
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHP1-RAD51 (FusionGDB2 ID:16610) |
Fusion Gene Summary for CHP1-RAD51 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHP1-RAD51 | Fusion gene ID: 16610 | Hgene | Tgene | Gene symbol | CHP1 | RAD51 | Gene ID | 26973 | 5888 |
| Gene name | cysteine and histidine rich domain containing 1 | RAD51 recombinase | |
| Synonyms | CHP1 | BRCC5|FANCR|HRAD51|HsRad51|HsT16930|MRMV2|RAD51A|RECA | |
| Cytomap | 11q14.3 | 15q15.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cysteine and histidine-rich domain-containing protein 1CHORD-containing protein 1CHP-1chord domain-containing protein 1cysteine and histidine-rich domain (CHORD) containing 1cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein | DNA repair protein RAD51 homolog 1BRCA1/BRCA2-containing complex, subunit 5RAD51 homolog ARecA, E. coli, homolog ofRecA-like proteinrecombination protein A | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q99653 | O15315 | |
| Ensembl transtripts involved in fusion gene | ENST00000558351, ENST00000334660, ENST00000560397, | ENST00000267868, ENST00000423169, ENST00000382643, ENST00000530766, ENST00000532743, ENST00000557850, | |
| Fusion gene scores | * DoF score | 11 X 13 X 7=1001 | 7 X 7 X 5=245 |
| # samples | 15 | 7 | |
| ** MAII score | log2(15/1001*10)=-2.73840756834011 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/245*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CHP1 [Title/Abstract] AND RAD51 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CHP1(41562816)-RAD51(41011003), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | RAD51 | GO:0000724 | double-strand break repair via homologous recombination | 16428451 |
| Tgene | RAD51 | GO:0006268 | DNA unwinding involved in DNA replication | 7988572 |
| Tgene | RAD51 | GO:0006974 | cellular response to DNA damage stimulus | 23509288|25585578 |
| Tgene | RAD51 | GO:0010569 | regulation of double-strand break repair via homologous recombination | 23754376 |
| Tgene | RAD51 | GO:0031297 | replication fork processing | 25585578 |
| Tgene | RAD51 | GO:0051106 | positive regulation of DNA ligation | 8929543 |
| Tgene | RAD51 | GO:0071479 | cellular response to ionizing radiation | 23509288|23754376 |
| Tgene | RAD51 | GO:0072757 | cellular response to camptothecin | 23509288 |
 Fusion gene breakpoints across CHP1 (5'-gene) Fusion gene breakpoints across CHP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across RAD51 (3'-gene) Fusion gene breakpoints across RAD51 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
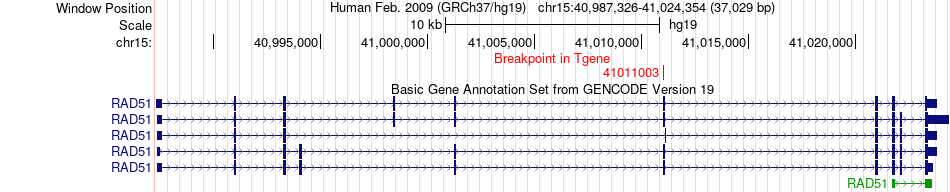 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-8085-01A | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
Top |
Fusion Gene ORF analysis for CHP1-RAD51 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000558351 | ENST00000267868 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 3UTR-3CDS | ENST00000558351 | ENST00000423169 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 3UTR-intron | ENST00000558351 | ENST00000382643 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 3UTR-intron | ENST00000558351 | ENST00000530766 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 3UTR-intron | ENST00000558351 | ENST00000532743 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 3UTR-intron | ENST00000558351 | ENST00000557850 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000334660 | ENST00000382643 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000334660 | ENST00000530766 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000334660 | ENST00000532743 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000334660 | ENST00000557850 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000560397 | ENST00000382643 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000560397 | ENST00000530766 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000560397 | ENST00000532743 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| 5CDS-intron | ENST00000560397 | ENST00000557850 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| In-frame | ENST00000334660 | ENST00000267868 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| In-frame | ENST00000334660 | ENST00000423169 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| In-frame | ENST00000560397 | ENST00000267868 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
| In-frame | ENST00000560397 | ENST00000423169 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000334660 | CHP1 | chr15 | 41562816 | + | ENST00000423169 | RAD51 | chr15 | 41011003 | + | 1528 | 651 | 63 | 1058 | 331 |
| ENST00000334660 | CHP1 | chr15 | 41562816 | + | ENST00000267868 | RAD51 | chr15 | 41011003 | + | 2214 | 651 | 63 | 1235 | 390 |
| ENST00000560397 | CHP1 | chr15 | 41562816 | + | ENST00000423169 | RAD51 | chr15 | 41011003 | + | 1378 | 501 | 0 | 908 | 302 |
| ENST00000560397 | CHP1 | chr15 | 41562816 | + | ENST00000267868 | RAD51 | chr15 | 41011003 | + | 2064 | 501 | 0 | 1085 | 361 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000334660 | ENST00000423169 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + | 0.00143779 | 0.9985623 |
| ENST00000334660 | ENST00000267868 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + | 0.000817646 | 0.9991823 |
| ENST00000560397 | ENST00000423169 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + | 0.001148936 | 0.99885106 |
| ENST00000560397 | ENST00000267868 | CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011003 | + | 0.00069662 | 0.99930334 |
Top |
Fusion Genomic Features for CHP1-RAD51 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011002 | + | 5.77E-10 | 1 |
| CHP1 | chr15 | 41562816 | + | RAD51 | chr15 | 41011002 | + | 5.77E-10 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
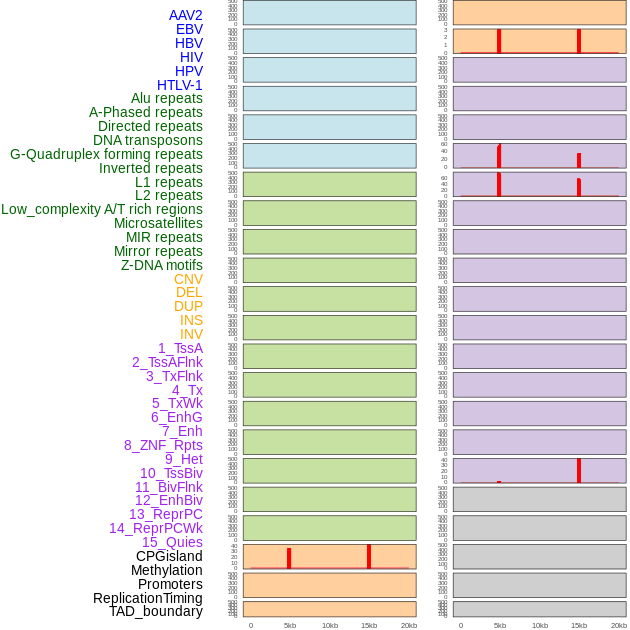 |
Top |
Fusion Protein Features for CHP1-RAD51 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:41562816/chr15:41011003) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHP1 | RAD51 |
| FUNCTION: Calcium-binding protein involved in different processes such as regulation of vesicular trafficking, plasma membrane Na(+)/H(+) exchanger and gene transcription. Involved in the constitutive exocytic membrane traffic. Mediates the association between microtubules and membrane-bound organelles of the endoplasmic reticulum and Golgi apparatus and is also required for the targeting and fusion of transcytotic vesicles (TCV) with the plasma membrane. Functions as an integral cofactor in cell pH regulation by controlling plasma membrane-type Na(+)/H(+) exchange activity. Affects the pH sensitivity of SLC9A1/NHE1 by increasing its sensitivity at acidic pH. Required for the stabilization and localization of SLC9A1/NHE1 at the plasma membrane. Inhibits serum- and GTPase-stimulated Na(+)/H(+) exchange. Plays a role as an inhibitor of ribosomal RNA transcription by repressing the nucleolar UBF1 transcriptional activity. May sequester UBF1 in the nucleoplasm and limit its translocation to the nucleolus. Associates to the ribosomal gene promoter. Acts as a negative regulator of the calcineurin/NFAT signaling pathway. Inhibits NFAT nuclear translocation and transcriptional activity by suppressing the calcium-dependent calcineurin phosphatase activity. Also negatively regulates the kinase activity of the apoptosis-induced kinase STK17B. Inhibits both STK17B auto- and substrate-phosphorylations in a calcium-dependent manner. {ECO:0000269|PubMed:10593895, ECO:0000269|PubMed:11350981, ECO:0000269|PubMed:15035633, ECO:0000269|PubMed:8901634}. | FUNCTION: Involved in the homologous recombination repair (HRR) pathway of double-stranded DNA breaks arising during DNA replication or induced by DNA-damaging agents. May promote the assembly of presynaptic RAD51 nucleoprotein filaments. Binds single-stranded DNA and double-stranded DNA and has DNA-dependent ATPase activity. Part of the RAD21 paralog protein complex BCDX2 which acts in the BRCA1-BRCA2-dependent HR pathway. Upon DNA damage, BCDX2 acts downstream of BRCA2 recruitment and upstream of RAD51 recruitment. BCDX2 binds predominantly to the intersection of the four duplex arms of the Holliday junction and to junction of replication forks. The BCDX2 complex was originally reported to bind single-stranded DNA, single-stranded gaps in duplex DNA and specifically to nicks in duplex DNA. The BCDX2 subcomplex RAD51B:RAD51C exhibits single-stranded DNA-dependent ATPase activity suggesting an involvement in early stages of the HR pathway. {ECO:0000269|PubMed:11751635, ECO:0000269|PubMed:11751636, ECO:0000269|PubMed:11842113, ECO:0000269|PubMed:12441335, ECO:0000269|PubMed:23108668, ECO:0000269|PubMed:23149936}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 123_134 | 137 | 196.0 | Calcium binding | Note=1 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 26_61 | 137 | 196.0 | Domain | EF-hand 1 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 66_101 | 137 | 196.0 | Domain | EF-hand 2 |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000557850 | 0 | 8 | 48_77 | 0 | 243.0 | Domain | Note=HhH | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000557850 | 0 | 8 | 127_134 | 0 | 243.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 164_175 | 137 | 196.0 | Calcium binding | Note=2 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 110_145 | 137 | 196.0 | Domain | EF-hand 3 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 151_186 | 137 | 196.0 | Domain | EF-hand 4 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 138_147 | 137 | 196.0 | Motif | Nuclear export signal 1 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 176_185 | 137 | 196.0 | Motif | Nuclear export signal 2 |
| Hgene | CHP1 | chr15:41562816 | chr15:41011003 | ENST00000334660 | + | 5 | 7 | 143_185 | 137 | 196.0 | Region | Note=Necessary for nuclear export signal |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000267868 | 4 | 10 | 48_77 | 145 | 340.0 | Domain | Note=HhH | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000382643 | 4 | 10 | 48_77 | 146 | 341.0 | Domain | Note=HhH | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000423169 | 4 | 9 | 48_77 | 145 | 281.0 | Domain | Note=HhH | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000532743 | 4 | 10 | 48_77 | 146 | 341.0 | Domain | Note=HhH | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000267868 | 4 | 10 | 127_134 | 145 | 340.0 | Nucleotide binding | ATP | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000382643 | 4 | 10 | 127_134 | 146 | 341.0 | Nucleotide binding | ATP | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000423169 | 4 | 9 | 127_134 | 145 | 281.0 | Nucleotide binding | ATP | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000532743 | 4 | 10 | 127_134 | 146 | 341.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for CHP1-RAD51 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16610_16610_1_CHP1-RAD51_CHP1_chr15_41562816_ENST00000334660_RAD51_chr15_41011003_ENST00000267868_length(transcript)=2214nt_BP=651nt CATTCGGGGCTGGAACTCGAACGCTGACGTTGGCTCCTGAGCCGGGCGCTGCCTCGCGCCTGATTGGCGGGTCCTCGGGGCCGCCCTCCC CAGCGCACCACCCCTGGGTTCCCTCCCGGGTCCGCAGTGGAAACACTGCCCTCTCCCTTCTTGACCCCTAGCCCTTCCTTCCCTCCCTCC TTCCCTCCTGTCGCCGTCTCTTCTGGCGCCGCTGCTCCCGGAGGAGCTCCCGGCACGGCGATGGGTTCTCGGGCCTCCACGTTACTGCGG GACGAAGAGCTCGAGGAGATCAAGAAGGAGACCGGCTTTTCCCACAGTCAAATCACTCGCCTCTACAGCCGGTTCACCAGCCTGGACAAA GGAGAGAATGGGACTCTCAGCCGGGAAGATTTCCAGAGGATTCCAGAACTTGCCATCAACCCACTGGGGGACCGGATCATCAATGCCTTC TTTCCAGAGGGAGAGGACCAGGTAAACTTCCGTGGATTCATGCGAACTTTGGCTCATTTCCGCCCCATTGAGGATAATGAAAAGAGCAAA GATGTGAATGGACCCGAACCACTCAACAGCCGAAGCAACAAACTGCACTTTGCTTTTCGACTATATGATTTGGATAAAGATGAAAAGATC TCCCGTGATGAGCTGTTACAGCTTCCCATTGACCGGGGTGGAGGTGAAGGAAAGGCCATGTACATTGACACTGAGGGTACCTTTAGGCCA GAACGGCTGCTGGCAGTGGCTGAGAGGTATGGTCTCTCTGGCAGTGATGTCCTGGATAATGTAGCATATGCTCGAGCGTTCAACACAGAC CACCAGACCCAGCTCCTTTATCAAGCATCAGCCATGATGGTAGAATCTAGGTATGCACTGCTTATTGTAGACAGTGCCACCGCCCTTTAC AGAACAGACTACTCGGGTCGAGGTGAGCTTTCAGCCAGGCAGATGCACTTGGCCAGGTTTCTGCGGATGCTTCTGCGACTCGCTGATGAG TTTGGTGTAGCAGTGGTAATCACTAATCAGGTGGTAGCTCAAGTGGATGGAGCAGCGATGTTTGCTGCTGATCCCAAAAAACCTATTGGA GGAAATATCATCGCCCATGCATCAACAACCAGATTGTATCTGAGGAAAGGAAGAGGGGAAACCAGAATCTGCAAAATCTACGACTCTCCC TGTCTTCCTGAAGCTGAAGCTATGTTCGCCATTAATGCAGATGGAGTGGGAGATGCCAAAGACTGAATCATTGGGTTTTTCCTCTGTTAA AAACCTTAAGTGCTGCAGCCTAATGAGAGTGCACTGCTCCCTGGGGTTCTCTACAGGCCTCTTCCTGTTGTGACTGCCAGGATAAAGCTT CCGGGAAAACAGCTATTATATCAGCTTTTCTGATGGTATAAACAGGAGACAGGTCAGTAGTCACAAACTGATCTAAAATGTTTATTCCTT CTGTAGTGTATTAATCTCTGTGTGTTTTCTTTGGTTTTGGAGGAGGGGTATGAAGTATCTTTGACATGGTGCCTTAGGAATGACTTGGGT TTAACAAGCTGTCTACTGGACAATCTTATGTTTCCAAGAGAACTAAAGCTGGAGAGACCTGACCCTTCTCTCACTTCTAAATTAATGGTA AAATAAAATGCCTCAGCTATGTAGCAAAGGGAATGGGTCTGCACAGATTCTTTTTTTCTGTCAGTAAAACTCTCAAGCAGGTTTTTAAGT TGTCTGTCTGAATGATCTTGTGTAAGGTTTTGGTTATGGAGTCTTGTGCCAAACCTACTAGGCCATTAGCCCTTCACCATCTACCTGCTT GGTCTTTCATTGCTAAGACTAACTCAAGATAATCCTAGAGTCTTAAAGCATTTCAGGCCAGTGTGGTGTCTTGCGCCTGTACTCCCAGCA CTTTGGGAGGCCGAGGCAGGTGGATCGCTTGAGCCCAGGAGTTTTAAGTCCAGCTTGGCCAAGGTGGTGAAATCCCATCTCTACAAAAAA TGCAGAACTTAATCTGGACACACTGTTACACGTGCCTGTAGTCCCAGCTACTCGATAGCCTGAGGTGGGAGAATCACTTAAGCCTGGAAG GTGGAAGTTGCAGTGAGTCGAGATTGCACTGCTGCATTCCAGCCAGGGTGACAGAGTGAGACCATGTTTCAAACAAGAAACATTTCAGAG >16610_16610_1_CHP1-RAD51_CHP1_chr15_41562816_ENST00000334660_RAD51_chr15_41011003_ENST00000267868_length(amino acids)=390AA_BP=26 MAGPRGRPPQRTTPGFPPGSAVETLPSPFLTPSPSFPPSFPPVAVSSGAAAPGGAPGTAMGSRASTLLRDEELEEIKKETGFSHSQITRL YSRFTSLDKGENGTLSREDFQRIPELAINPLGDRIINAFFPEGEDQVNFRGFMRTLAHFRPIEDNEKSKDVNGPEPLNSRSNKLHFAFRL YDLDKDEKISRDELLQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTDHQTQLLYQASAMMVESRYALL IVDSATALYRTDYSGRGELSARQMHLARFLRMLLRLADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGET -------------------------------------------------------------- >16610_16610_2_CHP1-RAD51_CHP1_chr15_41562816_ENST00000334660_RAD51_chr15_41011003_ENST00000423169_length(transcript)=1528nt_BP=651nt CATTCGGGGCTGGAACTCGAACGCTGACGTTGGCTCCTGAGCCGGGCGCTGCCTCGCGCCTGATTGGCGGGTCCTCGGGGCCGCCCTCCC CAGCGCACCACCCCTGGGTTCCCTCCCGGGTCCGCAGTGGAAACACTGCCCTCTCCCTTCTTGACCCCTAGCCCTTCCTTCCCTCCCTCC TTCCCTCCTGTCGCCGTCTCTTCTGGCGCCGCTGCTCCCGGAGGAGCTCCCGGCACGGCGATGGGTTCTCGGGCCTCCACGTTACTGCGG GACGAAGAGCTCGAGGAGATCAAGAAGGAGACCGGCTTTTCCCACAGTCAAATCACTCGCCTCTACAGCCGGTTCACCAGCCTGGACAAA GGAGAGAATGGGACTCTCAGCCGGGAAGATTTCCAGAGGATTCCAGAACTTGCCATCAACCCACTGGGGGACCGGATCATCAATGCCTTC TTTCCAGAGGGAGAGGACCAGGTAAACTTCCGTGGATTCATGCGAACTTTGGCTCATTTCCGCCCCATTGAGGATAATGAAAAGAGCAAA GATGTGAATGGACCCGAACCACTCAACAGCCGAAGCAACAAACTGCACTTTGCTTTTCGACTATATGATTTGGATAAAGATGAAAAGATC TCCCGTGATGAGCTGTTACAGCTTCCCATTGACCGGGGTGGAGGTGAAGGAAAGGCCATGTACATTGACACTGAGGGTACCTTTAGGCCA GAACGGCTGCTGGCAGTGGCTGAGAGGTATGGTCTCTCTGGCAGTGATGTCCTGGATAATGTAGCATATGCTCGAGCGTTCAACACAGAC CACCAGACCCAGCTCCTTTATCAAGCATCAGCCATGATGGTAGAATCTAGGTATGCACTGCTTATTGTAGACAGTGCCACCGCCCTTTAC AGAACAGACTACTCGGGTCGAGGTGAGCTTTCAGCCAGGCAGATGCACTTGGCCAGGTTTCTGCGGATGCTTCTGCGACTCGCTGATGAG ATTGTATCTGAGGAAAGGAAGAGGGGAAACCAGAATCTGCAAAATCTACGACTCTCCCTGTCTTCCTGAAGCTGAAGCTATGTTCGCCAT TAATGCAGATGGAGTGGGAGATGCCAAAGACTGAATCATTGGGTTTTTCCTCTGTTAAAAACCTTAAGTGCTGCAGCCTAATGAGAGTGC ACTGCTCCCTGGGGTTCTCTACAGGCCTCTTCCTGTTGTGACTGCCAGGATAAAGCTTCCGGGAAAACAGCTATTATATCAGCTTTTCTG ATGGTATAAACAGGAGACAGGTCAGTAGTCACAAACTGATCTAAAATGTTTATTCCTTCTGTAGTGTATTAATCTCTGTGTGTTTTCTTT GGTTTTGGAGGAGGGGTATGAAGTATCTTTGACATGGTGCCTTAGGAATGACTTGGGTTTAACAAGCTGTCTACTGGACAATCTTATGTT >16610_16610_2_CHP1-RAD51_CHP1_chr15_41562816_ENST00000334660_RAD51_chr15_41011003_ENST00000423169_length(amino acids)=331AA_BP=26 MAGPRGRPPQRTTPGFPPGSAVETLPSPFLTPSPSFPPSFPPVAVSSGAAAPGGAPGTAMGSRASTLLRDEELEEIKKETGFSHSQITRL YSRFTSLDKGENGTLSREDFQRIPELAINPLGDRIINAFFPEGEDQVNFRGFMRTLAHFRPIEDNEKSKDVNGPEPLNSRSNKLHFAFRL YDLDKDEKISRDELLQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTDHQTQLLYQASAMMVESRYALL -------------------------------------------------------------- >16610_16610_3_CHP1-RAD51_CHP1_chr15_41562816_ENST00000560397_RAD51_chr15_41011003_ENST00000267868_length(transcript)=2064nt_BP=501nt TTGACCCCTAGCCCTTCCTTCCCTCCCTCCTTCCCTCCTGTCGCCGTCTCTTCTGGCGCCGCTGCTCCCGGAGGAGCTCCCGGCACGGCG ATGGGTTCTCGGGCCTCCACGTTACTGCGGGACGAAGAGCTCGAGGAGATCAAGAAGGAGACCGGCTTTTCCCACAGTCAAATCACTCGC CTCTACAGCCGGTTCACCAGCCTGGACAAAGGAGAGAATGGGACTCTCAGCCGGGAAGATTTCCAGAGGATTCCAGAACTTGCCATCAAC CCACTGGGGGACCGGATCATCAATGCCTTCTTTCCAGAGGGAGAGGACCAGGTAAACTTCCGTGGATTCATGCGAACTTTGGCTCATTTC CGCCCCATTGAGGATAATGAAAAGAGCAAAGATGTGAATGGACCCGAACCACTCAACAGCCGAAGCAACAAACTGCACTTTGCTTTTCGA CTATATGATTTGGATAAAGATGAAAAGATCTCCCGTGATGAGCTGTTACAGCTTCCCATTGACCGGGGTGGAGGTGAAGGAAAGGCCATG TACATTGACACTGAGGGTACCTTTAGGCCAGAACGGCTGCTGGCAGTGGCTGAGAGGTATGGTCTCTCTGGCAGTGATGTCCTGGATAAT GTAGCATATGCTCGAGCGTTCAACACAGACCACCAGACCCAGCTCCTTTATCAAGCATCAGCCATGATGGTAGAATCTAGGTATGCACTG CTTATTGTAGACAGTGCCACCGCCCTTTACAGAACAGACTACTCGGGTCGAGGTGAGCTTTCAGCCAGGCAGATGCACTTGGCCAGGTTT CTGCGGATGCTTCTGCGACTCGCTGATGAGTTTGGTGTAGCAGTGGTAATCACTAATCAGGTGGTAGCTCAAGTGGATGGAGCAGCGATG TTTGCTGCTGATCCCAAAAAACCTATTGGAGGAAATATCATCGCCCATGCATCAACAACCAGATTGTATCTGAGGAAAGGAAGAGGGGAA ACCAGAATCTGCAAAATCTACGACTCTCCCTGTCTTCCTGAAGCTGAAGCTATGTTCGCCATTAATGCAGATGGAGTGGGAGATGCCAAA GACTGAATCATTGGGTTTTTCCTCTGTTAAAAACCTTAAGTGCTGCAGCCTAATGAGAGTGCACTGCTCCCTGGGGTTCTCTACAGGCCT CTTCCTGTTGTGACTGCCAGGATAAAGCTTCCGGGAAAACAGCTATTATATCAGCTTTTCTGATGGTATAAACAGGAGACAGGTCAGTAG TCACAAACTGATCTAAAATGTTTATTCCTTCTGTAGTGTATTAATCTCTGTGTGTTTTCTTTGGTTTTGGAGGAGGGGTATGAAGTATCT TTGACATGGTGCCTTAGGAATGACTTGGGTTTAACAAGCTGTCTACTGGACAATCTTATGTTTCCAAGAGAACTAAAGCTGGAGAGACCT GACCCTTCTCTCACTTCTAAATTAATGGTAAAATAAAATGCCTCAGCTATGTAGCAAAGGGAATGGGTCTGCACAGATTCTTTTTTTCTG TCAGTAAAACTCTCAAGCAGGTTTTTAAGTTGTCTGTCTGAATGATCTTGTGTAAGGTTTTGGTTATGGAGTCTTGTGCCAAACCTACTA GGCCATTAGCCCTTCACCATCTACCTGCTTGGTCTTTCATTGCTAAGACTAACTCAAGATAATCCTAGAGTCTTAAAGCATTTCAGGCCA GTGTGGTGTCTTGCGCCTGTACTCCCAGCACTTTGGGAGGCCGAGGCAGGTGGATCGCTTGAGCCCAGGAGTTTTAAGTCCAGCTTGGCC AAGGTGGTGAAATCCCATCTCTACAAAAAATGCAGAACTTAATCTGGACACACTGTTACACGTGCCTGTAGTCCCAGCTACTCGATAGCC TGAGGTGGGAGAATCACTTAAGCCTGGAAGGTGGAAGTTGCAGTGAGTCGAGATTGCACTGCTGCATTCCAGCCAGGGTGACAGAGTGAG >16610_16610_3_CHP1-RAD51_CHP1_chr15_41562816_ENST00000560397_RAD51_chr15_41011003_ENST00000267868_length(amino acids)=361AA_BP=165 LTPSPSFPPSFPPVAVSSGAAAPGGAPGTAMGSRASTLLRDEELEEIKKETGFSHSQITRLYSRFTSLDKGENGTLSREDFQRIPELAIN PLGDRIINAFFPEGEDQVNFRGFMRTLAHFRPIEDNEKSKDVNGPEPLNSRSNKLHFAFRLYDLDKDEKISRDELLQLPIDRGGGEGKAM YIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARF LRMLLRLADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRICKIYDSPCLPEAEAMFAINADGVGDAK -------------------------------------------------------------- >16610_16610_4_CHP1-RAD51_CHP1_chr15_41562816_ENST00000560397_RAD51_chr15_41011003_ENST00000423169_length(transcript)=1378nt_BP=501nt TTGACCCCTAGCCCTTCCTTCCCTCCCTCCTTCCCTCCTGTCGCCGTCTCTTCTGGCGCCGCTGCTCCCGGAGGAGCTCCCGGCACGGCG ATGGGTTCTCGGGCCTCCACGTTACTGCGGGACGAAGAGCTCGAGGAGATCAAGAAGGAGACCGGCTTTTCCCACAGTCAAATCACTCGC CTCTACAGCCGGTTCACCAGCCTGGACAAAGGAGAGAATGGGACTCTCAGCCGGGAAGATTTCCAGAGGATTCCAGAACTTGCCATCAAC CCACTGGGGGACCGGATCATCAATGCCTTCTTTCCAGAGGGAGAGGACCAGGTAAACTTCCGTGGATTCATGCGAACTTTGGCTCATTTC CGCCCCATTGAGGATAATGAAAAGAGCAAAGATGTGAATGGACCCGAACCACTCAACAGCCGAAGCAACAAACTGCACTTTGCTTTTCGA CTATATGATTTGGATAAAGATGAAAAGATCTCCCGTGATGAGCTGTTACAGCTTCCCATTGACCGGGGTGGAGGTGAAGGAAAGGCCATG TACATTGACACTGAGGGTACCTTTAGGCCAGAACGGCTGCTGGCAGTGGCTGAGAGGTATGGTCTCTCTGGCAGTGATGTCCTGGATAAT GTAGCATATGCTCGAGCGTTCAACACAGACCACCAGACCCAGCTCCTTTATCAAGCATCAGCCATGATGGTAGAATCTAGGTATGCACTG CTTATTGTAGACAGTGCCACCGCCCTTTACAGAACAGACTACTCGGGTCGAGGTGAGCTTTCAGCCAGGCAGATGCACTTGGCCAGGTTT CTGCGGATGCTTCTGCGACTCGCTGATGAGATTGTATCTGAGGAAAGGAAGAGGGGAAACCAGAATCTGCAAAATCTACGACTCTCCCTG TCTTCCTGAAGCTGAAGCTATGTTCGCCATTAATGCAGATGGAGTGGGAGATGCCAAAGACTGAATCATTGGGTTTTTCCTCTGTTAAAA ACCTTAAGTGCTGCAGCCTAATGAGAGTGCACTGCTCCCTGGGGTTCTCTACAGGCCTCTTCCTGTTGTGACTGCCAGGATAAAGCTTCC GGGAAAACAGCTATTATATCAGCTTTTCTGATGGTATAAACAGGAGACAGGTCAGTAGTCACAAACTGATCTAAAATGTTTATTCCTTCT GTAGTGTATTAATCTCTGTGTGTTTTCTTTGGTTTTGGAGGAGGGGTATGAAGTATCTTTGACATGGTGCCTTAGGAATGACTTGGGTTT AACAAGCTGTCTACTGGACAATCTTATGTTTCCAAGAGAACTAAAGCTGGAGAGACCTGACCCTTCTCTCACTTCTAAATTAATGGTAAA >16610_16610_4_CHP1-RAD51_CHP1_chr15_41562816_ENST00000560397_RAD51_chr15_41011003_ENST00000423169_length(amino acids)=302AA_BP=165 LTPSPSFPPSFPPVAVSSGAAAPGGAPGTAMGSRASTLLRDEELEEIKKETGFSHSQITRLYSRFTSLDKGENGTLSREDFQRIPELAIN PLGDRIINAFFPEGEDQVNFRGFMRTLAHFRPIEDNEKSKDVNGPEPLNSRSNKLHFAFRLYDLDKDEKISRDELLQLPIDRGGGEGKAM YIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHP1-RAD51 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000267868 | 4 | 10 | 184_257 | 145.0 | 340.0 | PALB2 | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000382643 | 4 | 10 | 184_257 | 146.0 | 341.0 | PALB2 | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000423169 | 4 | 9 | 184_257 | 145.0 | 281.0 | PALB2 | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000532743 | 4 | 10 | 184_257 | 146.0 | 341.0 | PALB2 | |
| Tgene | RAD51 | chr15:41562816 | chr15:41011003 | ENST00000557850 | 0 | 8 | 184_257 | 0 | 243.0 | PALB2 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHP1-RAD51 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHP1-RAD51 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
