
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHST9-BBS9 (FusionGDB2 ID:16716) |
Fusion Gene Summary for CHST9-BBS9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHST9-BBS9 | Fusion gene ID: 16716 | Hgene | Tgene | Gene symbol | CHST9 | BBS9 | Gene ID | 83539 | 27241 |
| Gene name | carbohydrate sulfotransferase 9 | Bardet-Biedl syndrome 9 | |
| Synonyms | GALNAC4ST-2|GalNAc4ST2 | B1|C18|D1|PTHB1 | |
| Cytomap | 18q11.2 | 7p14.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | carbohydrate sulfotransferase 9GalNAc-4-sulfotransferase 2N-acetylgalactosamine 4-O-sulfotransferase 2carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9galNAc-4-O-sulfotransferase 2 | protein PTHB1PTH-responsive osteosarcoma B1 proteinbardet-Biedl syndrome 9 proteinparathyroid hormone-responsive B1 gene protein | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q7L1S5 | Q3SYG4 | |
| Ensembl transtripts involved in fusion gene | ENST00000284224, ENST00000580774, ENST00000581714, | ENST00000425508, ENST00000482941, ENST00000242067, ENST00000350941, ENST00000354265, ENST00000355070, ENST00000396127, | |
| Fusion gene scores | * DoF score | 6 X 5 X 4=120 | 14 X 17 X 8=1904 |
| # samples | 7 | 19 | |
| ** MAII score | log2(7/120*10)=-0.777607578663552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/1904*10)=-3.324962154977 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CHST9 [Title/Abstract] AND BBS9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CHST9(24722653)-BBS9(33388680), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CHST9-BBS9 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CHST9 | GO:0006790 | sulfur compound metabolic process | 11445554 |
 Fusion gene breakpoints across CHST9 (5'-gene) Fusion gene breakpoints across CHST9 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
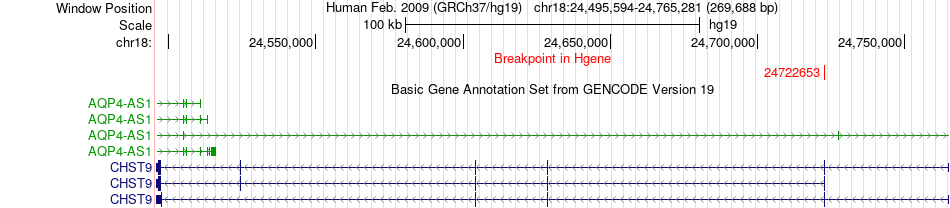 |
 Fusion gene breakpoints across BBS9 (3'-gene) Fusion gene breakpoints across BBS9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
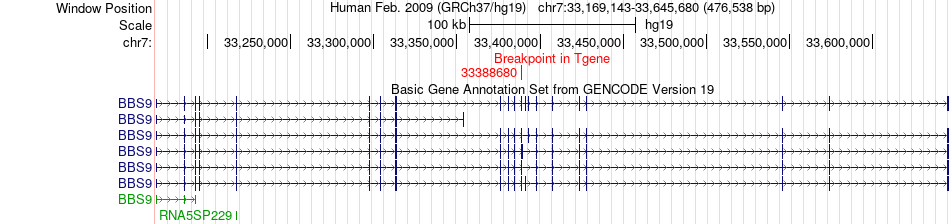 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-91-8499-01A | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
Top |
Fusion Gene ORF analysis for CHST9-BBS9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000284224 | ENST00000425508 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| 5CDS-intron | ENST00000284224 | ENST00000482941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| 5CDS-intron | ENST00000580774 | ENST00000425508 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| 5CDS-intron | ENST00000580774 | ENST00000482941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| 5CDS-intron | ENST00000581714 | ENST00000425508 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| 5CDS-intron | ENST00000581714 | ENST00000482941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000284224 | ENST00000242067 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000284224 | ENST00000350941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000284224 | ENST00000354265 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000284224 | ENST00000355070 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000284224 | ENST00000396127 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000580774 | ENST00000242067 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000580774 | ENST00000350941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000580774 | ENST00000354265 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000580774 | ENST00000355070 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| Frame-shift | ENST00000580774 | ENST00000396127 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| In-frame | ENST00000581714 | ENST00000242067 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| In-frame | ENST00000581714 | ENST00000350941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| In-frame | ENST00000581714 | ENST00000354265 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| In-frame | ENST00000581714 | ENST00000355070 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
| In-frame | ENST00000581714 | ENST00000396127 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000581714 | CHST9 | chr18 | 24722653 | - | ENST00000242067 | BBS9 | chr7 | 33388680 | + | 2527 | 350 | 296 | 1684 | 462 |
| ENST00000581714 | CHST9 | chr18 | 24722653 | - | ENST00000355070 | BBS9 | chr7 | 33388680 | + | 2512 | 350 | 296 | 1669 | 457 |
| ENST00000581714 | CHST9 | chr18 | 24722653 | - | ENST00000350941 | BBS9 | chr7 | 33388680 | + | 2407 | 350 | 296 | 1564 | 422 |
| ENST00000581714 | CHST9 | chr18 | 24722653 | - | ENST00000354265 | BBS9 | chr7 | 33388680 | + | 2422 | 350 | 296 | 1579 | 427 |
| ENST00000581714 | CHST9 | chr18 | 24722653 | - | ENST00000396127 | BBS9 | chr7 | 33388680 | + | 2422 | 350 | 296 | 1579 | 427 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000581714 | ENST00000242067 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + | 0.000691809 | 0.9993082 |
| ENST00000581714 | ENST00000355070 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + | 0.002344654 | 0.9976553 |
| ENST00000581714 | ENST00000350941 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + | 0.000642347 | 0.99935764 |
| ENST00000581714 | ENST00000354265 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + | 0.000623147 | 0.99937683 |
| ENST00000581714 | ENST00000396127 | CHST9 | chr18 | 24722653 | - | BBS9 | chr7 | 33388680 | + | 0.000666941 | 0.9993331 |
Top |
Fusion Genomic Features for CHST9-BBS9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CHST9 | chr18 | 24722652 | - | BBS9 | chr7 | 33388679 | + | 1.81E-05 | 0.999982 |
| CHST9 | chr18 | 24722652 | - | BBS9 | chr7 | 33388679 | + | 1.81E-05 | 0.999982 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
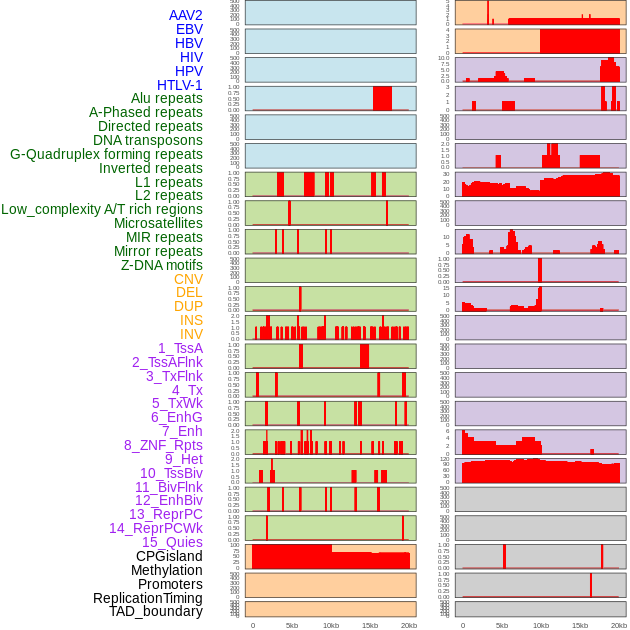 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
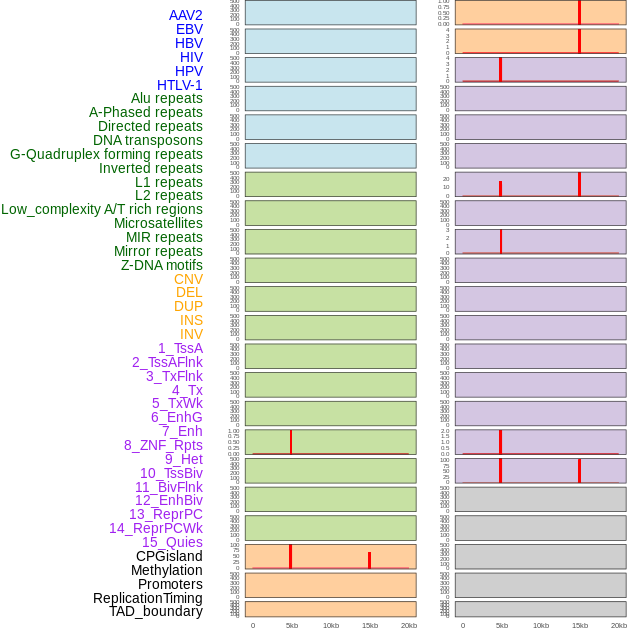 |
Top |
Fusion Protein Features for CHST9-BBS9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:24722653/chr7:33388680) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHST9 | BBS9 |
| FUNCTION: Catalyzes the transfer of sulfate to position 4 of non-reducing N-acetylgalactosamine (GalNAc) residues in both N-glycans and O-glycans. Participates in biosynthesis of glycoprotein hormones lutropin and thyrotropin, by mediating sulfation of their carbohydrate structures. Has a higher activity toward carbonic anhydrase VI than toward lutropin. Only active against terminal GalNAcbeta1,GalNAcbeta. Isoform 2, but not isoform 1, is active toward chondroitin. | FUNCTION: The BBSome complex is thought to function as a coat complex required for sorting of specific membrane proteins to the primary cilia. The BBSome complex is required for ciliogenesis but is dispensable for centriolar satellite function. This ciliogenic function is mediated in part by the Rab8 GDP/GTP exchange factor, which localizes to the basal body and contacts the BBSome. Rab8(GTP) enters the primary cilium and promotes extension of the ciliary membrane. Firstly the BBSome associates with the ciliary membrane and binds to RAB3IP/Rabin8, the guanosyl exchange factor (GEF) for Rab8 and then the Rab8-GTP localizes to the cilium and promotes docking and fusion of carrier vesicles to the base of the ciliary membrane. Required for proper BBSome complex assembly and its ciliary localization. {ECO:0000269|PubMed:17574030, ECO:0000269|PubMed:22072986}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000284224 | - | 2 | 6 | 1_12 | 40 | 444.0 | Topological domain | Cytoplasmic |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000580774 | - | 2 | 5 | 1_12 | 40 | 75.0 | Topological domain | Cytoplasmic |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000581714 | - | 1 | 5 | 1_12 | 40 | 444.0 | Topological domain | Cytoplasmic |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000284224 | - | 2 | 6 | 13_33 | 40 | 444.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000580774 | - | 2 | 5 | 13_33 | 40 | 75.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000581714 | - | 1 | 5 | 13_33 | 40 | 444.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000425508 | 0 | 10 | 1_407 | 0 | 311.0 | Region | Seven-bladed beta-propeller |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000284224 | - | 2 | 6 | 220_226 | 40 | 444.0 | Nucleotide binding | PAPS |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000284224 | - | 2 | 6 | 280_288 | 40 | 444.0 | Nucleotide binding | PAPS |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000580774 | - | 2 | 5 | 220_226 | 40 | 75.0 | Nucleotide binding | PAPS |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000580774 | - | 2 | 5 | 280_288 | 40 | 75.0 | Nucleotide binding | PAPS |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000581714 | - | 1 | 5 | 220_226 | 40 | 444.0 | Nucleotide binding | PAPS |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000581714 | - | 1 | 5 | 280_288 | 40 | 444.0 | Nucleotide binding | PAPS |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000284224 | - | 2 | 6 | 34_443 | 40 | 444.0 | Topological domain | Lumenal |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000580774 | - | 2 | 5 | 34_443 | 40 | 75.0 | Topological domain | Lumenal |
| Hgene | CHST9 | chr18:24722653 | chr7:33388680 | ENST00000581714 | - | 1 | 5 | 34_443 | 40 | 444.0 | Topological domain | Lumenal |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000242067 | 11 | 23 | 1_407 | 443 | 888.0 | Region | Seven-bladed beta-propeller | |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000350941 | 11 | 21 | 1_407 | 443 | 848.0 | Region | Seven-bladed beta-propeller | |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000396127 | 11 | 22 | 1_407 | 443 | 853.0 | Region | Seven-bladed beta-propeller |
Top |
Fusion Gene Sequence for CHST9-BBS9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16716_16716_1_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000242067_length(transcript)=2527nt_BP=350nt AGTCTTTTCTGTCTTCAGAGCATTAGTGTGCACATTAAGTCTTCAGCCTTGTTGAGATAGTGAATATTTATGCCGTATTTGAGAATTAAT ATTAGCAGAAATGTACAATGTAGATTTCTTTTTAAAATTCTAGGTTACTTTATGGAATTGGGCTCTTAGAGAACAAGAAAAGACTGAAGT TTTACGGGAAAACAAATCATGTGGTCTTCAGATTCTGAAATAAGGAGAAATGCAGCCATCTGAAATGGTCATGAACCCCAAACAAGTCTT CCTCTCTGTGCTGATATTTGGAGTAGCTGGGCTACTCCTCTTCATGTATTTGCAAGTCTGGATTGAAGAACAACATACAGGTCACACTGC AGAACAGAGTGATATTGCAAAAAGCCAAATTATCAGTCTACGTGCAACCACCATTAGAATTGACTTGTGATCAGTTCACCTTTGAATTTA TGACACCAGATTTGACTAGAACAGTAAGCTTTTCTGTTTATCTGAAAAGAAGTTATACACCATCAGAATTGGAAGGAAATGCTGTTGTTT CTTATTCCAGACCAACAGATCGAAATCCTGATGGCATTCCGCGAGTTATCCAATGTAAATTTAGACTTCCCCTAAAGTTAATTTGCCTAC CAGGTCAGCCTTCAAAAACTGCAAGCCACAAAATTACTATTGATACCAACAAATCTCCAGTCAGTCTTCTTAGTCTCTTCCCAGGTTTTG CCAGTCAGTCAGATGATGATCAGGTGAATGTAATGGGTTTTCACTTCTTAGGAGGTGCTCGAATTACTGTTCTTGCTTCCAAAACTTCTC AACGATATCGCATTCAGAGTGAACAATTTGAAGATCTTTGGCTCATAACCAATGAGCTTATTCTTCGCCTTCAAGAATATTTTGAAAAAC AGGGAGTCAAAGATTTTGCATGTTCTTTTTCGGGATCTATACCCCTTCAAGAATATTTTGAGTTGATTGATCATCATTTTGAGCTACGGA TAAATGGTGAAAAATTAGAAGAACTCTTATCTGAGAGAGCTGTACAATTTCGGGCCATTCAACGCCGGCTACTAGCAAGATTCAAAGATA AAACTCCTGCCCCTCTTCAACACCTGGACACCTTGTTAGATGGAACCTACAAGCAGGTAATTGCTCTAGCAGATGCAGTGGAGGAAAACC AAGGCAATCTGTTCCAGTCATTCACCAGGCTGAAGAGTGCCACCCATTTGGTGATTCTGCTGATCGCGCTGTGGCAGAAGCTTAGTGCTG ACCAGGTTGCTATTCTGGAAGCGGCATTTCTGCCGCTACAAGAAGACACTCAAGAATTGGGCTGGGAAGAAACGGTGGATGCCGCCATTT CCCACCTGTTGAAGACTTGCCTGTCGAAGAGTTCTAAGGAGCAGGCTTTGAACCTCAACAGCCAGCTGAACATACCCAAAGACACAAGCC AACTGAAGAAACATATCACCTTGCTCTGCGATAGATTATCCAAAGGTGGCCGTCTCTGCCTAAGTACCGATGCAGCAGCCCCACAGACCA TGGTCATGCCAGGTGGTTGTACTACAATCCCAGAGTCAGACCTAGAAGAAAGATCAGTAGAACAAGACTCTACAGAACTGTTTACCAACC ACAGACATCTCACTGCAGAGACACCCAGGCCTGAAGTTTCACCCCTCCAAGGAGTCTCGGAATAATTCAAGTAGAGTTGTTTGGTTGAGA GGAACATCCCCATCTCAAGGCCGAACCTGTGTGAACCTCATGCCAAGCACAGATATAGGGCTGGCGCAGGTGCTTCCTAAAGCTCACCTT CCTGGAGATGACATGCATAGAAAGAGGGGTTGGGACTTTTTACTTCACTAGGAGAACTTGTAACACCATGGGGAAGTCAGCTGAAACTTG TCTTGTTTTGCCAGGAAAGGAAGTAGTTGCCTTTGGTCATCCATCTGCTAATAGTCACAGAATACAGTGAAATGACATAGTTTTGGGTTA GATTTTATAATGCAAAGATTCAGATCCAAAATAATTTCATACCCCATTTTTTCACAGAATTCTTATATAGTAAATGTATCAAGTTTAATA AAGCATCTCATTGTCAAATAATATCTTGGATTTTATTTATAATTAGAGGGATTTATGAGTGATTGCTCTACATTATTTCTTCAAAGGAAA GGAAAGGAATTGAAGACTTTGCTACTCTCTGGTAAGACTTGAATGTGATTATTTTATAAATAAAAGAACCACTATGAAACTTTCATGTGA TTTGTTCTATTAAGCTAAACGTGGAAGAAGGGAAGTTTTCTAGTATTGTATGAAAATGTTTAAATATTTTCAAGGAAGCTAGTACATTAA CATCCCCTTTTCATTACCCCATTGGGTGGCACCTTTTAAAAATCCAGGTTATTAAAGGATTCACACTTTATTACAGTTAAAATTTAAGGT TTTTATGTGTAAGAAGTGAAAGAAACTTTGCTAATTTCAAGTTCGAATTACAGTTGTGAGATTGATTTATTTAAATAAAAATATGCCTTT >16716_16716_1_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000242067_length(amino acids)=462AA_BP=18 MGYSSSCICKSGLKNNIQVTLQNRVILQKAKLSVYVQPPLELTCDQFTFEFMTPDLTRTVSFSVYLKRSYTPSELEGNAVVSYSRPTDRN PDGIPRVIQCKFRLPLKLICLPGQPSKTASHKITIDTNKSPVSLLSLFPGFASQSDDDQVNVMGFHFLGGARITVLASKTSQRYRIQSEQ FEDLWLITNELILRLQEYFEKQGVKDFACSFSGSIPLQEYFELIDHHFELRINGEKLEELLSERAVQFRAIQRRLLARFKDKTPAPLQHL DTLLDGTYKQVIALADAVEENQGNLFQSFTRLKSATHLVILLIALWQKLSADQVAILEAAFLPLQEDTQELGWEETVDAAISHLLKTCLS KSSKEQALNLNSQLNIPKDTSQLKKHITLLCDRLSKGGRLCLSTDAAAPQTMVMPGGCTTIPESDLEERSVEQDSTELFTNHRHLTAETP -------------------------------------------------------------- >16716_16716_2_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000350941_length(transcript)=2407nt_BP=350nt AGTCTTTTCTGTCTTCAGAGCATTAGTGTGCACATTAAGTCTTCAGCCTTGTTGAGATAGTGAATATTTATGCCGTATTTGAGAATTAAT ATTAGCAGAAATGTACAATGTAGATTTCTTTTTAAAATTCTAGGTTACTTTATGGAATTGGGCTCTTAGAGAACAAGAAAAGACTGAAGT TTTACGGGAAAACAAATCATGTGGTCTTCAGATTCTGAAATAAGGAGAAATGCAGCCATCTGAAATGGTCATGAACCCCAAACAAGTCTT CCTCTCTGTGCTGATATTTGGAGTAGCTGGGCTACTCCTCTTCATGTATTTGCAAGTCTGGATTGAAGAACAACATACAGGTCACACTGC AGAACAGAGTGATATTGCAAAAAGCCAAATTATCAGTCTACGTGCAACCACCATTAGAATTGACTTGTGATCAGTTCACCTTTGAATTTA TGAGCATTCCGCGAGTTATCCAATGTAAATTTAGACTTCCCCTAAAGTTAATTTGCCTACCAGGTCAGCCTTCAAAAACTGCAAGCCACA AAATTACTATTGATACCAACAAATCTCCAGTCAGTCTTCTTAGTCTCTTCCCAGGTTTTGCCAGTCAGTCAGATGATGATCAGGTGAATG TAATGGGTTTTCACTTCTTAGGAGGTGCTCGAATTACTGTTCTTGCTTCCAAAACTTCTCAACGATATCGCATTCAGAGTGAACAATTTG AAGATCTTTGGCTCATAACCAATGAGCTTATTCTTCGCCTTCAAGAATATTTTGAAAAACAGGGAGTCAAAGATTTTGCATGTTCTTTTT CGGGATCTATACCCCTTCAAGAATATTTTGAGTTGATTGATCATCATTTTGAGCTACGGATAAATGGTGAAAAATTAGAAGAACTCTTAT CTGAGAGAGCTGTACAATTTCGGGCCATTCAACGCCGGCTACTAGCAAGATTCAAAGATAAAACTCCTGCCCCTCTTCAACACCTGGACA CCTTGTTAGATGGAACCTACAAGCAGGTAATTGCTCTAGCAGATGCAGTGGAGGAAAACCAAGGCAATCTGTTCCAGTCATTCACCAGGC TGAAGAGTGCCACCCATTTGGTGATTCTGCTGATCGCGCTGTGGCAGAAGCTTAGTGCTGACCAGGTTGCTATTCTGGAAGCGGCATTTC TGCCGCTACAAGAAGACACTCAAGAATTGGGCTGGGAAGAAACGGTGGATGCCGCCATTTCCCACCTGTTGAAGACTTGCCTGTCGAAGA GTTCTAAGGAGCAGGCTTTGAACCTCAACAGCCAGCTGAACATACCCAAAGACACAAGCCAACTGAAGAAACATATCACCTTGCTCTGCG ATAGATTATCCAAAGGTGGCCGTCTCTGCCTAAGTACCGATGCAGCAGCCCCACAGACCATGGTCATGCCAGGTGGTTGTACTACAATCC CAGAGTCAGACCTAGAAGAAAGATCAGTAGAACAAGACTCTACAGAACTGTTTACCAACCACAGACATCTCACTGCAGAGACACCCAGGC CTGAAGTTTCACCCCTCCAAGGAGTCTCGGAATAATTCAAGTAGAGTTGTTTGGTTGAGAGGAACATCCCCATCTCAAGGCCGAACCTGT GTGAACCTCATGCCAAGCACAGATATAGGGCTGGCGCAGGTGCTTCCTAAAGCTCACCTTCCTGGAGATGACATGCATAGAAAGAGGGGT TGGGACTTTTTACTTCACTAGGAGAACTTGTAACACCATGGGGAAGTCAGCTGAAACTTGTCTTGTTTTGCCAGGAAAGGAAGTAGTTGC CTTTGGTCATCCATCTGCTAATAGTCACAGAATACAGTGAAATGACATAGTTTTGGGTTAGATTTTATAATGCAAAGATTCAGATCCAAA ATAATTTCATACCCCATTTTTTCACAGAATTCTTATATAGTAAATGTATCAAGTTTAATAAAGCATCTCATTGTCAAATAATATCTTGGA TTTTATTTATAATTAGAGGGATTTATGAGTGATTGCTCTACATTATTTCTTCAAAGGAAAGGAAAGGAATTGAAGACTTTGCTACTCTCT GGTAAGACTTGAATGTGATTATTTTATAAATAAAAGAACCACTATGAAACTTTCATGTGATTTGTTCTATTAAGCTAAACGTGGAAGAAG GGAAGTTTTCTAGTATTGTATGAAAATGTTTAAATATTTTCAAGGAAGCTAGTACATTAACATCCCCTTTTCATTACCCCATTGGGTGGC ACCTTTTAAAAATCCAGGTTATTAAAGGATTCACACTTTATTACAGTTAAAATTTAAGGTTTTTATGTGTAAGAAGTGAAAGAAACTTTG >16716_16716_2_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000350941_length(amino acids)=422AA_BP=18 MGYSSSCICKSGLKNNIQVTLQNRVILQKAKLSVYVQPPLELTCDQFTFEFMSIPRVIQCKFRLPLKLICLPGQPSKTASHKITIDTNKS PVSLLSLFPGFASQSDDDQVNVMGFHFLGGARITVLASKTSQRYRIQSEQFEDLWLITNELILRLQEYFEKQGVKDFACSFSGSIPLQEY FELIDHHFELRINGEKLEELLSERAVQFRAIQRRLLARFKDKTPAPLQHLDTLLDGTYKQVIALADAVEENQGNLFQSFTRLKSATHLVI LLIALWQKLSADQVAILEAAFLPLQEDTQELGWEETVDAAISHLLKTCLSKSSKEQALNLNSQLNIPKDTSQLKKHITLLCDRLSKGGRL -------------------------------------------------------------- >16716_16716_3_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000354265_length(transcript)=2422nt_BP=350nt AGTCTTTTCTGTCTTCAGAGCATTAGTGTGCACATTAAGTCTTCAGCCTTGTTGAGATAGTGAATATTTATGCCGTATTTGAGAATTAAT ATTAGCAGAAATGTACAATGTAGATTTCTTTTTAAAATTCTAGGTTACTTTATGGAATTGGGCTCTTAGAGAACAAGAAAAGACTGAAGT TTTACGGGAAAACAAATCATGTGGTCTTCAGATTCTGAAATAAGGAGAAATGCAGCCATCTGAAATGGTCATGAACCCCAAACAAGTCTT CCTCTCTGTGCTGATATTTGGAGTAGCTGGGCTACTCCTCTTCATGTATTTGCAAGTCTGGATTGAAGAACAACATACAGGTCACACTGC AGAACAGAGTGATATTGCAAAAAGCCAAATTATCAGTCTACGTGCAACCACCATTAGAATTGACTTGTGATCAGTTCACCTTTGAATTTA TGAGTCTGAATGTTTTAGGCATTCCGCGAGTTATCCAATGTAAATTTAGACTTCCCCTAAAGTTAATTTGCCTACCAGGTCAGCCTTCAA AAACTGCAAGCCACAAAATTACTATTGATACCAACAAATCTCCAGTCAGTCTTCTTAGTCTCTTCCCAGGTTTTGCCAGTCAGTCAGATG ATGATCAGGTGAATGTAATGGGTTTTCACTTCTTAGGAGGTGCTCGAATTACTGTTCTTGCTTCCAAAACTTCTCAACGATATCGCATTC AGAGTGAACAATTTGAAGATCTTTGGCTCATAACCAATGAGCTTATTCTTCGCCTTCAAGAATATTTTGAAAAACAGGGAGTCAAAGATT TTGCATGTTCTTTTTCGGGATCTATACCCCTTCAAGAATATTTTGAGTTGATTGATCATCATTTTGAGCTACGGATAAATGGTGAAAAAT TAGAAGAACTCTTATCTGAGAGAGCTGTACAATTTCGGGCCATTCAACGCCGGCTACTAGCAAGATTCAAAGATAAAACTCCTGCCCCTC TTCAACACCTGGACACCTTGTTAGATGGAACCTACAAGCAGGTAATTGCTCTAGCAGATGCAGTGGAGGAAAACCAAGGCAATCTGTTCC AGTCATTCACCAGGCTGAAGAGTGCCACCCATTTGGTGATTCTGCTGATCGCGCTGTGGCAGAAGCTTAGTGCTGACCAGGTTGCTATTC TGGAAGCGGCATTTCTGCCGCTACAAGAAGACACTCAAGAATTGGGCTGGGAAGAAACGGTGGATGCCGCCATTTCCCACCTGTTGAAGA CTTGCCTGTCGAAGAGTTCTAAGGAGCAGGCTTTGAACCTCAACAGCCAGCTGAACATACCCAAAGACACAAGCCAACTGAAGAAACATA TCACCTTGCTCTGCGATAGATTATCCAAAGGTGGCCGTCTCTGCCTAAGTACCGATGCAGCAGCCCCACAGACCATGGTCATGCCAGGTG GTTGTACTACAATCCCAGAGTCAGACCTAGAAGAAAGATCAGTAGAACAAGACTCTACAGAACTGTTTACCAACCACAGACATCTCACTG CAGAGACACCCAGGCCTGAAGTTTCACCCCTCCAAGGAGTCTCGGAATAATTCAAGTAGAGTTGTTTGGTTGAGAGGAACATCCCCATCT CAAGGCCGAACCTGTGTGAACCTCATGCCAAGCACAGATATAGGGCTGGCGCAGGTGCTTCCTAAAGCTCACCTTCCTGGAGATGACATG CATAGAAAGAGGGGTTGGGACTTTTTACTTCACTAGGAGAACTTGTAACACCATGGGGAAGTCAGCTGAAACTTGTCTTGTTTTGCCAGG AAAGGAAGTAGTTGCCTTTGGTCATCCATCTGCTAATAGTCACAGAATACAGTGAAATGACATAGTTTTGGGTTAGATTTTATAATGCAA AGATTCAGATCCAAAATAATTTCATACCCCATTTTTTCACAGAATTCTTATATAGTAAATGTATCAAGTTTAATAAAGCATCTCATTGTC AAATAATATCTTGGATTTTATTTATAATTAGAGGGATTTATGAGTGATTGCTCTACATTATTTCTTCAAAGGAAAGGAAAGGAATTGAAG ACTTTGCTACTCTCTGGTAAGACTTGAATGTGATTATTTTATAAATAAAAGAACCACTATGAAACTTTCATGTGATTTGTTCTATTAAGC TAAACGTGGAAGAAGGGAAGTTTTCTAGTATTGTATGAAAATGTTTAAATATTTTCAAGGAAGCTAGTACATTAACATCCCCTTTTCATT ACCCCATTGGGTGGCACCTTTTAAAAATCCAGGTTATTAAAGGATTCACACTTTATTACAGTTAAAATTTAAGGTTTTTATGTGTAAGAA >16716_16716_3_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000354265_length(amino acids)=427AA_BP=18 MGYSSSCICKSGLKNNIQVTLQNRVILQKAKLSVYVQPPLELTCDQFTFEFMSLNVLGIPRVIQCKFRLPLKLICLPGQPSKTASHKITI DTNKSPVSLLSLFPGFASQSDDDQVNVMGFHFLGGARITVLASKTSQRYRIQSEQFEDLWLITNELILRLQEYFEKQGVKDFACSFSGSI PLQEYFELIDHHFELRINGEKLEELLSERAVQFRAIQRRLLARFKDKTPAPLQHLDTLLDGTYKQVIALADAVEENQGNLFQSFTRLKSA THLVILLIALWQKLSADQVAILEAAFLPLQEDTQELGWEETVDAAISHLLKTCLSKSSKEQALNLNSQLNIPKDTSQLKKHITLLCDRLS -------------------------------------------------------------- >16716_16716_4_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000355070_length(transcript)=2512nt_BP=350nt AGTCTTTTCTGTCTTCAGAGCATTAGTGTGCACATTAAGTCTTCAGCCTTGTTGAGATAGTGAATATTTATGCCGTATTTGAGAATTAAT ATTAGCAGAAATGTACAATGTAGATTTCTTTTTAAAATTCTAGGTTACTTTATGGAATTGGGCTCTTAGAGAACAAGAAAAGACTGAAGT TTTACGGGAAAACAAATCATGTGGTCTTCAGATTCTGAAATAAGGAGAAATGCAGCCATCTGAAATGGTCATGAACCCCAAACAAGTCTT CCTCTCTGTGCTGATATTTGGAGTAGCTGGGCTACTCCTCTTCATGTATTTGCAAGTCTGGATTGAAGAACAACATACAGGTCACACTGC AGAACAGAGTGATATTGCAAAAAGCCAAATTATCAGTCTACGTGCAACCACCATTAGAATTGACTTGTGATCAGTTCACCTTTGAATTTA TGACACCAGATTTGACTAGAACAGTAAGCTTTTCTGTTTATCTGAAAAGAAGTTATACACCATCAGAATTGGAAGGAAATGCTGTTGTTT CTTATTCCAGACCAACAGGCATTCCGCGAGTTATCCAATGTAAATTTAGACTTCCCCTAAAGTTAATTTGCCTACCAGGTCAGCCTTCAA AAACTGCAAGCCACAAAATTACTATTGATACCAACAAATCTCCAGTCAGTCTTCTTAGTCTCTTCCCAGGTTTTGCCAGTCAGTCAGATG ATGATCAGGTGAATGTAATGGGTTTTCACTTCTTAGGAGGTGCTCGAATTACTGTTCTTGCTTCCAAAACTTCTCAACGATATCGCATTC AGAGTGAACAATTTGAAGATCTTTGGCTCATAACCAATGAGCTTATTCTTCGCCTTCAAGAATATTTTGAAAAACAGGGAGTCAAAGATT TTGCATGTTCTTTTTCGGGATCTATACCCCTTCAAGAATATTTTGAGTTGATTGATCATCATTTTGAGCTACGGATAAATGGTGAAAAAT TAGAAGAACTCTTATCTGAGAGAGCTGTACAATTTCGGGCCATTCAACGCCGGCTACTAGCAAGATTCAAAGATAAAACTCCTGCCCCTC TTCAACACCTGGACACCTTGTTAGATGGAACCTACAAGCAGGTAATTGCTCTAGCAGATGCAGTGGAGGAAAACCAAGGCAATCTGTTCC AGTCATTCACCAGGCTGAAGAGTGCCACCCATTTGGTGATTCTGCTGATCGCGCTGTGGCAGAAGCTTAGTGCTGACCAGGTTGCTATTC TGGAAGCGGCATTTCTGCCGCTACAAGAAGACACTCAAGAATTGGGCTGGGAAGAAACGGTGGATGCCGCCATTTCCCACCTGTTGAAGA CTTGCCTGTCGAAGAGTTCTAAGGAGCAGGCTTTGAACCTCAACAGCCAGCTGAACATACCCAAAGACACAAGCCAACTGAAGAAACATA TCACCTTGCTCTGCGATAGATTATCCAAAGGTGGCCGTCTCTGCCTAAGTACCGATGCAGCAGCCCCACAGACCATGGTCATGCCAGGTG GTTGTACTACAATCCCAGAGTCAGACCTAGAAGAAAGATCAGTAGAACAAGACTCTACAGAACTGTTTACCAACCACAGACATCTCACTG CAGAGACACCCAGGCCTGAAGTTTCACCCCTCCAAGGAGTCTCGGAATAATTCAAGTAGAGTTGTTTGGTTGAGAGGAACATCCCCATCT CAAGGCCGAACCTGTGTGAACCTCATGCCAAGCACAGATATAGGGCTGGCGCAGGTGCTTCCTAAAGCTCACCTTCCTGGAGATGACATG CATAGAAAGAGGGGTTGGGACTTTTTACTTCACTAGGAGAACTTGTAACACCATGGGGAAGTCAGCTGAAACTTGTCTTGTTTTGCCAGG AAAGGAAGTAGTTGCCTTTGGTCATCCATCTGCTAATAGTCACAGAATACAGTGAAATGACATAGTTTTGGGTTAGATTTTATAATGCAA AGATTCAGATCCAAAATAATTTCATACCCCATTTTTTCACAGAATTCTTATATAGTAAATGTATCAAGTTTAATAAAGCATCTCATTGTC AAATAATATCTTGGATTTTATTTATAATTAGAGGGATTTATGAGTGATTGCTCTACATTATTTCTTCAAAGGAAAGGAAAGGAATTGAAG ACTTTGCTACTCTCTGGTAAGACTTGAATGTGATTATTTTATAAATAAAAGAACCACTATGAAACTTTCATGTGATTTGTTCTATTAAGC TAAACGTGGAAGAAGGGAAGTTTTCTAGTATTGTATGAAAATGTTTAAATATTTTCAAGGAAGCTAGTACATTAACATCCCCTTTTCATT ACCCCATTGGGTGGCACCTTTTAAAAATCCAGGTTATTAAAGGATTCACACTTTATTACAGTTAAAATTTAAGGTTTTTATGTGTAAGAA >16716_16716_4_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000355070_length(amino acids)=457AA_BP=18 MGYSSSCICKSGLKNNIQVTLQNRVILQKAKLSVYVQPPLELTCDQFTFEFMTPDLTRTVSFSVYLKRSYTPSELEGNAVVSYSRPTGIP RVIQCKFRLPLKLICLPGQPSKTASHKITIDTNKSPVSLLSLFPGFASQSDDDQVNVMGFHFLGGARITVLASKTSQRYRIQSEQFEDLW LITNELILRLQEYFEKQGVKDFACSFSGSIPLQEYFELIDHHFELRINGEKLEELLSERAVQFRAIQRRLLARFKDKTPAPLQHLDTLLD GTYKQVIALADAVEENQGNLFQSFTRLKSATHLVILLIALWQKLSADQVAILEAAFLPLQEDTQELGWEETVDAAISHLLKTCLSKSSKE QALNLNSQLNIPKDTSQLKKHITLLCDRLSKGGRLCLSTDAAAPQTMVMPGGCTTIPESDLEERSVEQDSTELFTNHRHLTAETPRPEVS -------------------------------------------------------------- >16716_16716_5_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000396127_length(transcript)=2422nt_BP=350nt AGTCTTTTCTGTCTTCAGAGCATTAGTGTGCACATTAAGTCTTCAGCCTTGTTGAGATAGTGAATATTTATGCCGTATTTGAGAATTAAT ATTAGCAGAAATGTACAATGTAGATTTCTTTTTAAAATTCTAGGTTACTTTATGGAATTGGGCTCTTAGAGAACAAGAAAAGACTGAAGT TTTACGGGAAAACAAATCATGTGGTCTTCAGATTCTGAAATAAGGAGAAATGCAGCCATCTGAAATGGTCATGAACCCCAAACAAGTCTT CCTCTCTGTGCTGATATTTGGAGTAGCTGGGCTACTCCTCTTCATGTATTTGCAAGTCTGGATTGAAGAACAACATACAGGTCACACTGC AGAACAGAGTGATATTGCAAAAAGCCAAATTATCAGTCTACGTGCAACCACCATTAGAATTGACTTGTGATCAGTTCACCTTTGAATTTA TGAATCGAAATCCTGATGGCATTCCGCGAGTTATCCAATGTAAATTTAGACTTCCCCTAAAGTTAATTTGCCTACCAGGTCAGCCTTCAA AAACTGCAAGCCACAAAATTACTATTGATACCAACAAATCTCCAGTCAGTCTTCTTAGTCTCTTCCCAGGTTTTGCCAGTCAGTCAGATG ATGATCAGGTGAATGTAATGGGTTTTCACTTCTTAGGAGGTGCTCGAATTACTGTTCTTGCTTCCAAAACTTCTCAACGATATCGCATTC AGAGTGAACAATTTGAAGATCTTTGGCTCATAACCAATGAGCTTATTCTTCGCCTTCAAGAATATTTTGAAAAACAGGGAGTCAAAGATT TTGCATGTTCTTTTTCGGGATCTATACCCCTTCAAGAATATTTTGAGTTGATTGATCATCATTTTGAGCTACGGATAAATGGTGAAAAAT TAGAAGAACTCTTATCTGAGAGAGCTGTACAATTTCGGGCCATTCAACGCCGGCTACTAGCAAGATTCAAAGATAAAACTCCTGCCCCTC TTCAACACCTGGACACCTTGTTAGATGGAACCTACAAGCAGGTAATTGCTCTAGCAGATGCAGTGGAGGAAAACCAAGGCAATCTGTTCC AGTCATTCACCAGGCTGAAGAGTGCCACCCATTTGGTGATTCTGCTGATCGCGCTGTGGCAGAAGCTTAGTGCTGACCAGGTTGCTATTC TGGAAGCGGCATTTCTGCCGCTACAAGAAGACACTCAAGAATTGGGCTGGGAAGAAACGGTGGATGCCGCCATTTCCCACCTGTTGAAGA CTTGCCTGTCGAAGAGTTCTAAGGAGCAGGCTTTGAACCTCAACAGCCAGCTGAACATACCCAAAGACACAAGCCAACTGAAGAAACATA TCACCTTGCTCTGCGATAGATTATCCAAAGGTGGCCGTCTCTGCCTAAGTACCGATGCAGCAGCCCCACAGACCATGGTCATGCCAGGTG GTTGTACTACAATCCCAGAGTCAGACCTAGAAGAAAGATCAGTAGAACAAGACTCTACAGAACTGTTTACCAACCACAGACATCTCACTG CAGAGACACCCAGGCCTGAAGTTTCACCCCTCCAAGGAGTCTCGGAATAATTCAAGTAGAGTTGTTTGGTTGAGAGGAACATCCCCATCT CAAGGCCGAACCTGTGTGAACCTCATGCCAAGCACAGATATAGGGCTGGCGCAGGTGCTTCCTAAAGCTCACCTTCCTGGAGATGACATG CATAGAAAGAGGGGTTGGGACTTTTTACTTCACTAGGAGAACTTGTAACACCATGGGGAAGTCAGCTGAAACTTGTCTTGTTTTGCCAGG AAAGGAAGTAGTTGCCTTTGGTCATCCATCTGCTAATAGTCACAGAATACAGTGAAATGACATAGTTTTGGGTTAGATTTTATAATGCAA AGATTCAGATCCAAAATAATTTCATACCCCATTTTTTCACAGAATTCTTATATAGTAAATGTATCAAGTTTAATAAAGCATCTCATTGTC AAATAATATCTTGGATTTTATTTATAATTAGAGGGATTTATGAGTGATTGCTCTACATTATTTCTTCAAAGGAAAGGAAAGGAATTGAAG ACTTTGCTACTCTCTGGTAAGACTTGAATGTGATTATTTTATAAATAAAAGAACCACTATGAAACTTTCATGTGATTTGTTCTATTAAGC TAAACGTGGAAGAAGGGAAGTTTTCTAGTATTGTATGAAAATGTTTAAATATTTTCAAGGAAGCTAGTACATTAACATCCCCTTTTCATT ACCCCATTGGGTGGCACCTTTTAAAAATCCAGGTTATTAAAGGATTCACACTTTATTACAGTTAAAATTTAAGGTTTTTATGTGTAAGAA >16716_16716_5_CHST9-BBS9_CHST9_chr18_24722653_ENST00000581714_BBS9_chr7_33388680_ENST00000396127_length(amino acids)=427AA_BP=18 MGYSSSCICKSGLKNNIQVTLQNRVILQKAKLSVYVQPPLELTCDQFTFEFMNRNPDGIPRVIQCKFRLPLKLICLPGQPSKTASHKITI DTNKSPVSLLSLFPGFASQSDDDQVNVMGFHFLGGARITVLASKTSQRYRIQSEQFEDLWLITNELILRLQEYFEKQGVKDFACSFSGSI PLQEYFELIDHHFELRINGEKLEELLSERAVQFRAIQRRLLARFKDKTPAPLQHLDTLLDGTYKQVIALADAVEENQGNLFQSFTRLKSA THLVILLIALWQKLSADQVAILEAAFLPLQEDTQELGWEETVDAAISHLLKTCLSKSSKEQALNLNSQLNIPKDTSQLKKHITLLCDRLS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHST9-BBS9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000242067 | 11 | 23 | 685_765 | 443.0 | 888.0 | LZTL1 | |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000350941 | 11 | 21 | 685_765 | 443.0 | 848.0 | LZTL1 | |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000396127 | 11 | 22 | 685_765 | 443.0 | 853.0 | LZTL1 | |
| Tgene | BBS9 | chr18:24722653 | chr7:33388680 | ENST00000425508 | 0 | 10 | 685_765 | 0 | 311.0 | LZTL1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHST9-BBS9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHST9-BBS9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
