
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CHTF18-RHOT2 (FusionGDB2 ID:16743) |
Fusion Gene Summary for CHTF18-RHOT2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CHTF18-RHOT2 | Fusion gene ID: 16743 | Hgene | Tgene | Gene symbol | CHTF18 | RHOT2 | Gene ID | 63922 | 89941 |
| Gene name | chromosome transmission fidelity factor 18 | ras homolog family member T2 | |
| Synonyms | C16orf41|C321D2.2|C321D2.3|C321D2.4|CHL12|Ctf18|RUVBL | ARHT2|C16orf39|MIRO-2|MIRO2|RASL | |
| Cytomap | 16p13.3 | 16p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | chromosome transmission fidelity protein 18 homologC321D2.3 (novel protein)C321D2.4 (novel protein)CTF18, chromosome transmission fidelity factor 18 homologhomolog of yeast CHL12some homology with holliday junction DNA helicase RUVB like | mitochondrial Rho GTPase 2mitochondrial Rho (MIRO) GTPase 2ras homolog gene family, member T2 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q8WVB6 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000262315, ENST00000317063, ENST00000455171, ENST00000491530, | ENST00000569943, ENST00000315082, | |
| Fusion gene scores | * DoF score | 1 X 2 X 1=2 | 7 X 8 X 5=280 |
| # samples | 2 | 8 | |
| ** MAII score | log2(2/2*10)=3.32192809488736 | log2(8/280*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CHTF18 [Title/Abstract] AND RHOT2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CHTF18(841967)-RHOT2(718670), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CHTF18 | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 12930902 |
 Fusion gene breakpoints across CHTF18 (5'-gene) Fusion gene breakpoints across CHTF18 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
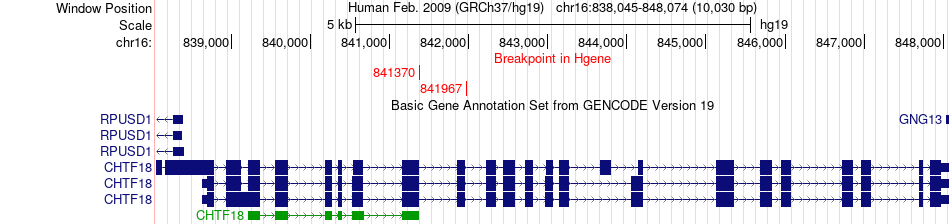 |
 Fusion gene breakpoints across RHOT2 (3'-gene) Fusion gene breakpoints across RHOT2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
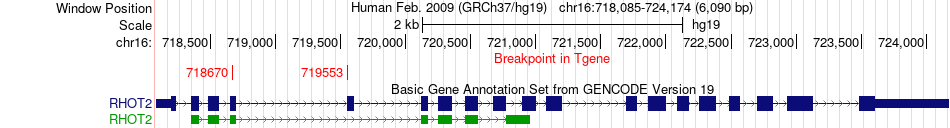 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-A452-01A | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| ChimerDB4 | STAD | TCGA-BR-A4QI-01A | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
Top |
Fusion Gene ORF analysis for CHTF18-RHOT2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000262315 | ENST00000569943 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| 5CDS-intron | ENST00000317063 | ENST00000569943 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| 5CDS-intron | ENST00000455171 | ENST00000569943 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| 5CDS-intron | ENST00000491530 | ENST00000569943 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| In-frame | ENST00000262315 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| In-frame | ENST00000317063 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| In-frame | ENST00000455171 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| In-frame | ENST00000491530 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + |
| intron-3CDS | ENST00000262315 | ENST00000315082 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3CDS | ENST00000317063 | ENST00000315082 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3CDS | ENST00000455171 | ENST00000315082 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3CDS | ENST00000491530 | ENST00000315082 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3UTR | ENST00000262315 | ENST00000569943 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3UTR | ENST00000317063 | ENST00000569943 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3UTR | ENST00000455171 | ENST00000569943 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
| intron-3UTR | ENST00000491530 | ENST00000569943 | CHTF18 | chr16 | 841967 | + | RHOT2 | chr16 | 718670 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000317063 | CHTF18 | chr16 | 841370 | + | ENST00000315082 | RHOT2 | chr16 | 719553 | + | 3892 | 1689 | 0 | 3323 | 1107 |
| ENST00000262315 | CHTF18 | chr16 | 841370 | + | ENST00000315082 | RHOT2 | chr16 | 719553 | + | 3370 | 1167 | 63 | 2801 | 912 |
| ENST00000455171 | CHTF18 | chr16 | 841370 | + | ENST00000315082 | RHOT2 | chr16 | 719553 | + | 3452 | 1249 | 61 | 2883 | 940 |
| ENST00000491530 | CHTF18 | chr16 | 841370 | + | ENST00000315082 | RHOT2 | chr16 | 719553 | + | 3022 | 819 | 624 | 2453 | 609 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000317063 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + | 0.006854458 | 0.9931456 |
| ENST00000262315 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + | 0.006903247 | 0.99309677 |
| ENST00000455171 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + | 0.005056501 | 0.99494344 |
| ENST00000491530 | ENST00000315082 | CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719553 | + | 0.038706824 | 0.9612932 |
Top |
Fusion Genomic Features for CHTF18-RHOT2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719552 | + | 0.06755225 | 0.9324478 |
| CHTF18 | chr16 | 841370 | + | RHOT2 | chr16 | 719552 | + | 0.06755225 | 0.9324478 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
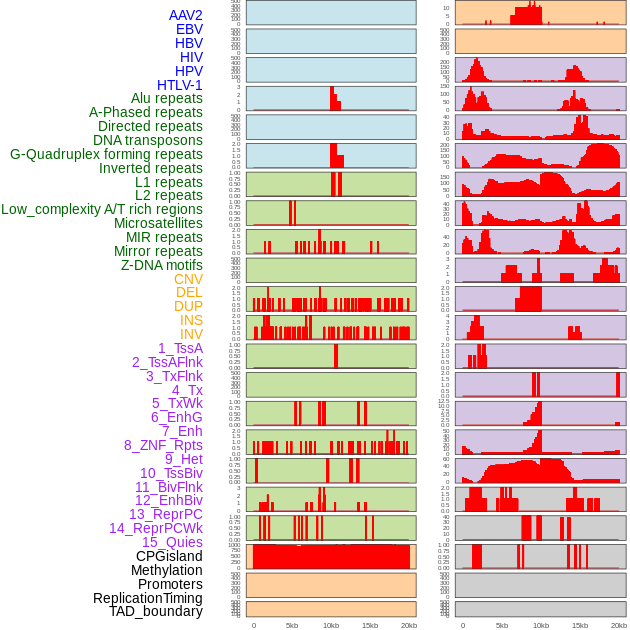 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
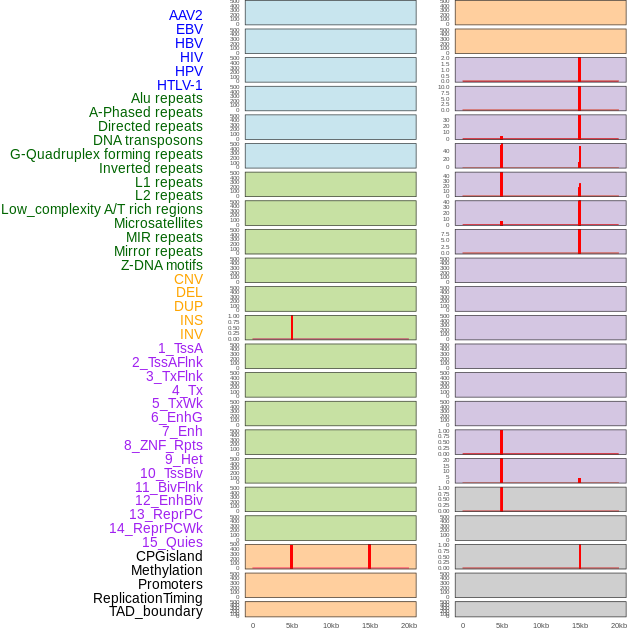 |
Top |
Fusion Protein Features for CHTF18-RHOT2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:841967/chr16:718670) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHTF18 | . |
| FUNCTION: Chromosome cohesion factor involved in sister chromatid cohesion and fidelity of chromosome transmission. Component of one of the cell nuclear antigen loader complexes, CTF18-replication factor C (CTF18-RFC), which consists of CTF18, CTF8, DCC1, RFC2, RFC3, RFC4 and RFC5. The CTF18-RFC complex binds to single-stranded and primed DNAs and has weak ATPase activity that is stimulated by the presence of primed DNA, replication protein A (RPA) and by proliferating cell nuclear antigen (PCNA). The CTF18-RFC complex catalyzes the ATP-dependent loading of PCNA onto primed and gapped DNA. Interacts with and stimulates DNA polymerase POLH. During DNA repair synthesis, involved in loading DNA polymerase POLE at the sites of local damage (PubMed:20227374). {ECO:0000269|PubMed:12766176, ECO:0000269|PubMed:12930902, ECO:0000269|PubMed:17545166, ECO:0000269|PubMed:20227374}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHTF18 | chr16:841370 | chr16:719553 | ENST00000455171 | + | 7 | 21 | 374_381 | 396 | 1004.0 | Nucleotide binding | ATP |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 197_208 | 74 | 619.0 | Calcium binding | 1 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 317_328 | 74 | 619.0 | Calcium binding | 2 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 184_219 | 74 | 619.0 | Domain | EF-hand 1 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 304_339 | 74 | 619.0 | Domain | EF-hand 2 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 414_576 | 74 | 619.0 | Domain | Miro 2 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 118_121 | 74 | 619.0 | Nucleotide binding | GTP 1 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 423_430 | 74 | 619.0 | Nucleotide binding | GTP 2 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 459_463 | 74 | 619.0 | Nucleotide binding | GTP 2 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 524_527 | 74 | 619.0 | Nucleotide binding | GTP 2 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 616_618 | 74 | 619.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 593_615 | 74 | 619.0 | Transmembrane | Helical%3B Anchor for type IV membrane protein |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHTF18 | chr16:841370 | chr16:719553 | ENST00000262315 | + | 8 | 22 | 374_381 | 368 | 976.0 | Nucleotide binding | ATP |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 2_168 | 74 | 619.0 | Domain | Miro 1 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 11_18 | 74 | 619.0 | Nucleotide binding | GTP 1 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 57_61 | 74 | 619.0 | Nucleotide binding | GTP 1 | |
| Tgene | RHOT2 | chr16:841370 | chr16:719553 | ENST00000315082 | 3 | 19 | 1_592 | 74 | 619.0 | Topological domain | Cytoplasmic |
Top |
Fusion Gene Sequence for CHTF18-RHOT2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16743_16743_1_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000262315_RHOT2_chr16_719553_ENST00000315082_length(transcript)=3370nt_BP=1167nt GCAGTGCGCGACGGCGGCGGCGGCGCGGGAGGTTCGGAGCGGGAGCTCGGGCTCGCGGACGGTATGGAGGACTACGAGCAGGAGCTGTGC GGCGTCGAGGATGATTTCCACAACCAGTTCGCGGCCGAGCTGGAGGTGCTGGCAGAGCTGGAAGGGGCGTCGACTCCGTCGCCCTCCGGG GTCCCCCTGTTCACCGCGGGCCGACCCCCGCGGACGTTCGAGGAGGCCCTTGCCAGAGGGGACGCGGCCTCCAGTCCCGCCCCAGCCGCA TCTGTGGGCAGCAGCCAGGGCGGCGCCAGGAAGAGGCAGGTGGACGCCGACCTGCAGCCGGCCGGGTCCCTGCCCCACGCCCCCAGGATC AAACGGCCTAGGCTGCAGGTGGTCAAGAGGCTGAACTTCAGATCGGAGGAGATGGAGGAGCCGCCCCCTCCCGACTCCTCGCCGACGGAC ATCACCCCGCCGCCGAGCCCTGAGGACCTCGCAGAGCTTTGGGGCCACGGAGTCTCAGAAGCTGCTGCCGACGTGGGTCTCACACGGGCC TCACCAGCTGCCCGCAATCCCGTCCTGAGGCGGCCCCCCATCTTGGAGGACTACGTCCACGTGACATCCACGGAGGGCGTCCGGGCTTAT CTGGTGCTGCGTGCTGACCCCATGGCCCCGGGGGTGCAGGGCTCTCTCCTCCACGTCCCATGGCGAGGCGGTGGCCAGCTGGACCTGCTG GGTGTGTCCTTAGCCTCCCTGAAGAAGCAGGTCGACGGCGAGCGGCGGGAGCGGCTGCTTCAGGAGGCCCAGAAGCTTTCAGACACCCTG CACAGTCTCAGGTCGGGGGAGGAGGAGGCAGCCCAGCCCTTGGGGGCCCCTGAGGAGGAGCCGACTGACGGTCAAGACGCCTCCAGTCAC TGCCTCTGGGTGGATGAGTTTGCACCCCGCCACTACACGGAGCTGCTCAGTGATGACTTCACCAACCGCTGCCTGCTCAAGTGGCTGAAG TTGTGGGACCTGGTGGTGTTTGGCCACGAGAGGCCTTCCCGGAAGCCCAGGCCCAGTGTTGAGCCGGCCCGGGTCAGCAAGGAGGCCACA GCCCCAGGCAAGTGGAAGAGCCACGAACAGGTGCTGGAGGAGATGCTGGAGGCTGGGCTGGACCCGAGCCAGCGACCGAAGCAGAAGGCA AACGTGGTGTGTGTGGTGTATGACGTCTCTGAGGAGGCCACCATTGAGAAGATTCGAACTAAGTGGATCCCACTGGTGAATGGGGGGACC ACGCAGGGGCCCAGGGTGCCCATCATCCTAGTGGGCAACAAGTCAGACCTGCGGTCGGGGAGCTCCATGGAGGCCGTGCTCCCCATCATG AGCCAGTTTCCCGAGATTGAGACCTGCGTGGAGTGTTCGGCCAAGAACCTGAGGAACATCTCAGAGCTGTTCTACTACGCCCAGAAGGCC GTCCTGCATCCCACAGCCCCCCTCTATGACCCTGAGGCCAAGCAGTTGAGGCCCGCGTGCGCCCAGGCGCTGACGCGCATCTTCAGGCTC TCAGATCAGGACCTGGACCAGGCGCTCAGTGACGAAGAGCTCAACGCTTTCCAGAAATCCTGCTTTGGGCACCCCCTGGCCCCGCAGGCC CTGGAGGACGTGAAGACGGTGGTGTGCAGGAACGTGGCGGGCGGCGTGCGGGAGGACCGGCTGACCCTGGATGGTTTCCTCTTCCTGAAC ACGCTCTTCATCCAGCGCGGCCGGCACGAGACCACCTGGACCATCCTGCGGCGCTTCGGCTACAGCGATGCCCTGGAGCTGACTGCGGAC TATCTCTCCCCTCTGATCCACGTGCCCCCCGGCTGCAGCACGGAGCTCAACCACCTTGGCTACCAGTTTGTGCAGAGAGTGTTTGAGAAG CACGACCAGGACCGCGACGGCGCCCTCTCGCCCGTGGAGCTGCAAAGCCTTTTCAGTGTGTTCCCAGCAGCGCCCTGGGGCCCCGAGCTC CCACGCACAGTCCGCACAGAGGCCGGCCGGTTGCCCCTGCACGGATACCTCTGCCAGTGGACCCTGGTGACCTACCTGGACGTCCGGAGC TGCCTTGGACACCTAGGCTACCTGGGCTACCCCACCCTCTGTGAGCAGGACCAGGCCCATGCCATCACAGTCACTCGTGAGAAGAGGCTG GACCAGGAGAAGGGACAGACGCAGCGGAGCGTCCTCCTGTGCAAGGTGGTAGGGGCCCGTGGAGTGGGCAAGTCTGCCTTCCTGCAGGCC TTTCTCGGCCGCGGCCTGGGGCACCAGGACACGAGGGAGCAGCCTCCCGGCTACGCCATCGACACGGTGCAGGTCAATGGACAGGAGAAG TACTTGATCCTCTGTGAGGTGGGCACAGATGGTCTGCTGGCCACATCGCTGGACGCCACCTGTGACGTTGCCTGCTTGATGTTTGATGGC AGTGACCCAAAGTCCTTTGCACATTGTGCCAGCGTCTACAAGCACCATTACATGGACGGGCAGACCCCCTGCCTCTTTGTCTCCTCCAAG GCCGACCTGCCCGAAGGTGTCGCGGTGTCTGGCCCATCACCGGCCGAGTTTTGCCGCAAGCACCGGCTACCCGCTCCCGTGCCGTTCTCC TGTGCTGGCCCAGCCGAGCCCAGCACCACCATCTTCACCCAGCTCGCCACCATGGCCGCCTTCCCACATTTGGTCCACGCAGAGCTGCAT CCCTCTTCCTTCTGGCTCCGGGGGCTGCTGGGGGTTGTCGGGGCCGCCGTGGCCGCAGTCCTCAGCTTCTCACTCTACAGGGTCCTGGTG AAGAGCCAGTGAGGCCCCTGGTACCCAAGCCCCCTCCCCTGACCTGGGTGTGCCTCGCTGCTGGGGCTCTGCAGGGGCAGCACAGCTGGG GTGCAGGCCAGGCTGCCACTCCGGGAACGCCTTTGCGCCGGGACTTTTTGTTTCTGAAGGCAGTCGATCTGCAGCGGGGCCTTATGCTGC CATGCACTGCCCTGGCTCCTGCCGGACCCCCAGGGTGGGCCGTGGCAGGTGGCTGAGCAGGAGCTCCCAAGTGCCGGCCACCGCTGTCAG GGATTGCCCACCCCTGGGCATCATGTGTGTGGGGCCGGGGAGCACAGGTGTGGGAGCTGGTGACCCCAGACCCAGAATTCTCAGGGCTCT ACCCCCCTTTCCTGGTCCTAGGTGGCCAGTGGGTATGAGGAGGGCTGGAAGGCAGAGCTTTGGGCCAAAAGCAGGCGTTGGGGGGTCCCC CCTCAAGTTTGGAGCCGTTTCCGTGGTTGTAGCAGAGGACCGGAGGTTGGGTTCCTGATTAAACTTCACTGTGTGTTTTCTATCTCGGAT >16743_16743_1_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000262315_RHOT2_chr16_719553_ENST00000315082_length(amino acids)=912AA_BP=368 MEDYEQELCGVEDDFHNQFAAELEVLAELEGASTPSPSGVPLFTAGRPPRTFEEALARGDAASSPAPAASVGSSQGGARKRQVDADLQPA GSLPHAPRIKRPRLQVVKRLNFRSEEMEEPPPPDSSPTDITPPPSPEDLAELWGHGVSEAAADVGLTRASPAARNPVLRRPPILEDYVHV TSTEGVRAYLVLRADPMAPGVQGSLLHVPWRGGGQLDLLGVSLASLKKQVDGERRERLLQEAQKLSDTLHSLRSGEEEAAQPLGAPEEEP TDGQDASSHCLWVDEFAPRHYTELLSDDFTNRCLLKWLKLWDLVVFGHERPSRKPRPSVEPARVSKEATAPGKWKSHEQVLEEMLEAGLD PSQRPKQKANVVCVVYDVSEEATIEKIRTKWIPLVNGGTTQGPRVPIILVGNKSDLRSGSSMEAVLPIMSQFPEIETCVECSAKNLRNIS ELFYYAQKAVLHPTAPLYDPEAKQLRPACAQALTRIFRLSDQDLDQALSDEELNAFQKSCFGHPLAPQALEDVKTVVCRNVAGGVREDRL TLDGFLFLNTLFIQRGRHETTWTILRRFGYSDALELTADYLSPLIHVPPGCSTELNHLGYQFVQRVFEKHDQDRDGALSPVELQSLFSVF PAAPWGPELPRTVRTEAGRLPLHGYLCQWTLVTYLDVRSCLGHLGYLGYPTLCEQDQAHAITVTREKRLDQEKGQTQRSVLLCKVVGARG VGKSAFLQAFLGRGLGHQDTREQPPGYAIDTVQVNGQEKYLILCEVGTDGLLATSLDATCDVACLMFDGSDPKSFAHCASVYKHHYMDGQ TPCLFVSSKADLPEGVAVSGPSPAEFCRKHRLPAPVPFSCAGPAEPSTTIFTQLATMAAFPHLVHAELHPSSFWLRGLLGVVGAAVAAVL -------------------------------------------------------------- >16743_16743_2_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000317063_RHOT2_chr16_719553_ENST00000315082_length(transcript)=3892nt_BP=1689nt ATGCACGTGAATGCCTCTGCCCGACGGTGGGGGCGGGGAGGGGCTGCAGCACTACCAGGCCCGGCAGCCAGTCCGCGCAGACCCGCGCCC CGCACAGGCCCGGTGGGCAGGCCGACCCTGGCTCCGAGCCCCTCCCGCGCCCACGCCCACGCCAAGACCAACCTGCTGCCGGGCCGTGCA GTCCAGGCCCCCGATGCCGTGCCCGGCGCCGGCTCCAGCCGCGCGCCCGCGCGCTGGCGACCCAGAGACCCTGGAAGCTCCGCTCCGGAC GCCTGGCAGCGCTTCCGCCCTGCACCGCTGCGCGCGCAGCCCCGCCCCCCCCCCCGGCCCGCCCCCGGCCCGCCCCCGCCGCGGAAGGGC GGCTGCGGCGGCCAATGGCGAGCTCCCCAGGACAGGACGCGGGGGCTGCGGAGCAGGAACTCGCCCCGCCCACCCCTCGCGCAGCCCCGC CTCCGCCACGCCAACCAATGGCGCGCGCCGCCGGGCACGCCGGCTGCTGATTGGCGGCGCCCGGCACGCTCGGGGCGGGCAGTGCGCGAC GGCGGCGGCGGCGCGGGAGGTTCGGAGCGGGAGCTCGGGCTCGCGGACGGTATGGAGGACTACGAGCAGGAGCTGTGCGGCGTCGAGGAT GATTTCCACAACCAGTTCGCGGCCGAGCTGGAGGTGCTGGCAGAGCTGGAAGGGGCGTCGACTCCGTCGCCCTCCGGGGTCCCCCTGTTC ACCGCGGGCCGACCCCCGCGGACGTTCGAGGAGGCCCTTGCCAGAGGGGACGCGGCCTCCAGTCCCGCCCCAGCCGCATCTGTGGGCAGC AGCCAGGGCGGCGCCAGGAAGAGGCAGGTGGACGCCGACCTGCAGCCGGCCGGGTCCCTGCCCCACGCCCCCAGGATCAAACGGCCTAGG CTGCAGGTGGTCAAGAGGCTGAACTTCAGATCGGAGGAGATGGAGGAGCCGCCCCCTCCCGACTCCTCGCCGACGGACATCACCCCGCCG CCGAGCCCTGAGGACCTCGCAGAGCTTTGGGGCCACGGGTTCTCAGAAGCTGCTGCCGACGTGGGTCTCACACGGGCCTCACCAGCTGCC CGCAATCCCGTCCTGAGGCGGCCCCCCATCTTGGAGGACTACGTCCACGTGACATCCACGGAGGGCGTCCGGGCTTATCTGGTGCTGCGT GCTGACCCCATGGCCCCGGGGGTGCAGGGCTCTCTCCTCCACGTCCCATGGCGAGGCGGTGGCCAGCTGGACCTGCTGGGTGTGTCCTTA GCCTCCCTGAAGAAGCAGGTCGACGGCGAGCGGCGGGAGCGGCTGCTTCAGGAGGCCCAGAAGCTTTCAGACACCCTGCACAGGTCGGGG GAGGAGGAGGCAGCCCAGCCCTTGGGGGCCCCTGAGGAGGAGCCGACTGACGGTCAAGACGCCTCCAGTCACTGCCTCTGGGTGGATGAG TTTGCACCCCGCCACTACACGGAGCTGCTCAGTGATGACTTCACCAACCGCTGCCTGCTCAAGTGGCTGAAGTTGTGGGACCTGGTGGTG TTTGGCCACGAGAGGCCTTCCCGGAAGCCCAGGCCCAGTGTTGAGCCGGCCCGGGTCAGCAAGGAGGCCACAGCCCCAGGCAAGTGGAAG AGCCACGAACAGGTGCTGGAGGAGATGCTGGAGGCTGGGCTGGACCCGAGCCAGCGACCGAAGCAGAAGGCAAACGTGGTGTGTGTGGTG TATGACGTCTCTGAGGAGGCCACCATTGAGAAGATTCGAACTAAGTGGATCCCACTGGTGAATGGGGGGACCACGCAGGGGCCCAGGGTG CCCATCATCCTAGTGGGCAACAAGTCAGACCTGCGGTCGGGGAGCTCCATGGAGGCCGTGCTCCCCATCATGAGCCAGTTTCCCGAGATT GAGACCTGCGTGGAGTGTTCGGCCAAGAACCTGAGGAACATCTCAGAGCTGTTCTACTACGCCCAGAAGGCCGTCCTGCATCCCACAGCC CCCCTCTATGACCCTGAGGCCAAGCAGTTGAGGCCCGCGTGCGCCCAGGCGCTGACGCGCATCTTCAGGCTCTCAGATCAGGACCTGGAC CAGGCGCTCAGTGACGAAGAGCTCAACGCTTTCCAGAAATCCTGCTTTGGGCACCCCCTGGCCCCGCAGGCCCTGGAGGACGTGAAGACG GTGGTGTGCAGGAACGTGGCGGGCGGCGTGCGGGAGGACCGGCTGACCCTGGATGGTTTCCTCTTCCTGAACACGCTCTTCATCCAGCGC GGCCGGCACGAGACCACCTGGACCATCCTGCGGCGCTTCGGCTACAGCGATGCCCTGGAGCTGACTGCGGACTATCTCTCCCCTCTGATC CACGTGCCCCCCGGCTGCAGCACGGAGCTCAACCACCTTGGCTACCAGTTTGTGCAGAGAGTGTTTGAGAAGCACGACCAGGACCGCGAC GGCGCCCTCTCGCCCGTGGAGCTGCAAAGCCTTTTCAGTGTGTTCCCAGCAGCGCCCTGGGGCCCCGAGCTCCCACGCACAGTCCGCACA GAGGCCGGCCGGTTGCCCCTGCACGGATACCTCTGCCAGTGGACCCTGGTGACCTACCTGGACGTCCGGAGCTGCCTTGGACACCTAGGC TACCTGGGCTACCCCACCCTCTGTGAGCAGGACCAGGCCCATGCCATCACAGTCACTCGTGAGAAGAGGCTGGACCAGGAGAAGGGACAG ACGCAGCGGAGCGTCCTCCTGTGCAAGGTGGTAGGGGCCCGTGGAGTGGGCAAGTCTGCCTTCCTGCAGGCCTTTCTCGGCCGCGGCCTG GGGCACCAGGACACGAGGGAGCAGCCTCCCGGCTACGCCATCGACACGGTGCAGGTCAATGGACAGGAGAAGTACTTGATCCTCTGTGAG GTGGGCACAGATGGTCTGCTGGCCACATCGCTGGACGCCACCTGTGACGTTGCCTGCTTGATGTTTGATGGCAGTGACCCAAAGTCCTTT GCACATTGTGCCAGCGTCTACAAGCACCATTACATGGACGGGCAGACCCCCTGCCTCTTTGTCTCCTCCAAGGCCGACCTGCCCGAAGGT GTCGCGGTGTCTGGCCCATCACCGGCCGAGTTTTGCCGCAAGCACCGGCTACCCGCTCCCGTGCCGTTCTCCTGTGCTGGCCCAGCCGAG CCCAGCACCACCATCTTCACCCAGCTCGCCACCATGGCCGCCTTCCCACATTTGGTCCACGCAGAGCTGCATCCCTCTTCCTTCTGGCTC CGGGGGCTGCTGGGGGTTGTCGGGGCCGCCGTGGCCGCAGTCCTCAGCTTCTCACTCTACAGGGTCCTGGTGAAGAGCCAGTGAGGCCCC TGGTACCCAAGCCCCCTCCCCTGACCTGGGTGTGCCTCGCTGCTGGGGCTCTGCAGGGGCAGCACAGCTGGGGTGCAGGCCAGGCTGCCA CTCCGGGAACGCCTTTGCGCCGGGACTTTTTGTTTCTGAAGGCAGTCGATCTGCAGCGGGGCCTTATGCTGCCATGCACTGCCCTGGCTC CTGCCGGACCCCCAGGGTGGGCCGTGGCAGGTGGCTGAGCAGGAGCTCCCAAGTGCCGGCCACCGCTGTCAGGGATTGCCCACCCCTGGG CATCATGTGTGTGGGGCCGGGGAGCACAGGTGTGGGAGCTGGTGACCCCAGACCCAGAATTCTCAGGGCTCTACCCCCCTTTCCTGGTCC TAGGTGGCCAGTGGGTATGAGGAGGGCTGGAAGGCAGAGCTTTGGGCCAAAAGCAGGCGTTGGGGGGTCCCCCCTCAAGTTTGGAGCCGT TTCCGTGGTTGTAGCAGAGGACCGGAGGTTGGGTTCCTGATTAAACTTCACTGTGTGTTTTCTATCTCGGATCCCAGTCTCTGAAGACAA >16743_16743_2_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000317063_RHOT2_chr16_719553_ENST00000315082_length(amino acids)=1107AA_BP=10 MHVNASARRWGRGGAAALPGPAASPRRPAPRTGPVGRPTLAPSPSRAHAHAKTNLLPGRAVQAPDAVPGAGSSRAPARWRPRDPGSSAPD AWQRFRPAPLRAQPRPPPRPAPGPPPPRKGGCGGQWRAPQDRTRGLRSRNSPRPPLAQPRLRHANQWRAPPGTPAADWRRPARSGRAVRD GGGGAGGSERELGLADGMEDYEQELCGVEDDFHNQFAAELEVLAELEGASTPSPSGVPLFTAGRPPRTFEEALARGDAASSPAPAASVGS SQGGARKRQVDADLQPAGSLPHAPRIKRPRLQVVKRLNFRSEEMEEPPPPDSSPTDITPPPSPEDLAELWGHGFSEAAADVGLTRASPAA RNPVLRRPPILEDYVHVTSTEGVRAYLVLRADPMAPGVQGSLLHVPWRGGGQLDLLGVSLASLKKQVDGERRERLLQEAQKLSDTLHRSG EEEAAQPLGAPEEEPTDGQDASSHCLWVDEFAPRHYTELLSDDFTNRCLLKWLKLWDLVVFGHERPSRKPRPSVEPARVSKEATAPGKWK SHEQVLEEMLEAGLDPSQRPKQKANVVCVVYDVSEEATIEKIRTKWIPLVNGGTTQGPRVPIILVGNKSDLRSGSSMEAVLPIMSQFPEI ETCVECSAKNLRNISELFYYAQKAVLHPTAPLYDPEAKQLRPACAQALTRIFRLSDQDLDQALSDEELNAFQKSCFGHPLAPQALEDVKT VVCRNVAGGVREDRLTLDGFLFLNTLFIQRGRHETTWTILRRFGYSDALELTADYLSPLIHVPPGCSTELNHLGYQFVQRVFEKHDQDRD GALSPVELQSLFSVFPAAPWGPELPRTVRTEAGRLPLHGYLCQWTLVTYLDVRSCLGHLGYLGYPTLCEQDQAHAITVTREKRLDQEKGQ TQRSVLLCKVVGARGVGKSAFLQAFLGRGLGHQDTREQPPGYAIDTVQVNGQEKYLILCEVGTDGLLATSLDATCDVACLMFDGSDPKSF AHCASVYKHHYMDGQTPCLFVSSKADLPEGVAVSGPSPAEFCRKHRLPAPVPFSCAGPAEPSTTIFTQLATMAAFPHLVHAELHPSSFWL -------------------------------------------------------------- >16743_16743_3_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000455171_RHOT2_chr16_719553_ENST00000315082_length(transcript)=3452nt_BP=1249nt AGTGCGCGACGGCGGCGGCGGCGCGGGAGGTTCGGAGCGGGAGCTCGGGCTCGCGGACGGTATGGAGGACTACGAGCAGGAGCTGTGCGG CGTCGAGGATGATTTCCACAACCAGTTCGCGGCCGAGCTGGAGGTGCTGGCAGAGCTGGAAGGGGCGTCGACTCCGTCGCCCTCCGGGGT CCCCCTGTTCACCGCGGGCCGACCCCCGCGGACGTTCGAGGAGGCCCTTGCCAGAGGGGACGCGGCCTCCAGTCCCGCCCCAGCCGCATC TGTGGGCAGCAGCCAGGGCGGCGCCAGGAAGAGGCAGGTGGACGCCGACCTGCAGCCGGCCGGGTCCCTGCCCCACGGTAGGTTGGCGGC ATTGCCCCAGGGCCTCCGGAGTGGGCGCGAGGCTGAGGAGGCCTCTGGTTCCCTGCATGTGTCTCCCCCAGCCCCCAGGATCAAACGGCC TAGGCTGCAGGTGGTCAAGAGGCTGAACTTCAGATCGGAGGAGATGGAGGAGCCGCCCCCTCCCGACTCCTCGCCGACGGACATCACCCC GCCGCCGAGCCCTGAGGACCTCGCAGAGCTTTGGGGCCACGGAGTCTCAGAAGCTGCTGCCGACGTGGGTCTCACACGGGCCTCACCAGC TGCCCGCAATCCCGTCCTGAGGCGGCCCCCCATCTTGGAGGACTACGTCCACGTGACATCCACGGAGGGCGTCCGGGCTTATCTGGTGCT GCGTGCTGACCCCATGGCCCCGGGGGTGCAGGGCTCTCTCCTCCACGTCCCATGGCGAGGCGGTGGCCAGCTGGACCTGCTGGGTGTGTC CTTAGCCTCCCTGAAGAAGCAGGTCGACGGCGAGCGGCGGGAGCGGCTGCTTCAGGAGGCCCAGAAGCTTTCAGACACCCTGCACAGTCT CAGGTCGGGGGAGGAGGAGGCAGCCCAGCCCTTGGGGGCCCCTGAGGAGGAGCCGACTGACGGTCAAGACGCCTCCAGTCACTGCCTCTG GGTGGATGAGTTTGCACCCCGCCACTACACGGAGCTGCTCAGTGATGACTTCACCAACCGCTGCCTGCTCAAGTGGCTGAAGTTGTGGGA CCTGGTGGTGTTTGGCCACGAGAGGCCTTCCCGGAAGCCCAGGCCCAGTGTTGAGCCGGCCCGGGTCAGCAAGGAGGCCACAGCCCCAGG CAAGTGGAAGAGCCACGAACAGGTGCTGGAGGAGATGCTGGAGGCTGGGCTGGACCCGAGCCAGCGACCGAAGCAGAAGGCAAACGTGGT GTGTGTGGTGTATGACGTCTCTGAGGAGGCCACCATTGAGAAGATTCGAACTAAGTGGATCCCACTGGTGAATGGGGGGACCACGCAGGG GCCCAGGGTGCCCATCATCCTAGTGGGCAACAAGTCAGACCTGCGGTCGGGGAGCTCCATGGAGGCCGTGCTCCCCATCATGAGCCAGTT TCCCGAGATTGAGACCTGCGTGGAGTGTTCGGCCAAGAACCTGAGGAACATCTCAGAGCTGTTCTACTACGCCCAGAAGGCCGTCCTGCA TCCCACAGCCCCCCTCTATGACCCTGAGGCCAAGCAGTTGAGGCCCGCGTGCGCCCAGGCGCTGACGCGCATCTTCAGGCTCTCAGATCA GGACCTGGACCAGGCGCTCAGTGACGAAGAGCTCAACGCTTTCCAGAAATCCTGCTTTGGGCACCCCCTGGCCCCGCAGGCCCTGGAGGA CGTGAAGACGGTGGTGTGCAGGAACGTGGCGGGCGGCGTGCGGGAGGACCGGCTGACCCTGGATGGTTTCCTCTTCCTGAACACGCTCTT CATCCAGCGCGGCCGGCACGAGACCACCTGGACCATCCTGCGGCGCTTCGGCTACAGCGATGCCCTGGAGCTGACTGCGGACTATCTCTC CCCTCTGATCCACGTGCCCCCCGGCTGCAGCACGGAGCTCAACCACCTTGGCTACCAGTTTGTGCAGAGAGTGTTTGAGAAGCACGACCA GGACCGCGACGGCGCCCTCTCGCCCGTGGAGCTGCAAAGCCTTTTCAGTGTGTTCCCAGCAGCGCCCTGGGGCCCCGAGCTCCCACGCAC AGTCCGCACAGAGGCCGGCCGGTTGCCCCTGCACGGATACCTCTGCCAGTGGACCCTGGTGACCTACCTGGACGTCCGGAGCTGCCTTGG ACACCTAGGCTACCTGGGCTACCCCACCCTCTGTGAGCAGGACCAGGCCCATGCCATCACAGTCACTCGTGAGAAGAGGCTGGACCAGGA GAAGGGACAGACGCAGCGGAGCGTCCTCCTGTGCAAGGTGGTAGGGGCCCGTGGAGTGGGCAAGTCTGCCTTCCTGCAGGCCTTTCTCGG CCGCGGCCTGGGGCACCAGGACACGAGGGAGCAGCCTCCCGGCTACGCCATCGACACGGTGCAGGTCAATGGACAGGAGAAGTACTTGAT CCTCTGTGAGGTGGGCACAGATGGTCTGCTGGCCACATCGCTGGACGCCACCTGTGACGTTGCCTGCTTGATGTTTGATGGCAGTGACCC AAAGTCCTTTGCACATTGTGCCAGCGTCTACAAGCACCATTACATGGACGGGCAGACCCCCTGCCTCTTTGTCTCCTCCAAGGCCGACCT GCCCGAAGGTGTCGCGGTGTCTGGCCCATCACCGGCCGAGTTTTGCCGCAAGCACCGGCTACCCGCTCCCGTGCCGTTCTCCTGTGCTGG CCCAGCCGAGCCCAGCACCACCATCTTCACCCAGCTCGCCACCATGGCCGCCTTCCCACATTTGGTCCACGCAGAGCTGCATCCCTCTTC CTTCTGGCTCCGGGGGCTGCTGGGGGTTGTCGGGGCCGCCGTGGCCGCAGTCCTCAGCTTCTCACTCTACAGGGTCCTGGTGAAGAGCCA GTGAGGCCCCTGGTACCCAAGCCCCCTCCCCTGACCTGGGTGTGCCTCGCTGCTGGGGCTCTGCAGGGGCAGCACAGCTGGGGTGCAGGC CAGGCTGCCACTCCGGGAACGCCTTTGCGCCGGGACTTTTTGTTTCTGAAGGCAGTCGATCTGCAGCGGGGCCTTATGCTGCCATGCACT GCCCTGGCTCCTGCCGGACCCCCAGGGTGGGCCGTGGCAGGTGGCTGAGCAGGAGCTCCCAAGTGCCGGCCACCGCTGTCAGGGATTGCC CACCCCTGGGCATCATGTGTGTGGGGCCGGGGAGCACAGGTGTGGGAGCTGGTGACCCCAGACCCAGAATTCTCAGGGCTCTACCCCCCT TTCCTGGTCCTAGGTGGCCAGTGGGTATGAGGAGGGCTGGAAGGCAGAGCTTTGGGCCAAAAGCAGGCGTTGGGGGGTCCCCCCTCAAGT TTGGAGCCGTTTCCGTGGTTGTAGCAGAGGACCGGAGGTTGGGTTCCTGATTAAACTTCACTGTGTGTTTTCTATCTCGGATCCCAGTCT >16743_16743_3_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000455171_RHOT2_chr16_719553_ENST00000315082_length(amino acids)=940AA_BP=396 MEDYEQELCGVEDDFHNQFAAELEVLAELEGASTPSPSGVPLFTAGRPPRTFEEALARGDAASSPAPAASVGSSQGGARKRQVDADLQPA GSLPHGRLAALPQGLRSGREAEEASGSLHVSPPAPRIKRPRLQVVKRLNFRSEEMEEPPPPDSSPTDITPPPSPEDLAELWGHGVSEAAA DVGLTRASPAARNPVLRRPPILEDYVHVTSTEGVRAYLVLRADPMAPGVQGSLLHVPWRGGGQLDLLGVSLASLKKQVDGERRERLLQEA QKLSDTLHSLRSGEEEAAQPLGAPEEEPTDGQDASSHCLWVDEFAPRHYTELLSDDFTNRCLLKWLKLWDLVVFGHERPSRKPRPSVEPA RVSKEATAPGKWKSHEQVLEEMLEAGLDPSQRPKQKANVVCVVYDVSEEATIEKIRTKWIPLVNGGTTQGPRVPIILVGNKSDLRSGSSM EAVLPIMSQFPEIETCVECSAKNLRNISELFYYAQKAVLHPTAPLYDPEAKQLRPACAQALTRIFRLSDQDLDQALSDEELNAFQKSCFG HPLAPQALEDVKTVVCRNVAGGVREDRLTLDGFLFLNTLFIQRGRHETTWTILRRFGYSDALELTADYLSPLIHVPPGCSTELNHLGYQF VQRVFEKHDQDRDGALSPVELQSLFSVFPAAPWGPELPRTVRTEAGRLPLHGYLCQWTLVTYLDVRSCLGHLGYLGYPTLCEQDQAHAIT VTREKRLDQEKGQTQRSVLLCKVVGARGVGKSAFLQAFLGRGLGHQDTREQPPGYAIDTVQVNGQEKYLILCEVGTDGLLATSLDATCDV ACLMFDGSDPKSFAHCASVYKHHYMDGQTPCLFVSSKADLPEGVAVSGPSPAEFCRKHRLPAPVPFSCAGPAEPSTTIFTQLATMAAFPH -------------------------------------------------------------- >16743_16743_4_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000491530_RHOT2_chr16_719553_ENST00000315082_length(transcript)=3022nt_BP=819nt CAGGATCAAACGGCCTAGGCTGCAGGTGGTCAAGAGGCTGAACTTCAGATCGGAGGAGATGGAGGAGCCGCCCCCTCCCGACTCCTCGCC GACGGACATCACCCCGCCGCCGAGCCCTGAGGACCTCGCAGAGCTTTGGGGCCACGGAGTCTCAGAAGCTGCTGCCGACGTGGGTCTCAC ACGGGCCTCACCAGCTGCCCGCAATCCCGTCCTGAGGCGGCCCCCCATCTTGGAGGACTACGTCCACGTGACATCCACGGAGGGCGTCCG GGCTTATCTGGTGCTGCGTGCTGACCCCATGGCCCCGGGGGTGCAGGGCTCTCTCCTCCACGTCCCATGGCGAGGCGGTGGCCAGCTGGA CCTGCTGGGTGTGTCCTTAGCCTCCCTGAAGAAGCAGGTCGACGGCGAGCGGCGGGAGCGGCTGCTTCAGGAGGCCCAGAAGCTTTCAGA CACCCTGCACAGTCTCAGGTCGGGGGAGGAGGAGGCAGCCCAGCCCTTGGGGGCCCCTGAGGAGGAGCCGACTGACGGTCAAGACGCCTC CAGTCACTGCCTCTGGGTGGATGAGTTTGCACCCCGCCACTACACGGAGCTGCTCAGTGATGACGTGAGTTCACCAACCGCTGCCTGCTC AAGTGGCTGAAGTTGTGGGACCTGGTGGTGTTTGGCCACGAGAGGCCTTCCCGGAAGCCCAGGCCCAGTGTTGAGCCGGCCCGGGTCAGC AAGGAGGCCACAGCCCCAGGCAAGTGGAAGAGCCACGAACAGGTGCTGGAGGAGATGCTGGAGGCTGGGCTGGACCCGAGCCAGCGACCG AAGCAGAAGGCAAACGTGGTGTGTGTGGTGTATGACGTCTCTGAGGAGGCCACCATTGAGAAGATTCGAACTAAGTGGATCCCACTGGTG AATGGGGGGACCACGCAGGGGCCCAGGGTGCCCATCATCCTAGTGGGCAACAAGTCAGACCTGCGGTCGGGGAGCTCCATGGAGGCCGTG CTCCCCATCATGAGCCAGTTTCCCGAGATTGAGACCTGCGTGGAGTGTTCGGCCAAGAACCTGAGGAACATCTCAGAGCTGTTCTACTAC GCCCAGAAGGCCGTCCTGCATCCCACAGCCCCCCTCTATGACCCTGAGGCCAAGCAGTTGAGGCCCGCGTGCGCCCAGGCGCTGACGCGC ATCTTCAGGCTCTCAGATCAGGACCTGGACCAGGCGCTCAGTGACGAAGAGCTCAACGCTTTCCAGAAATCCTGCTTTGGGCACCCCCTG GCCCCGCAGGCCCTGGAGGACGTGAAGACGGTGGTGTGCAGGAACGTGGCGGGCGGCGTGCGGGAGGACCGGCTGACCCTGGATGGTTTC CTCTTCCTGAACACGCTCTTCATCCAGCGCGGCCGGCACGAGACCACCTGGACCATCCTGCGGCGCTTCGGCTACAGCGATGCCCTGGAG CTGACTGCGGACTATCTCTCCCCTCTGATCCACGTGCCCCCCGGCTGCAGCACGGAGCTCAACCACCTTGGCTACCAGTTTGTGCAGAGA GTGTTTGAGAAGCACGACCAGGACCGCGACGGCGCCCTCTCGCCCGTGGAGCTGCAAAGCCTTTTCAGTGTGTTCCCAGCAGCGCCCTGG GGCCCCGAGCTCCCACGCACAGTCCGCACAGAGGCCGGCCGGTTGCCCCTGCACGGATACCTCTGCCAGTGGACCCTGGTGACCTACCTG GACGTCCGGAGCTGCCTTGGACACCTAGGCTACCTGGGCTACCCCACCCTCTGTGAGCAGGACCAGGCCCATGCCATCACAGTCACTCGT GAGAAGAGGCTGGACCAGGAGAAGGGACAGACGCAGCGGAGCGTCCTCCTGTGCAAGGTGGTAGGGGCCCGTGGAGTGGGCAAGTCTGCC TTCCTGCAGGCCTTTCTCGGCCGCGGCCTGGGGCACCAGGACACGAGGGAGCAGCCTCCCGGCTACGCCATCGACACGGTGCAGGTCAAT GGACAGGAGAAGTACTTGATCCTCTGTGAGGTGGGCACAGATGGTCTGCTGGCCACATCGCTGGACGCCACCTGTGACGTTGCCTGCTTG ATGTTTGATGGCAGTGACCCAAAGTCCTTTGCACATTGTGCCAGCGTCTACAAGCACCATTACATGGACGGGCAGACCCCCTGCCTCTTT GTCTCCTCCAAGGCCGACCTGCCCGAAGGTGTCGCGGTGTCTGGCCCATCACCGGCCGAGTTTTGCCGCAAGCACCGGCTACCCGCTCCC GTGCCGTTCTCCTGTGCTGGCCCAGCCGAGCCCAGCACCACCATCTTCACCCAGCTCGCCACCATGGCCGCCTTCCCACATTTGGTCCAC GCAGAGCTGCATCCCTCTTCCTTCTGGCTCCGGGGGCTGCTGGGGGTTGTCGGGGCCGCCGTGGCCGCAGTCCTCAGCTTCTCACTCTAC AGGGTCCTGGTGAAGAGCCAGTGAGGCCCCTGGTACCCAAGCCCCCTCCCCTGACCTGGGTGTGCCTCGCTGCTGGGGCTCTGCAGGGGC AGCACAGCTGGGGTGCAGGCCAGGCTGCCACTCCGGGAACGCCTTTGCGCCGGGACTTTTTGTTTCTGAAGGCAGTCGATCTGCAGCGGG GCCTTATGCTGCCATGCACTGCCCTGGCTCCTGCCGGACCCCCAGGGTGGGCCGTGGCAGGTGGCTGAGCAGGAGCTCCCAAGTGCCGGC CACCGCTGTCAGGGATTGCCCACCCCTGGGCATCATGTGTGTGGGGCCGGGGAGCACAGGTGTGGGAGCTGGTGACCCCAGACCCAGAAT TCTCAGGGCTCTACCCCCCTTTCCTGGTCCTAGGTGGCCAGTGGGTATGAGGAGGGCTGGAAGGCAGAGCTTTGGGCCAAAAGCAGGCGT TGGGGGGTCCCCCCTCAAGTTTGGAGCCGTTTCCGTGGTTGTAGCAGAGGACCGGAGGTTGGGTTCCTGATTAAACTTCACTGTGTGTTT >16743_16743_4_CHTF18-RHOT2_CHTF18_chr16_841370_ENST00000491530_RHOT2_chr16_719553_ENST00000315082_length(amino acids)=609AA_BP=65 MLKWLKLWDLVVFGHERPSRKPRPSVEPARVSKEATAPGKWKSHEQVLEEMLEAGLDPSQRPKQKANVVCVVYDVSEEATIEKIRTKWIP LVNGGTTQGPRVPIILVGNKSDLRSGSSMEAVLPIMSQFPEIETCVECSAKNLRNISELFYYAQKAVLHPTAPLYDPEAKQLRPACAQAL TRIFRLSDQDLDQALSDEELNAFQKSCFGHPLAPQALEDVKTVVCRNVAGGVREDRLTLDGFLFLNTLFIQRGRHETTWTILRRFGYSDA LELTADYLSPLIHVPPGCSTELNHLGYQFVQRVFEKHDQDRDGALSPVELQSLFSVFPAAPWGPELPRTVRTEAGRLPLHGYLCQWTLVT YLDVRSCLGHLGYLGYPTLCEQDQAHAITVTREKRLDQEKGQTQRSVLLCKVVGARGVGKSAFLQAFLGRGLGHQDTREQPPGYAIDTVQ VNGQEKYLILCEVGTDGLLATSLDATCDVACLMFDGSDPKSFAHCASVYKHHYMDGQTPCLFVSSKADLPEGVAVSGPSPAEFCRKHRLP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CHTF18-RHOT2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CHTF18-RHOT2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CHTF18-RHOT2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
