
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CIRBP-PKNOX1 (FusionGDB2 ID:16835) |
Fusion Gene Summary for CIRBP-PKNOX1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CIRBP-PKNOX1 | Fusion gene ID: 16835 | Hgene | Tgene | Gene symbol | CIRBP | PKNOX1 | Gene ID | 1153 | 5316 |
| Gene name | cold inducible RNA binding protein | PBX/knotted 1 homeobox 1 | |
| Synonyms | CIRP | PREP1|pkonx1c | |
| Cytomap | 19p13.3 | 21q22.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cold-inducible RNA-binding proteinA18 hnRNPglycine-rich RNA binding proteintesticular tissue protein Li 39 | homeobox protein PKNOX1PBX/knotted homeobox 1Pbx regulating protein-1homeobox protein PREP-1human homeobox-containing protein | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q14011 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000320936, ENST00000444172, ENST00000585630, ENST00000586472, ENST00000586773, ENST00000587323, ENST00000587896, ENST00000588030, ENST00000588090, ENST00000588230, ENST00000589235, ENST00000589660, ENST00000589686, ENST00000589710, ENST00000591935, ENST00000413636, ENST00000590188, | ENST00000607150, ENST00000291547, ENST00000432907, | |
| Fusion gene scores | * DoF score | 12 X 7 X 6=504 | 13 X 10 X 6=780 |
| # samples | 13 | 15 | |
| ** MAII score | log2(13/504*10)=-1.95491211047146 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/780*10)=-2.37851162325373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CIRBP [Title/Abstract] AND PKNOX1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CIRBP(1271411)-PKNOX1(44450156), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CIRBP-PKNOX1 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. CIRBP-PKNOX1 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CIRBP | GO:0009411 | response to UV | 11574538 |
| Hgene | CIRBP | GO:0045727 | positive regulation of translation | 11574538 |
| Hgene | CIRBP | GO:0048255 | mRNA stabilization | 11574538 |
 Fusion gene breakpoints across CIRBP (5'-gene) Fusion gene breakpoints across CIRBP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
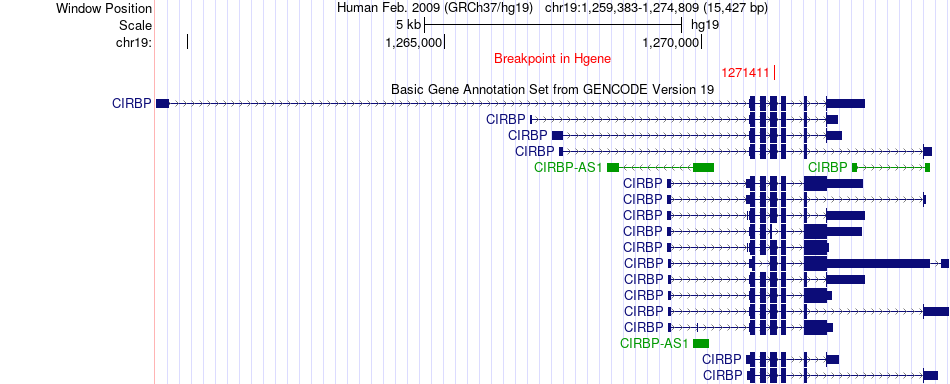 |
 Fusion gene breakpoints across PKNOX1 (3'-gene) Fusion gene breakpoints across PKNOX1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
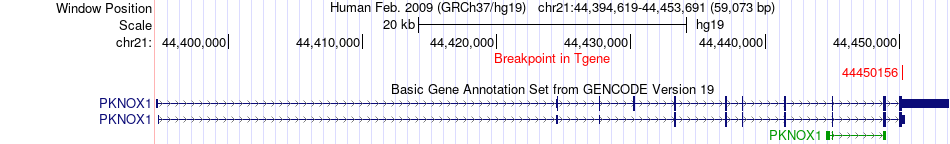 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | DA500684 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
Top |
Fusion Gene ORF analysis for CIRBP-PKNOX1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000320936 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000444172 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000585630 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000586472 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000586773 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000587323 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000587896 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000588030 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000588090 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000588230 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000589235 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000589660 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000589686 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000589710 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| 5CDS-intron | ENST00000591935 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000320936 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000444172 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000585630 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000586472 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000586773 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000587323 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000587896 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000588030 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000588090 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000588230 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000589235 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000589660 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000589686 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000589710 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| Frame-shift | ENST00000591935 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000320936 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000444172 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000585630 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000586472 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000586773 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000587323 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000587896 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000588030 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000588090 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000588230 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000589235 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000589660 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000589686 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000589710 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| In-frame | ENST00000591935 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| intron-3CDS | ENST00000413636 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| intron-3CDS | ENST00000413636 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| intron-3CDS | ENST00000590188 | ENST00000291547 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| intron-3CDS | ENST00000590188 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| intron-intron | ENST00000413636 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
| intron-intron | ENST00000590188 | ENST00000607150 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000588030 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 810 | 525 | 26 | 601 | 191 |
| ENST00000589660 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 605 | 320 | 55 | 396 | 113 |
| ENST00000588090 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 761 | 476 | 118 | 552 | 144 |
| ENST00000591935 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 620 | 335 | 46 | 411 | 121 |
| ENST00000589710 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 697 | 412 | 132 | 488 | 118 |
| ENST00000588230 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 635 | 350 | 64 | 426 | 120 |
| ENST00000586472 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 635 | 350 | 64 | 426 | 120 |
| ENST00000589686 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 696 | 411 | 131 | 487 | 118 |
| ENST00000444172 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 525 | 240 | 125 | 316 | 63 |
| ENST00000587323 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 630 | 345 | 59 | 421 | 120 |
| ENST00000320936 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 630 | 345 | 59 | 421 | 120 |
| ENST00000587896 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 621 | 336 | 11 | 412 | 133 |
| ENST00000589235 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 617 | 332 | 46 | 408 | 120 |
| ENST00000585630 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 618 | 333 | 53 | 409 | 118 |
| ENST00000586773 | CIRBP | chr19 | 1271411 | + | ENST00000432907 | PKNOX1 | chr21 | 44450156 | + | 599 | 314 | 34 | 390 | 118 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000588030 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.46627837 | 0.5337217 |
| ENST00000589660 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.3026048 | 0.6973952 |
| ENST00000588090 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.52368414 | 0.47631583 |
| ENST00000591935 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.5232948 | 0.4767052 |
| ENST00000589710 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.46131095 | 0.538689 |
| ENST00000588230 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.4540677 | 0.5459323 |
| ENST00000586472 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.4540677 | 0.5459323 |
| ENST00000589686 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.43979123 | 0.5602088 |
| ENST00000444172 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.882474 | 0.11752604 |
| ENST00000587323 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.45305884 | 0.54694116 |
| ENST00000320936 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.45305884 | 0.54694116 |
| ENST00000587896 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.4769931 | 0.5230069 |
| ENST00000589235 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.43894953 | 0.5610505 |
| ENST00000585630 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.3642256 | 0.6357744 |
| ENST00000586773 | ENST00000432907 | CIRBP | chr19 | 1271411 | + | PKNOX1 | chr21 | 44450156 | + | 0.36507106 | 0.63492894 |
Top |
Fusion Genomic Features for CIRBP-PKNOX1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
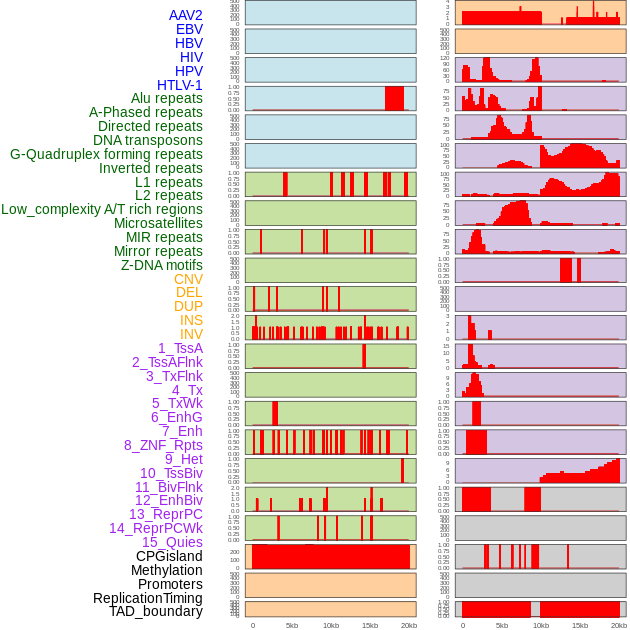 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CIRBP-PKNOX1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:1271411/chr21:44450156) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CIRBP | . |
| FUNCTION: Cold-inducible mRNA binding protein that plays a protective role in the genotoxic stress response by stabilizing transcripts of genes involved in cell survival. Acts as a translational activator. Seems to play an essential role in cold-induced suppression of cell proliferation. Binds specifically to the 3'-untranslated regions (3'-UTRs) of stress-responsive transcripts RPA2 and TXN. Acts as a translational repressor (By similarity). Promotes assembly of stress granules (SGs), when overexpressed. {ECO:0000250, ECO:0000269|PubMed:11574538, ECO:0000269|PubMed:16513844}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PKNOX1 | chr19:1271411 | chr21:44450156 | ENST00000291547 | 0 | 11 | 259_321 | 0 | 437.0 | DNA binding | Homeobox%3B TALE-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000320936 | + | 1 | 7 | 92_158 | 0 | 173.0 | Compositional bias | Note=Gly-rich |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000585630 | + | 1 | 6 | 92_158 | 0 | 173.0 | Compositional bias | Note=Gly-rich |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000586472 | + | 1 | 8 | 92_158 | 0 | 173.0 | Compositional bias | Note=Gly-rich |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000588030 | + | 1 | 7 | 92_158 | 0 | 173.0 | Compositional bias | Note=Gly-rich |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000588090 | + | 1 | 7 | 92_158 | 0 | 173.0 | Compositional bias | Note=Gly-rich |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000589660 | + | 1 | 7 | 92_158 | 0 | 173.0 | Compositional bias | Note=Gly-rich |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000320936 | + | 1 | 7 | 6_84 | 0 | 173.0 | Domain | RRM |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000585630 | + | 1 | 6 | 6_84 | 0 | 173.0 | Domain | RRM |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000586472 | + | 1 | 8 | 6_84 | 0 | 173.0 | Domain | RRM |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000588030 | + | 1 | 7 | 6_84 | 0 | 173.0 | Domain | RRM |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000588090 | + | 1 | 7 | 6_84 | 0 | 173.0 | Domain | RRM |
| Hgene | CIRBP | chr19:1271411 | chr21:44450156 | ENST00000589660 | + | 1 | 7 | 6_84 | 0 | 173.0 | Domain | RRM |
Top |
Fusion Gene Sequence for CIRBP-PKNOX1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16835_16835_1_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000320936_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=630nt_BP=345nt TCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGCCGCCATGGCATCAG ATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTG AAGTGGTGGTTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCA TGATGGCCATGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACAT CAGCGGGCTGGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTT TACACAGTTTTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCG GTGCAGCGACTTCTTTCAAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGTGCGTGTGTGTGGATTTTTAAAGAAATTCTTT >16835_16835_1_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000320936_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=120AA_BP=96 MPDSVAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRG -------------------------------------------------------------- >16835_16835_2_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000444172_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=525nt_BP=240nt ACTCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGCCGCCATGGCATC AGATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTC TGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTCTT GGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTATTT CTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTCTT >16835_16835_2_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000444172_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=63AA_BP=39 -------------------------------------------------------------- >16835_16835_3_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000585630_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=618nt_BP=333nt GGAAGCGGGGAGAGGTGGTGAGAGATGACCCCTGTTGAGGATTTCATTGGTGTCTGCCACAGGCCGCCATGGCATCAGATGAAGGCAAAC TTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTG TGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGA ATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGT CTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTA TTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTT >16835_16835_3_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000585630_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=118AA_BP=94 MPQAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRGRG -------------------------------------------------------------- >16835_16835_4_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000586472_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=635nt_BP=350nt CTCACTCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGCCGCCATGGC ATCAGATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGAT CTCTGAAGTGGTGGTTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGA TGCCATGATGGCCATGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCC CACATCAGCGGGCTGGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAA CATTTTACACAGTTTTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTT TGCCGGTGCAGCGACTTCTTTCAAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGTGCGTGTGTGTGGATTTTTAAAGAAAT >16835_16835_4_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000586472_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=120AA_BP=96 MPDSVAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRG -------------------------------------------------------------- >16835_16835_5_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000586773_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=599nt_BP=314nt GAGAGATGACCCCTGTTGAGGATTTCATTGGTGTCTGCCACAGGCCGCCATGGCATCAGATGAAGGCAAACTTTTTGTTGGAGGGCTGAG TTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTGTGAAAGACAGGGAGACCCA GAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGAATGGGAAGGGGGGCCGGGG CTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTCTTGGAGAACAGTGACTCC CTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTATTTCTAATATGTTTTATAT GTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTCTTTCAAGTGTGTGTGTGT >16835_16835_5_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000586773_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=118AA_BP=94 MPQAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRGRG -------------------------------------------------------------- >16835_16835_6_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000587323_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=630nt_BP=345nt TCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGCCGCCATGGCATCAG ATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTG AAGTGGTGGTTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCA TGATGGCCATGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACAT CAGCGGGCTGGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTT TACACAGTTTTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCG GTGCAGCGACTTCTTTCAAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGTGCGTGTGTGTGGATTTTTAAAGAAATTCTTT >16835_16835_6_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000587323_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=120AA_BP=96 MPDSVAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRG -------------------------------------------------------------- >16835_16835_7_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000587896_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=621nt_BP=336nt GGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGTGTGGCCGCCATGGCATCAGATGAAGGCA AACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGG TTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCA TGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCT GGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTT TTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGA >16835_16835_7_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000587896_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=133AA_BP=109 MWCAVFPLASGTCPTQCVAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGK -------------------------------------------------------------- >16835_16835_8_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000588030_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=810nt_BP=525nt AGTCCCTTAGGGTGTGGGGCGAGGGCCTGCCTGCCTGCTTCGTGGCCTGGGCAGGACCCTGGCCCCCTCAGGACCTGCCAGTATGGGGGA GACGCGACCACCAGCTCTCCCCAGCCACAGGAGTCGGGGACTCCCACTCTGCCACACCCACCTGCTCCCCTTCCTGGAGTCTCACATCTG TGTGGCTTGCTGCAGGGAGATCACCAGGCCGACTGGGAGGACCTAGGACTCCGCAGGGTACCTTGGGCTGGCAGGCCGCCATGGCATCAG ATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTG AAGTGGTGGTTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCA TGATGGCCATGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACAT CAGCGGGCTGGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTT TACACAGTTTTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCG GTGCAGCGACTTCTTTCAAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGTGCGTGTGTGTGGATTTTTAAAGAAATTCTTT >16835_16835_8_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000588030_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=191AA_BP=167 MPACFVAWAGPWPPQDLPVWGRRDHQLSPATGVGDSHSATPTCSPSWSLTSVWLAAGRSPGRLGGPRTPQGTLGWQAAMASDEGKLFVGG LSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRGRGFSRGPLPTSAGWSWRTV -------------------------------------------------------------- >16835_16835_9_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000588090_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=761nt_BP=476nt AAACGCCGCGGACGGAGCGAGGCCGCAGCGCCCCCTGGTGTCCGCAACGGGCAGCGCGTCCCGGTCGGGTCCCGCATGGTCAGTGGCCAG CAGGGGTGGATGAAGGGGTCCCAGCCGCCTGAGACCCCCGGTGGGGACACAGGATCGCGGTGAGATACCTGGGAGTCACGGGTGCTCGCA GACGCAGCTGACCCCAGAGGCACAGGCCGCCATGGCATCAGATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCA GTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGG GTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCG AGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAG CAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGC ACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTCTTTCAAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT >16835_16835_9_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000588090_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=144AA_BP=120 MRPPVGTQDRGEIPGSHGCSQTQLTPEAQAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDD -------------------------------------------------------------- >16835_16835_10_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000588230_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=635nt_BP=350nt CTCACTCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGCCGCCATGGC ATCAGATGAAGGCAAACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGAT CTCTGAAGTGGTGGTTGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGA TGCCATGATGGCCATGAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCC CACATCAGCGGGCTGGTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAA CATTTTACACAGTTTTATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTT TGCCGGTGCAGCGACTTCTTTCAAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGTGTGTGCGTGTGTGTGGATTTTTAAAGAAAT >16835_16835_10_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000588230_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=120AA_BP=96 MPDSVAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRG -------------------------------------------------------------- >16835_16835_11_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589235_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=617nt_BP=332nt GGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGCCGCCATGGCATCAGATGAAGGCAAACT TTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTGT GAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGAA TGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTC TTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTAT TTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTC >16835_16835_11_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589235_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=120AA_BP=96 MPDSVAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRG -------------------------------------------------------------- >16835_16835_12_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589660_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=605nt_BP=320nt CGACGCCCCGCAGGGGAGGAGGCCCGGGAGGAGTATCAGGGTGCAGGAGGCCGCCATGGCATCAGATGAAGGCAAACTTTTTGTTGGAGG GCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTGTGAAAGACAGGGA GACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGAATGGGAAGGGGGG CCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTCTTGGAGAACAGT GACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTATTTCTAATATGTT TTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTCTTTCAAGTGTGT >16835_16835_12_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589660_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=113AA_BP=89 MASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRGRGFSRGP -------------------------------------------------------------- >16835_16835_13_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589686_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=696nt_BP=411nt CTCACTCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGAAGCGGGGAG AGGTGGTGAGAGATGACCCCTGTTGAGGATTTCATTGGTGTCTGCCACAGGCCGCCATGGCATCAGATGAAGGCAAACTTTTTGTTGGAG GGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTGTGAAAGACAGGG AGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGAATGGGAAGGGGG GCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTCTTGGAGAACAG TGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTATTTCTAATATGT TTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTCTTTCAAGTGTG >16835_16835_13_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589686_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=118AA_BP=94 MPQAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRGRG -------------------------------------------------------------- >16835_16835_14_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589710_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=697nt_BP=412nt CCTCACTCGCGCGTTAGGAGGCTCGGGTCGTTGTGGTGCGCTGTCTTCCCGCTTGCGTCAGGGACCTGCCCGACTCAGTGGAAGCGGGGA GAGGTGGTGAGAGATGACCCCTGTTGAGGATTTCATTGGTGTCTGCCACAGGCCGCCATGGCATCAGATGAAGGCAAACTTTTTGTTGGA GGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGTTGTGAAAGACAGG GAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCATGAATGGGAAGGGG GGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTGGTCTTGGAGAACA GTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTTTATTTCTAATATG TTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGACTTCTTTCAAGTGT >16835_16835_14_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000589710_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=118AA_BP=94 MPQAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGRGRG -------------------------------------------------------------- >16835_16835_15_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000591935_PKNOX1_chr21_44450156_ENST00000432907_length(transcript)=620nt_BP=335nt GGATCGCGGTGAGATACCTGGGAGTCACGGGTGCTCGCAGACGCAGCTGACCCCAGAGGCACAGGCCGCCATGGCATCAGATGAAGGCAA ACTTTTTGTTGGAGGGCTGAGTTTTGACACCAATGAGCAGTCGCTGGAGCAGGTCTTCTCAAAGTACGGACAGATCTCTGAAGTGGTGGT TGTGAAAGACAGGGAGACCCAGAGATCTCGGGGATTTGGGTTTGTCACCTTTGAGAACATTGACGACGCTAAGGATGCCATGATGGCCAT GAATGGGAAGGGGGGCCGGGGCTTCTTCCGTGGGGGCCGAGGACGGGGCCGTGGGTTCTCTAGAGGGCCCCTGCCCACATCAGCGGGCTG GTCTTGGAGAACAGTGACTCCCTGCAGTAGGGGCAGGAGCAGACGCACCTGACTTTTTGGAGTTTGCACAGCAAACATTTTACACAGTTT TATTTCTAATATGTTTTATATGTAGATATAGAAGAGTGCACTTTTGTATTTCATAGTAAGCTTAAAGCGCGTCTTTGCCGGTGCAGCGAC >16835_16835_15_CIRBP-PKNOX1_CIRBP_chr19_1271411_ENST00000591935_PKNOX1_chr21_44450156_ENST00000432907_length(amino acids)=121AA_BP=97 MTPEAQAAMASDEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAMNGKGGRGFFRGGRGR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CIRBP-PKNOX1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CIRBP-PKNOX1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CIRBP-PKNOX1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
