
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHMP5-CTNNA1 (FusionGDB2 ID:16566) |
Fusion Gene Summary for CHMP5-CTNNA1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHMP5-CTNNA1 | Fusion gene ID: 16566 | Hgene | Tgene | Gene symbol | CHMP5 | CTNNA1 | Gene ID | 51510 | 1495 |
| Gene name | charged multivesicular body protein 5 | catenin alpha 1 | |
| Synonyms | C9orf83|CGI-34|HSPC177|PNAS-2|SNF7DC2|Vps60 | CAP102|MDPT2 | |
| Cytomap | 9p13.3 | 5q31.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | charged multivesicular body protein 5SNF7 domain containing 2SNF7 domain-containing protein 2apoptosis-related protein PNAS-2chromatin-modifying protein 5hVps60vacuolar protein sorting-associated protein 60 | catenin alpha-1alpha-E-catenincatenin (cadherin-associated protein), alpha 1, 102kDaepididymis secretory sperm binding proteinrenal carcinoma antigen NY-REN-13 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NZZ3 | P35221 | |
| Ensembl transtripts involved in fusion gene | ENST00000487080, ENST00000223500, ENST00000419016, | ENST00000302763, ENST00000355078, ENST00000518825, ENST00000520400, ENST00000540387, | |
| Fusion gene scores | * DoF score | 5 X 4 X 4=80 | 16 X 20 X 7=2240 |
| # samples | 6 | 22 | |
| ** MAII score | log2(6/80*10)=-0.415037499278844 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/2240*10)=-3.34792330342031 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CHMP5 [Title/Abstract] AND CTNNA1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CHMP5(33278223)-CTNNA1(138264934), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CTNNA1 | GO:0071681 | cellular response to indole-3-methanol | 10868478 |
 Fusion gene breakpoints across CHMP5 (5'-gene) Fusion gene breakpoints across CHMP5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
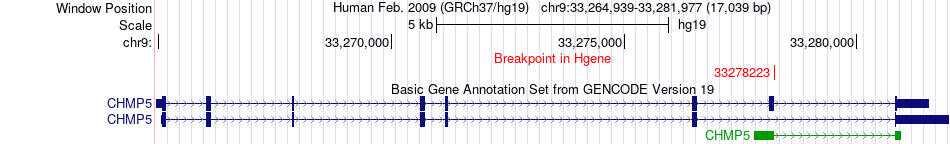 |
 Fusion gene breakpoints across CTNNA1 (3'-gene) Fusion gene breakpoints across CTNNA1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
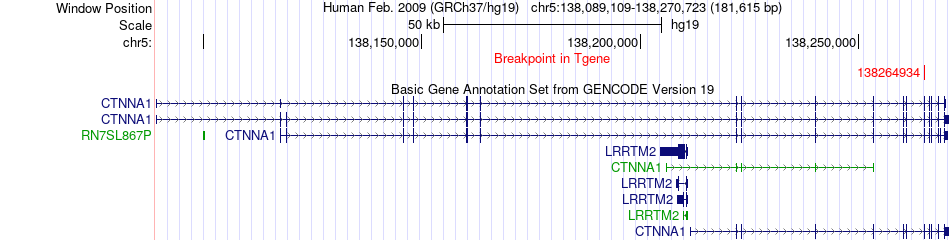 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 61N | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
Top |
Fusion Gene ORF analysis for CHMP5-CTNNA1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000487080 | ENST00000302763 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| 3UTR-3CDS | ENST00000487080 | ENST00000355078 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| 3UTR-3CDS | ENST00000487080 | ENST00000518825 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| 3UTR-intron | ENST00000487080 | ENST00000520400 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| 3UTR-intron | ENST00000487080 | ENST00000540387 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| 5CDS-intron | ENST00000223500 | ENST00000520400 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| 5CDS-intron | ENST00000223500 | ENST00000540387 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| In-frame | ENST00000223500 | ENST00000302763 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| In-frame | ENST00000223500 | ENST00000355078 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| In-frame | ENST00000223500 | ENST00000518825 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| intron-3CDS | ENST00000419016 | ENST00000302763 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| intron-3CDS | ENST00000419016 | ENST00000355078 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| intron-3CDS | ENST00000419016 | ENST00000518825 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| intron-intron | ENST00000419016 | ENST00000520400 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
| intron-intron | ENST00000419016 | ENST00000540387 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000223500 | CHMP5 | chr9 | 33278223 | + | ENST00000355078 | CTNNA1 | chr5 | 138264934 | + | 1692 | 746 | 128 | 1567 | 479 |
| ENST00000223500 | CHMP5 | chr9 | 33278223 | + | ENST00000302763 | CTNNA1 | chr5 | 138264934 | + | 2513 | 746 | 128 | 1567 | 479 |
| ENST00000223500 | CHMP5 | chr9 | 33278223 | + | ENST00000518825 | CTNNA1 | chr5 | 138264934 | + | 2381 | 746 | 128 | 1372 | 414 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000223500 | ENST00000355078 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + | 0.001854548 | 0.99814546 |
| ENST00000223500 | ENST00000302763 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + | 0.001089741 | 0.9989103 |
| ENST00000223500 | ENST00000518825 | CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + | 0.001591107 | 0.998409 |
Top |
Fusion Genomic Features for CHMP5-CTNNA1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + | 8.06E-07 | 0.99999917 |
| CHMP5 | chr9 | 33278223 | + | CTNNA1 | chr5 | 138264934 | + | 8.06E-07 | 0.99999917 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
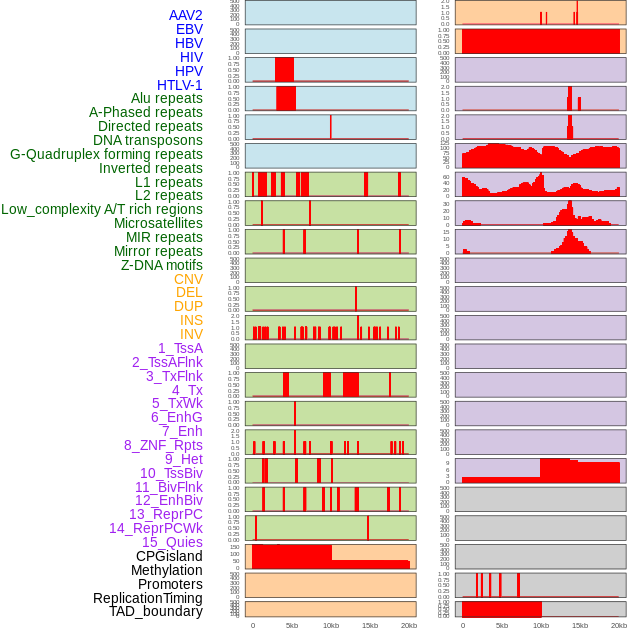 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
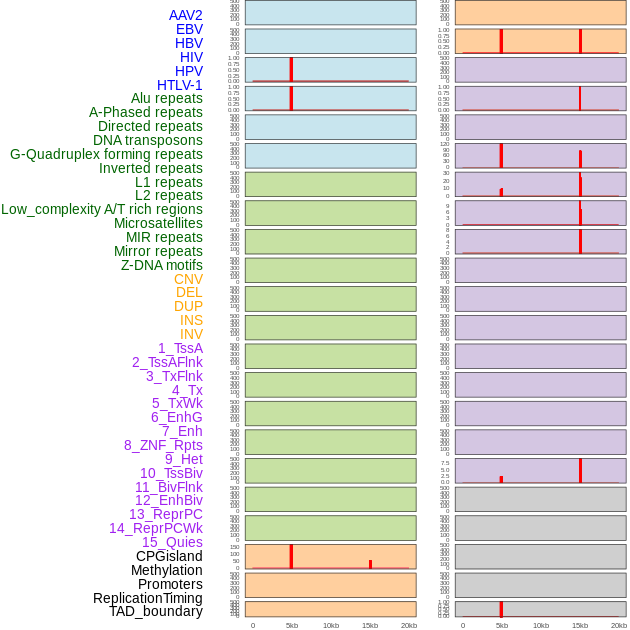 |
Top |
Fusion Protein Features for CHMP5-CTNNA1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:33278223/chr5:138264934) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHMP5 | CTNNA1 |
| FUNCTION: Probable peripherally associated component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and mostly are delivered to lysosomes enabling degradation of membrane proteins, such as stimulated growth factor receptors, lysosomal enzymes and lipids. The MVB pathway appears to require the sequential function of ESCRT-O, -I,-II and -III complexes. ESCRT-III proteins mostly dissociate from the invaginating membrane before the ILV is released. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis and the budding of enveloped viruses (HIV-1 and other lentiviruses). ESCRT-III proteins are believed to mediate the necessary vesicle extrusion and/or membrane fission activities, possibly in conjunction with the AAA ATPase VPS4. Involved in HIV-1 p6- and p9-dependent virus release. {ECO:0000269|PubMed:14519844}. | FUNCTION: Associates with the cytoplasmic domain of a variety of cadherins. The association of catenins to cadherins produces a complex which is linked to the actin filament network, and which seems to be of primary importance for cadherins cell-adhesion properties. Can associate with both E- and N-cadherins. Originally believed to be a stable component of E-cadherin/catenin adhesion complexes and to mediate the linkage of cadherins to the actin cytoskeleton at adherens junctions. In contrast, cortical actin was found to be much more dynamic than E-cadherin/catenin complexes and CTNNA1 was shown not to bind to F-actin when assembled in the complex suggesting a different linkage between actin and adherens junctions components. The homodimeric form may regulate actin filament assembly and inhibit actin branching by competing with the Arp2/3 complex for binding to actin filaments. Involved in the regulation of WWTR1/TAZ, YAP1 and TGFB1-dependent SMAD2 and SMAD3 nuclear accumulation (By similarity). May play a crucial role in cell differentiation. {ECO:0000250|UniProtKB:P26231, ECO:0000269|PubMed:25653389}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHMP5 | chr9:33278223 | chr5:138264934 | ENST00000223500 | + | 7 | 8 | 26_179 | 203 | 220.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHMP5 | chr9:33278223 | chr5:138264934 | ENST00000419016 | + | 1 | 7 | 26_179 | 0 | 172.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | CTNNA1 | chr9:33278223 | chr5:138264934 | ENST00000302763 | 12 | 18 | 2_228 | 633 | 907.0 | Region | Note=Involved in homodimerization | |
| Tgene | CTNNA1 | chr9:33278223 | chr5:138264934 | ENST00000540387 | 6 | 12 | 2_228 | 263 | 537.0 | Region | Note=Involved in homodimerization |
Top |
Fusion Gene Sequence for CHMP5-CTNNA1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16566_16566_1_CHMP5-CTNNA1_CHMP5_chr9_33278223_ENST00000223500_CTNNA1_chr5_138264934_ENST00000302763_length(transcript)=2513nt_BP=746nt AATCCGGAAAAAGAATCGGGAAACGCCAGGAGGCATATTGCGCTTGCGCACGGAGGGGCCGGAAGTCGAGGCGGGAGTGACTCTGCTTCC GTTTCTGGTTTTGCTCTAGTGTTTGGGTTTCTTCGCGGCTGCTCAAGATGAACCGACTCTTCGGGAAAGCGAAACCCAAGGCTCCGCCGC CCAGCCTGACTGACTGCATTGGCACGGTGGACAGTAGAGCAGAATCCATTGACAAGAAGATTTCTCGATTGGATGCTGAGCTAGTGAAGT ATAAGGATCAGATCAAGAAGATGAGAGAGGGTCCTGCAAAGAATATGGTCAAGCAGAAAGCCTTGCGAGTTTTAAAGCAAAAGAGGATGT ATGAGCAGCAGCGGGACAATCTTGCCCAACAGTCATTCAACATGGAACAAGCCAATTATACCATCCAGTCTTTGAAGGACACCAAGACCA CGGTTGATGCTATGAAACTGGGAGTAAAGGAAATGAAGAAGGCATACAAGCAAGTGAAGATCGACCAGATTGAGGATTTACAAGACCAGC TAGAGGATATGATGGAAGATGCAAATGAAATCCAAGAAGCACTGAGTCGCAGTTATGGCACCCCAGAACTGGATGAAGATGATTTAGAAG CAGAGTTGGATGCACTAGGTGATGAGCTTCTGGCTGATGAAGACAGTTCTTATTTGGATGAGGCAGCATCTGCACCTGCAATTCCAGAAG GTGTTCCCACTGATACAAAAAACAAGACCCCTGAGGAGTTGGATGACTCTGACTTTGAGACAGAAGATTTTGATGTCAGAAGCAGGACGA GCGTCCAGACAGAAGACGATCAGCTGATAGCTGGCCAGAGTGCCCGGGCGATCATGGCTCAGCTTCCCCAGGAGCAAAAAGCGAAGATTG CGGAACAGGTGGCCAGCTTCCAGGAAGAAAAGAGCAAGCTGGATGCTGAAGTGTCCAAATGGGACGACAGTGGCAATGACATCATTGTGC TGGCCAAGCAGATGTGCATGATTATGATGGAGATGACAGACTTTACCCGAGGTAAAGGACCACTCAAAAATACATCGGATGTCATCAGTG CTGCCAAGAAAATTGCTGAGGCAGGATCCAGGATGGACAAGCTTGGCCGCACCATTGCAGACCATTGCCCCGACTCGGCTTGCAAGCAGG ACCTGCTGGCCTACCTGCAACGCATCGCCCTCTACTGCCACCAGCTGAACATCTGCAGCAAGGTCAAGGCCGAGGTGCAGAATCTCGGCG GGGAGCTTGTTGTCTCTGGGGTGGACAGCGCCATGTCCCTGATCCAGGCAGCCAAGAACTTGATGAATGCTGTGGTGCAGACAGTGAAGG CATCCTACGTCGCCTCTACCAAATACCAAAAGTCACAGGGTATGGCTTCCCTCAACCTTCCTGCTGTGTCATGGAAGATGAAGGCACCAG AGAAAAAGCCATTGGTGAAGAGAGAGAAACAGGATGAGACACAGACCAAGATTAAACGGGCATCTCAGAAGAAGCACGTGAACCCGGTGC AGGCCCTCAGCGAGTTCAAAGCTATGGACAGCATCTAAGTCTGCCCAGGCCGGCCGCCCCCACCCCTCGGGGCTCCTGAATATCAGTCAC TGTTCGTCACTCAAATGAATTTGCTAAATACAACACTGATACTAGATTCCACAGGGAAATGGGCAGACTGAACCAGTCCAGGTGGTGAAT TTTCCAAGAACATAGTTTAAGTTGATTAAAAATGCTTTTAGAATGCAGGAGCCTACTTCTAGCTGTATTTTTTGTATGCTTAAATAAAAA TAAAAATTCATAACCAAAGAGAATCCCACATTAGCTTGTTAGTAATGCTCTGACCAAGCCGAGATGCCCATTCTCTTAGTGATGGCGGCG TTAGGGTTTGAGAGAAGGGAATTTGGCTCAACTTCAGTTGAGAGGGTGCAGTCCAGACAGCTTGACTGCTTTTAAATGACCAAAGATGAC CTGTGGTAAGCAACCTGGGCATCTTAGGAAGCAGTCCCTGGAGAAGGCATGTTCCCAGAAAGGTCTCTGGAGGGACAAACTCACTCAGTA AAACATAATGTATCATGAAGAAAACTGATTCTCTATGACATGAAATGAAAATTTTAATGCATTGTTATAATTACTAATGTACGCTGCTGC AGGACATTAATAAAGTTGCTTTTTTAGGCTACAGTGTCTCGATGCCATAATCAGAACACACTTTTTTTCCTCTTTCTCCCAGCTTCAAAT GCAAATTCATCATTGGGCTCACTTCTAATAACTGCAGTGTTTCCCGCCTTGGGCTTGCAGCAGAAAAACCTGACAACATAGTGTTTGCTA AGGCAGTAATTTAGACTTTACCTTATTTGTGATTACTGTAGTGATTGATTGATTGATTACTATTAACTACAAGGTATAATTTACTATCAC >16566_16566_1_CHMP5-CTNNA1_CHMP5_chr9_33278223_ENST00000223500_CTNNA1_chr5_138264934_ENST00000302763_length(amino acids)=479AA_BP=206 MLKMNRLFGKAKPKAPPPSLTDCIGTVDSRAESIDKKISRLDAELVKYKDQIKKMREGPAKNMVKQKALRVLKQKRMYEQQRDNLAQQSF NMEQANYTIQSLKDTKTTVDAMKLGVKEMKKAYKQVKIDQIEDLQDQLEDMMEDANEIQEALSRSYGTPELDEDDLEAELDALGDELLAD EDSSYLDEAASAPAIPEGVPTDTKNKTPEELDDSDFETEDFDVRSRTSVQTEDDQLIAGQSARAIMAQLPQEQKAKIAEQVASFQEEKSK LDAEVSKWDDSGNDIIVLAKQMCMIMMEMTDFTRGKGPLKNTSDVISAAKKIAEAGSRMDKLGRTIADHCPDSACKQDLLAYLQRIALYC HQLNICSKVKAEVQNLGGELVVSGVDSAMSLIQAAKNLMNAVVQTVKASYVASTKYQKSQGMASLNLPAVSWKMKAPEKKPLVKREKQDE -------------------------------------------------------------- >16566_16566_2_CHMP5-CTNNA1_CHMP5_chr9_33278223_ENST00000223500_CTNNA1_chr5_138264934_ENST00000355078_length(transcript)=1692nt_BP=746nt AATCCGGAAAAAGAATCGGGAAACGCCAGGAGGCATATTGCGCTTGCGCACGGAGGGGCCGGAAGTCGAGGCGGGAGTGACTCTGCTTCC GTTTCTGGTTTTGCTCTAGTGTTTGGGTTTCTTCGCGGCTGCTCAAGATGAACCGACTCTTCGGGAAAGCGAAACCCAAGGCTCCGCCGC CCAGCCTGACTGACTGCATTGGCACGGTGGACAGTAGAGCAGAATCCATTGACAAGAAGATTTCTCGATTGGATGCTGAGCTAGTGAAGT ATAAGGATCAGATCAAGAAGATGAGAGAGGGTCCTGCAAAGAATATGGTCAAGCAGAAAGCCTTGCGAGTTTTAAAGCAAAAGAGGATGT ATGAGCAGCAGCGGGACAATCTTGCCCAACAGTCATTCAACATGGAACAAGCCAATTATACCATCCAGTCTTTGAAGGACACCAAGACCA CGGTTGATGCTATGAAACTGGGAGTAAAGGAAATGAAGAAGGCATACAAGCAAGTGAAGATCGACCAGATTGAGGATTTACAAGACCAGC TAGAGGATATGATGGAAGATGCAAATGAAATCCAAGAAGCACTGAGTCGCAGTTATGGCACCCCAGAACTGGATGAAGATGATTTAGAAG CAGAGTTGGATGCACTAGGTGATGAGCTTCTGGCTGATGAAGACAGTTCTTATTTGGATGAGGCAGCATCTGCACCTGCAATTCCAGAAG GTGTTCCCACTGATACAAAAAACAAGACCCCTGAGGAGTTGGATGACTCTGACTTTGAGACAGAAGATTTTGATGTCAGAAGCAGGACGA GCGTCCAGACAGAAGACGATCAGCTGATAGCTGGCCAGAGTGCCCGGGCGATCATGGCTCAGCTTCCCCAGGAGCAAAAAGCGAAGATTG CGGAACAGGTGGCCAGCTTCCAGGAAGAAAAGAGCAAGCTGGATGCTGAAGTGTCCAAATGGGACGACAGTGGCAATGACATCATTGTGC TGGCCAAGCAGATGTGCATGATTATGATGGAGATGACAGACTTTACCCGAGGTAAAGGACCACTCAAAAATACATCGGATGTCATCAGTG CTGCCAAGAAAATTGCTGAGGCAGGATCCAGGATGGACAAGCTTGGCCGCACCATTGCAGACCATTGCCCCGACTCGGCTTGCAAGCAGG ACCTGCTGGCCTACCTGCAACGCATCGCCCTCTACTGCCACCAGCTGAACATCTGCAGCAAGGTCAAGGCCGAGGTGCAGAATCTCGGCG GGGAGCTTGTTGTCTCTGGGGTGGACAGCGCCATGTCCCTGATCCAGGCAGCCAAGAACTTGATGAATGCTGTGGTGCAGACAGTGAAGG CATCCTACGTCGCCTCTACCAAATACCAAAAGTCACAGGGTATGGCTTCCCTCAACCTTCCTGCTGTGTCATGGAAGATGAAGGCACCAG AGAAAAAGCCATTGGTGAAGAGAGAGAAACAGGATGAGACACAGACCAAGATTAAACGGGCATCTCAGAAGAAGCACGTGAACCCGGTGC AGGCCCTCAGCGAGTTCAAAGCTATGGACAGCATCTAAGTCTGCCCAGGCCGGCCGCCCCCACCCCTCGGGGCTCCTGAATATCAGTCAC >16566_16566_2_CHMP5-CTNNA1_CHMP5_chr9_33278223_ENST00000223500_CTNNA1_chr5_138264934_ENST00000355078_length(amino acids)=479AA_BP=206 MLKMNRLFGKAKPKAPPPSLTDCIGTVDSRAESIDKKISRLDAELVKYKDQIKKMREGPAKNMVKQKALRVLKQKRMYEQQRDNLAQQSF NMEQANYTIQSLKDTKTTVDAMKLGVKEMKKAYKQVKIDQIEDLQDQLEDMMEDANEIQEALSRSYGTPELDEDDLEAELDALGDELLAD EDSSYLDEAASAPAIPEGVPTDTKNKTPEELDDSDFETEDFDVRSRTSVQTEDDQLIAGQSARAIMAQLPQEQKAKIAEQVASFQEEKSK LDAEVSKWDDSGNDIIVLAKQMCMIMMEMTDFTRGKGPLKNTSDVISAAKKIAEAGSRMDKLGRTIADHCPDSACKQDLLAYLQRIALYC HQLNICSKVKAEVQNLGGELVVSGVDSAMSLIQAAKNLMNAVVQTVKASYVASTKYQKSQGMASLNLPAVSWKMKAPEKKPLVKREKQDE -------------------------------------------------------------- >16566_16566_3_CHMP5-CTNNA1_CHMP5_chr9_33278223_ENST00000223500_CTNNA1_chr5_138264934_ENST00000518825_length(transcript)=2381nt_BP=746nt AATCCGGAAAAAGAATCGGGAAACGCCAGGAGGCATATTGCGCTTGCGCACGGAGGGGCCGGAAGTCGAGGCGGGAGTGACTCTGCTTCC GTTTCTGGTTTTGCTCTAGTGTTTGGGTTTCTTCGCGGCTGCTCAAGATGAACCGACTCTTCGGGAAAGCGAAACCCAAGGCTCCGCCGC CCAGCCTGACTGACTGCATTGGCACGGTGGACAGTAGAGCAGAATCCATTGACAAGAAGATTTCTCGATTGGATGCTGAGCTAGTGAAGT ATAAGGATCAGATCAAGAAGATGAGAGAGGGTCCTGCAAAGAATATGGTCAAGCAGAAAGCCTTGCGAGTTTTAAAGCAAAAGAGGATGT ATGAGCAGCAGCGGGACAATCTTGCCCAACAGTCATTCAACATGGAACAAGCCAATTATACCATCCAGTCTTTGAAGGACACCAAGACCA CGGTTGATGCTATGAAACTGGGAGTAAAGGAAATGAAGAAGGCATACAAGCAAGTGAAGATCGACCAGATTGAGGATTTACAAGACCAGC TAGAGGATATGATGGAAGATGCAAATGAAATCCAAGAAGCACTGAGTCGCAGTTATGGCACCCCAGAACTGGATGAAGATGATTTAGAAG CAGAGTTGGATGCACTAGGTGATGAGCTTCTGGCTGATGAAGACAGTTCTTATTTGGATGAGGCAGCATCTGCACCTGCAATTCCAGAAG GTGTTCCCACTGATACAAAAAACAAGACCCCTGAGGAGTTGGATGACTCTGACTTTGAGACAGAAGATTTTGATGTCAGAAGCAGGACGA GCGTCCAGACAGAAGACGATCAGCTGATAGCTGGCCAGAGTGCCCGGGCGATCATGGCTCAGCTTCCCCAGGAGCAAAAAGCGAAGATTG CGGAACAGGTGGCCAGCTTCCAGGAAGAAAAGAGCAAGCTGGATGCTGAAGTGTCCAAATGGGACGACAGTGGCAATGACATCATTGTGC TGGCCAAGCAGATGTGCATGATTATGATGGAGATGACAGACTTTACCCGAGGTAAAGGACCACTCAAAAATACATCGGATGTCATCAGTG CTGCCAAGAAAATTGCTGAGGCAGGATCCAGGATGGACAAGCTTGGCCGCACCATTGCAGACCATTGCCCCGACTCGGCTTGCAAGCAGG ACCTGCTGGCCTACCTGCAACGCATCGCCCTCTACTGCCACCAGCTGAACATCTGCAGCAAGGTCAAGGCCGAGGTGCAGAATCTCGGCG GGGAGCTTGTTGTCTCTGGGAACTGTGACACCTGCGGGGCACTGCAAGGGCTGAAAGGCTGGCCTCCTCCCCTTTGCTGGCCACTCACTG GGTGGACAGCGCCATGTCCCTGATCCAGGCAGCCAAGAACTTGATGAATGCTGTGGTGCAGACAGTGAAGGCATCCTACGTCGCCTCTAC CAAATACCAAAAGTCACAGGGTATGGCTTCCCTCAACCTTCCTGCTGTGTCATGGAAGATGAAGGCACCAGAGAAAAAGCCATTGGTGAA GAGAGAGAAACAGGATGAGACACAGACCAAGATTAAACGGGCATCTCAGAAGAAGCACGTGAACCCGGTGCAGGCCCTCAGCGAGTTCAA AGCTATGGACAGCATCTAAGTCTGCCCAGGCCGGCCGCCCCCACCCCTCGGGGCTCCTGAATATCAGTCACTGTTCGTCACTCAAATGAA TTTGCTAAATACAACACTGATACTAGATTCCACAGGGAAATGGGCAGACTGAACCAGTCCAGGTGGTGAATTTTCCAAGAACATAGTTTA AGTTGATTAAAAATGCTTTTAGAATGCAGGAGCCTACTTCTAGCTGTATTTTTTGTATGCTTAAATAAAAATAAAAATTCATAACCAAAG AGAATCCCACATTAGCTTGTTAGTAATGCTCTGACCAAGCCGAGATGCCCATTCTCTTAGTGATGGCGGCGTTAGGGTTTGAGAGAAGGG AATTTGGCTCAACTTCAGTTGAGAGGGTGCAGTCCAGACAGCTTGACTGCTTTTAAATGACCAAAGATGACCTGTGGTAAGCAACCTGGG CATCTTAGGAAGCAGTCCCTGGAGAAGGCATGTTCCCAGAAAGGTCTCTGGAGGGACAAACTCACTCAGTAAAACATAATGTATCATGAA GAAAACTGATTCTCTATGACATGAAATGAAAATTTTAATGCATTGTTATAATTACTAATGTACGCTGCTGCAGGACATTAATAAAGTTGC TTTTTTAGGCTACAGTGTCTCGATGCCATAATCAGAACACACTTTTTTTCCTCTTTCTCCCAGCTTCAAATGCAAATTCATCATTGGGCT >16566_16566_3_CHMP5-CTNNA1_CHMP5_chr9_33278223_ENST00000223500_CTNNA1_chr5_138264934_ENST00000518825_length(amino acids)=414AA_BP=206 MLKMNRLFGKAKPKAPPPSLTDCIGTVDSRAESIDKKISRLDAELVKYKDQIKKMREGPAKNMVKQKALRVLKQKRMYEQQRDNLAQQSF NMEQANYTIQSLKDTKTTVDAMKLGVKEMKKAYKQVKIDQIEDLQDQLEDMMEDANEIQEALSRSYGTPELDEDDLEAELDALGDELLAD EDSSYLDEAASAPAIPEGVPTDTKNKTPEELDDSDFETEDFDVRSRTSVQTEDDQLIAGQSARAIMAQLPQEQKAKIAEQVASFQEEKSK LDAEVSKWDDSGNDIIVLAKQMCMIMMEMTDFTRGKGPLKNTSDVISAAKKIAEAGSRMDKLGRTIADHCPDSACKQDLLAYLQRIALYC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHMP5-CTNNA1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | CHMP5 | chr9:33278223 | chr5:138264934 | ENST00000223500 | + | 7 | 8 | 121_158 | 203.0 | 220.0 | VTA1 |
| Tgene | CTNNA1 | chr9:33278223 | chr5:138264934 | ENST00000540387 | 6 | 12 | 325_394 | 263.0 | 537.0 | alpha-actinin |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CHMP5 | chr9:33278223 | chr5:138264934 | ENST00000419016 | + | 1 | 7 | 121_158 | 0 | 172.0 | VTA1 |
| Tgene | CTNNA1 | chr9:33278223 | chr5:138264934 | ENST00000302763 | 12 | 18 | 325_394 | 633.0 | 907.0 | alpha-actinin | |
| Tgene | CTNNA1 | chr9:33278223 | chr5:138264934 | ENST00000302763 | 12 | 18 | 97_148 | 633.0 | 907.0 | JUP and CTNNB1 | |
| Tgene | CTNNA1 | chr9:33278223 | chr5:138264934 | ENST00000540387 | 6 | 12 | 97_148 | 263.0 | 537.0 | JUP and CTNNB1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHMP5-CTNNA1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHMP5-CTNNA1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
