >16537_16537_2_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000263856_EPB41L5_chr2_120900578_ENST00000443902_length(amino acids)=169AA_BP=15 MGLFGKTQEKPPKELEEVVKLTEKCLNNVIESPGLNVMRVPPDFKSNILKAQVEAVHKVTKEDSLLSHKNANVQDAATNSAVLNENNVPL -------------------------------------------------------------- >16537_16537_3_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000409727_EPB41L5_chr2_120900578_ENST00000263713_length(transcript)=4886nt_BP=71nt AAGGGGCAAGACGGGTTCAGTTCGTCATGGGGCTGTTTGGAAAGACCCAGGAGAAGCCGCCCAAAGAACTGGAGGAAGTGGTGAAGTTGA CTGAGAAATGCCTTAATAATGTCATTGAGAGCCCAGGATTGAATGTCATGAGAGTTCCTCCTGACTTCAAGAGTAACATTTTGAAGGCTC AAGTAGAAGCAGTGCATAAGGTTACAAAAGAAGATAGCTTATTAAGTCATAAAAATGCCAATGTTCAGGATGCTGCCACAAACAGTGCTG TGTTAAATGAGAATAATGTGCCCCTCCCCAAAGAGTCTCTTGAGACTCTGATGCTTATCACACCTGCCGACAGTGGTTCTGTTCTAAAGG AAGCTACAGATGAATTGGATGCCTTGCTTGCATCTCTAACTGAGAATCTAATTGATCACACAGTTGCACCTCAGGTGTCTTCCACATCCA TGATCACACCCCGGTGGATTGTTCCGCAGAGTGGTGCCATGTCTAATGGACTTGCGGGATGTGAAATGCTTTTGACAGGGAAGGAGGGAC ATGGTAATAAAGATGGAATCTCACTGATCTCTCCCCCAGCGCCATTCTTGGTAGATGCTGTGACCAGCTCTGGTCCCATTTTGGCAGAAG AAGCTGTCCTGAAGCAGAAGTGTTTACTGACCACTGAGCTCTGAGGGCCTGTAGCTGGAATACGCATCTCTCCAGCATTCCGTCCTGGGA TCCGTTTCAGCTAGAATATGTTGGATTCAGGAGCTTGTCCATTATTTGTAGGTAAAAAAAGCTGCACGTAGATTTGACTTCAACTCCGTA AAAAAGACAGCTGTATTTTCCGTCCAACTGGAATTGTTGAATCACACTGCATAGCTGCCCAAAAGAGAGTGTTTGGTCTTGAACTTTCTA TACTTTTATAAATGTTACAAATTCCCGAAAGAAGGGAATTTCTTTTTCTGGGGTTTCCTTCAAACTCTTGGCTCCACCTAGCGGTTCTAT TTGTTCATAACAACTTCATAACAAGCCTGCCTCTGGTAGTCAACAGCCTTTTGAAAGCTATTTCCATCTAGTATCAGGGTGAGAGCATCC TTGATCTGGCTGCCTGTTAGAGAAATTGCACTTTTCCTGACTTACCTAGAAATCAAGAATTTAGGAAATTAATGTGGACACTATAAAGGC AGACTTAGGGCCAACTTTTTTTTTTTTTTACAATTATTACAACACTAAAGAGAAGTTTAGAATATAGAGAGTTTTTAAATGTCTCCCATT CTTTTGATTTCTTACTGTACTGGCTATCTTAATATTTCAAGTTTACATCAAGATAAACCCTGAGAAGAACTACGGAGAAATCAAATAAAA TCCTGTCATATTTTTTTCACCCTGCCTTTCCACAGGAAGCACTCACAGGCACCACACACGTATCATGTAACTTATCAGTGGGGTGGGTTA CTGTTGAAGAGACCCTGGGGCATTTACCTCAGGCATCTGCACTCCTCCGAGCCCGGTGGAGAATGCAGGCTGCTGTAGTCTCAGGTAATG AAGGCACAGCACAGCAGTACTCCACATTGTTTCCTATTTGGACATAGACTTCATTTCCTTTCAGTATAAGCTGAATAAATTTAGAGCTTT CAAACTGGAAAAAAAAATGAAACAAAACAAATACACCAAGACCAAAATAGGCAATAGGAACAGGGGTGAAGGGATGTTGTTTCTTAAATA CCTACCATGTATGAAGCTATACACAGCATATACCGAAAGAACCTGCATTGCACTAGGAATTCTGTGTTAGTTTAAAAGAGATCTCTAAAA CTTCCCCATCCCTTTGGGCCTGTACAAAGAAATTCTGGATGTTAAAATAATATATACTCATCACAGAAAAATAAAGTATAGCAATGTCCA TCTGTAATTCTAATACCCAGAAATAACGCTATTTAGCTTTATAATTTTCGAATGAACAAGGTAAACCTCGGTTGCCATGGGGAAGAAGGA TGATGTGGAAACCATATTGGTAAAGTTGTTAATCCCTCGTTATGGAGAACTGATCTTAAGCTATACCTCCTGGAATTTGCTTTCTAGTTT TCTGTCCTGCAATATGTATATAATTAAGCACTAATTTGTACTGCTTAGCATAAAAGAACATCCAGTCTTAGATCCTTAAAACTTCATGGA TTGGACTTTCCTGGGCTCCTTATAACATAATCGTGTGTCCAGGCAAACGCACACTAGTGTCTGACTGGAAAGCTCAGGAATTTTAATCTT GCACTGTTTCCCAGGGAGCTGTAGTGATTGGAACCCACGTTTGCACAAAACATTTTTGCAGAAGGAAAGTCAACACTTCTTGCTGGCTGC CTCCCCTTAGCCATTATGCTAAAAACAGCTTCTGAGTTTCACTGGTGGGGCTCTTGCCAGTTCTTAATTATAGGACATATTTTCTCAAAG CTGAAGGTGACACCTAGAACCAGGGGCTTGACCCAGGACATGATGGAATGAGCATCAAATTTTCAGTGTCTTGGCAACCGTAGATGTCCT ACAGGGTTACCGTTGTGCTGCTCACCACAGAGCAGCTGAGGCATTATGCCTTGGAAGACCTAAATCTCCCATCCAGTTCAGGAGGTGACA ACATCCTTATTTTAAACTTCCTAAAATTAGGAATTAGGTAGTTGGACATAGTCTGTGACCTTTATGTCGTTGGATACCTGTATTCTTGAC AGTTAGAATATTGGTAGGGACTTTGTTAAAATTCACTTGAATTTCAAGCTCAGAGGAAACTTTGTCTCATGCCCTGACATGAAGTGGCAA ACACGGAAGTTCATACTTGAATGCTGAATTGGCCCCGACAGATTAAATGCGTGTTGGGGATTGGTTTCCTGTCATAGCTGCTGCTGCTGC CATGCGCAGAGCTGCTGTAACAGCTCTTCCTGTTCTGCTCCCCTGAGAACAGTGTGGTGGGGAGAGGCAGGGCTGAGGTGGTCTACGAAT GTGGCAGGTAGGGAAGGGGAGATGTCTGTCTCTTGAGAAGAGAGAGGCATGTGTGCCGGCATCCTTGATGGGTTCAAAGAGAAAGGTTGG AGATGATAGTGGGTGAGAAGCAGGCTGGTGAGACTGGGCTGAGGTTGGAGAAGGGGCAGCCGGGGGCACTGCTGAGGGTTTGCCGTGCAC GCCTCGGACGGAGCACGGTGGGGTGGCGGGGAGCAGATAGTGCTGCCTCCCTGCGGGCAGACAGGAGAGGAACCTCAACTCAGTCCATTT CATAGCCCTGATAGGGGAAGTGGGAGTTGACAGGATGGTTTAAAATAAGACGTGAAGGTTTCAGTTACCTGCTCTAGACTTTGCCTGAGA ACTTGTAAATTAATCAGTGAGACCTAATTTGTGACATGTCAGTAGCATCATCTTTTGACACACAGGAGGTCATGGTCATTTCATTCCTAC TCTTCAGGAGACACTGCTGAACAGAGGAATGATTCTGTTCCTTGTGTGCTTACTTCCTTAACATTTATACATTGTTTTAAGAAAAAACTT TTAAAAATATTTCTTATAGTCTCCTAACATTTGTCTCTAGCCTTTGCCTTTGTACAATCACAGATATCCTATGGAGATTTAAGGATGAAA GCCCTGAGTTGTTCTTGGGTTCTTGGATCTGGACTACTTGTTATCTTATGCTTCTCACTTCTGGCTAAAACTTGCACCTCTTCTTCTCTT AGCTAAGCCCCAAAATGAAGATTTCCTTCAGAAGTCTTGTTAGCAGAATTATTTATCAGTCACAGAGAGAAAAATCTGCTATTTTTCTAA GTAAGAGTCTCGAGAAGCAGAGTTTTTGTCTTGTCATTGAGAGGAGTCAGCAGTCTTGTTCTGTAAAGGACCAGAGATGGTAAATACTGT CCCACTCAGCTCTGCTGGCGCAGTACAGCAGCAGCAGCCCCAGCACAGCTGTGTTCCTGCGGAGTCCCCTTTACAAAGCCGCTGACCCCT GATGTGAAACTTTGTAGAGCAGCAGAGTGGCTGCGTGAAACGGGAGGCTGGCAGGTCCTCAGATAGGTTCTGCAGTGTTACCTGTCACTT GGAGGCAGCCAACACTTCTGGACATTGCATCCTTATTCACACATGTGGCAGCTGAACGAGGTGCTGTTGTGGGTGTCTCAGCTCTGAGGG TCTTTGTGAGCTCCCACTGTTGTGGGTGTCTCAGCTCTGAGGGTCTTTGTGAGCTCCCACTGTTGTGGGTGTCTCAGCTCTGAGGGTCTT TGTGAGTTCCCACCAACTTTTAATTATTCATGCCCTTGACCATGTGGTTGCTTGGAGACCTGGGGTCTTCTGCAGACTGAAGAAGACACA TTTCTAGATTATTTGTCCTTTTTATCCTCTCAAAAATTTAAACACTGTACCTCTTCAGTGGTCAGAAGAAAGTTGGAAACTTTTCCTACA TATTAGGCGTTAGTATGAGGACATTTGTTTGAATTATAGAAATTTGCCCTGAGCTGAACTGGGTTGTGTTAACACATTGGTAGAGCTATG ATTCCTTCCCAGTTCTAAGAGATACGATCTGTAAGTCCCTATGTCACCACATTGCTTGAGATGATCATTCAGTTACTTGTCAGGATTTCT CCTCTTCAGAGAGATTTTTTTTTATAGCACAGATTCCTTTGCCCCTTTTATCTCCTTATCTGGATATGATAAGTGGTTATGAGGGTCTCA CTAACTATTTTGTGTTTACCTTTTATATGTGTAAAACTTTGCAGTAGCATTTAAAGTGTAATTTATTTTTCTATCAAGTGCACTATTCAT TTAGTGTGTTCCAGTTTTATATGACTTGTATTAGAAACACTGCACTGAGTTGTTTGTACACTGAAATGAGAACTCTAGATGTAACTCTAT >16537_16537_3_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000409727_EPB41L5_chr2_120900578_ENST00000263713_length(amino acids)=215AA_BP=15 MGLFGKTQEKPPKELEEVVKLTEKCLNNVIESPGLNVMRVPPDFKSNILKAQVEAVHKVTKEDSLLSHKNANVQDAATNSAVLNENNVPL PKESLETLMLITPADSGSVLKEATDELDALLASLTENLIDHTVAPQVSSTSMITPRWIVPQSGAMSNGLAGCEMLLTGKEGHGNKDGISL -------------------------------------------------------------- >16537_16537_4_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000409727_EPB41L5_chr2_120900578_ENST00000443902_length(transcript)=1497nt_BP=71nt AAGGGGCAAGACGGGTTCAGTTCGTCATGGGGCTGTTTGGAAAGACCCAGGAGAAGCCGCCCAAAGAACTGGAGGAAGTGGTGAAGTTGA CTGAGAAATGCCTTAATAATGTCATTGAGAGCCCAGGATTGAATGTCATGAGAGTTCCTCCTGACTTCAAGAGTAACATTTTGAAGGCTC AAGTAGAAGCAGTGCATAAGGTTACAAAAGAAGATAGCTTATTAAGTCATAAAAATGCCAATGTTCAGGATGCTGCCACAAACAGTGCTG TGTTAAATGAGAATAATGTGCCCCTCCCCAAAGAGTCTCTTGAGACTCTGATGCTTATCACACCTGCCGACAGTGGTTCTGTTCTAAAGG AAGCTACAGATGAATTGGATGCCTTGCTTGCATCTCTAACTGAGAATCTAATTGATCACACAGTTGCACCTCAGGTGTCTTCCACATCCA TGATCACACCCCGGTGGATTGTTCCGCTCTGGTCCCATTTTGGCAGAAGAAGCTGTCCTGAAGCAGAAGTGTTTACTGACCACTGAGCTC TGAGGGCCTGTAGCTGGAATACGCATCTCTCCAGCATTCCGTCCTGGGATCCGTTTCAGCTAGAATATGTTGGATTCAGGAGCTTGTCCA TTATTTGTAGGTAAAAAAAGCTGCACGTAGATTTGACTTCAACTCCGTAAAAAAGACAGCTGTATTTTCCGTCCAACTGGAATTGTTGAA TCACACTGCATAGCTGCCCAAAAGAGAGTGTTTGGTCTTGAACTTTCTATACTTTTATAAATGTTACAAATTCCCGAAAGAAGGGAATTT CTTTTTCTGGGGTTTCCTTCAAACTCTTGGCTCCACCTAGCGGTTCTATTTGTTCATAACAACTTCATAACAAGCCTGCCTCTGGTAGTC AACAGCCTTTTGAAAGCTATTTCCATCTAGTATCAGGGTGAGAGCATCCTTGATCTGGCTGCCTGTTAGAGAAATTGCACTTTTCCTGAC TTACCTAGAAATCAAGAATTTAGGAAATTAATGTGGACACTATAAAGGCAGACTTAGGGCCAACTTTTTTTTTTTTTTACAATTATTACA ACACTAAAGAGAAGTTTAGAATATAGAGAGTTTTTAAATGTCTCCCATTCTTTTGATTTCTTACTGTACTGGCTATCTTAATATTTCAAG TTTACATCAAGATAAACCCTGAGAAGAACTACGGAGAAATCAAATAAAATCCTGTCATATTTTTTTCACCCTGCCTTTCCACAGGAAGCA CTCACAGGCACCACACACGTATCATGTAACTTATCAGTGGGGTGGGTTACTGTTGAAGAGACCCTGGGGCATTTACCTCAGGCATCTGCA CTCCTCCGAGCCCGGTGGAGAATGCAGGCTGCTGTAGTCTCAGGTAATGAAGGCACAGCACAGCAGTACTCCACATTGTTTCCTATTTGG >16537_16537_4_CHMP3-EPB41L5_CHMP3_chr2_86790426_ENST00000409727_EPB41L5_chr2_120900578_ENST00000443902_length(amino acids)=169AA_BP=15 MGLFGKTQEKPPKELEEVVKLTEKCLNNVIESPGLNVMRVPPDFKSNILKAQVEAVHKVTKEDSLLSHKNANVQDAATNSAVLNENNVPL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHMP3-EPB41L5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000263856 | - | 1 | 6 | 203_207 | 15.0 | 223.0 | STAMBP |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409225 | - | 1 | 5 | 203_207 | 0 | 157.0 | STAMBP |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409727 | - | 1 | 6 | 203_207 | 15.0 | 183.0 | STAMBP |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000439940 | - | 1 | 8 | 203_207 | 0 | 252.0 | STAMBP |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000263856 | - | 1 | 6 | 151_222 | 15.0 | 223.0 | VPS4A |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409225 | - | 1 | 5 | 151_222 | 0 | 157.0 | VPS4A |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000409727 | - | 1 | 6 | 151_222 | 15.0 | 183.0 | VPS4A |
| Hgene | CHMP3 | chr2:86790426 | chr2:120900578 | ENST00000439940 | - | 1 | 8 | 151_222 | 0 | 252.0 | VPS4A |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHMP3-EPB41L5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHMP3-EPB41L5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
