
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CLASP2-BRAT1 (FusionGDB2 ID:16941) |
Fusion Gene Summary for CLASP2-BRAT1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CLASP2-BRAT1 | Fusion gene ID: 16941 | Hgene | Tgene | Gene symbol | CLASP2 | BRAT1 | Gene ID | 23122 | 221927 |
| Gene name | cytoplasmic linker associated protein 2 | BRCA1 associated ATM activator 1 | |
| Synonyms | - | BAAT1|C7orf27|NEDCAS|RMFSL | |
| Cytomap | 3p22.3 | 7p22.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CLIP-associating protein 2CLIP-associating protein CLASP2multiple asters (Mast)-like homolog 2protein Orbit homolog 2 | BRCA1-associated ATM activator 1BRCA1-associated protein required for ATM activation protein 1HEAT repeat-containing protein C7orf27 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O75122 | Q6PJG6 | |
| Ensembl transtripts involved in fusion gene | ENST00000539981, ENST00000307312, ENST00000359576, ENST00000399362, ENST00000461133, ENST00000468888, ENST00000480013, ENST00000313350, ENST00000333778, ENST00000482896, ENST00000487200, | ENST00000340611, ENST00000473879, | |
| Fusion gene scores | * DoF score | 9 X 9 X 6=486 | 6 X 3 X 5=90 |
| # samples | 9 | 6 | |
| ** MAII score | log2(9/486*10)=-2.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/90*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CLASP2 [Title/Abstract] AND BRAT1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CLASP2(33557515)-BRAT1(2583596), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CLASP2-BRAT1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CLASP2-BRAT1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CLASP2 (5'-gene) Fusion gene breakpoints across CLASP2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
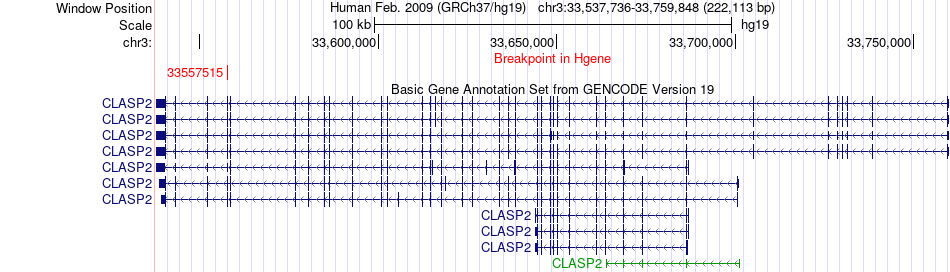 |
 Fusion gene breakpoints across BRAT1 (3'-gene) Fusion gene breakpoints across BRAT1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
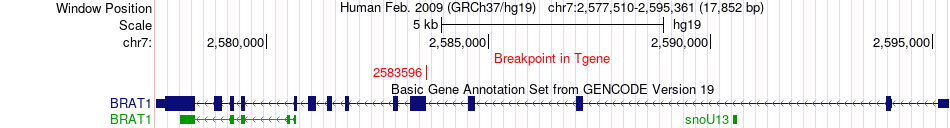 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-28-1753-01A | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
Top |
Fusion Gene ORF analysis for CLASP2-BRAT1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000539981 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 3UTR-intron | ENST00000539981 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 5CDS-intron | ENST00000307312 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 5CDS-intron | ENST00000359576 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 5CDS-intron | ENST00000399362 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 5CDS-intron | ENST00000461133 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 5CDS-intron | ENST00000468888 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| 5CDS-intron | ENST00000480013 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| Frame-shift | ENST00000359576 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| Frame-shift | ENST00000399362 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| Frame-shift | ENST00000461133 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| Frame-shift | ENST00000468888 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| Frame-shift | ENST00000480013 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| In-frame | ENST00000307312 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-3CDS | ENST00000313350 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-3CDS | ENST00000333778 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-3CDS | ENST00000482896 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-3CDS | ENST00000487200 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-intron | ENST00000313350 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-intron | ENST00000333778 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-intron | ENST00000482896 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
| intron-intron | ENST00000487200 | ENST00000473879 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000307312 | CLASP2 | chr3 | 33557515 | - | ENST00000340611 | BRAT1 | chr7 | 2583596 | - | 6692 | 4464 | 291 | 4688 | 1465 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000307312 | ENST00000340611 | CLASP2 | chr3 | 33557515 | - | BRAT1 | chr7 | 2583596 | - | 0.002757772 | 0.9972422 |
Top |
Fusion Genomic Features for CLASP2-BRAT1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CLASP2-BRAT1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:33557515/chr7:2583596) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CLASP2 | BRAT1 |
| FUNCTION: Microtubule plus-end tracking protein that promotes the stabilization of dynamic microtubules (PubMed:26003921). Involved in the nucleation of noncentrosomal microtubules originating from the trans-Golgi network (TGN). Required for the polarization of the cytoplasmic microtubule arrays in migrating cells towards the leading edge of the cell. May act at the cell cortex to enhance the frequency of rescue of depolymerizing microtubules by attaching their plus-ends to cortical platforms composed of ERC1 and PHLDB2 (PubMed:16824950). This cortical microtubule stabilizing activity is regulated at least in part by phosphatidylinositol 3-kinase signaling. Also performs a similar stabilizing function at the kinetochore which is essential for the bipolar alignment of chromosomes on the mitotic spindle (PubMed:16866869, PubMed:16914514). Acts as a mediator of ERBB2-dependent stabilization of microtubules at the cell cortex. {ECO:0000269|PubMed:11290329, ECO:0000269|PubMed:15631994, ECO:0000269|PubMed:16824950, ECO:0000269|PubMed:16866869, ECO:0000269|PubMed:16914514, ECO:0000269|PubMed:17543864, ECO:0000269|PubMed:20937854, ECO:0000269|PubMed:26003921}. | FUNCTION: Involved in DNA damage response; activates kinases ATM, SMC1A and PRKDC by modulating their phosphorylation status following ionizing radiation (IR) stress (PubMed:16452482, PubMed:22977523). Plays a role in regulating mitochondrial function and cell proliferation (PubMed:25070371). Required for protein stability of MTOR and MTOR-related proteins, and cell cycle progress by growth factors (PubMed:25657994). {ECO:0000269|PubMed:16452482, ECO:0000269|PubMed:22977523, ECO:0000269|PubMed:25070371, ECO:0000269|PubMed:25657994}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | BRAT1 | chr3:33557515 | chr7:2583596 | ENST00000340611 | 3 | 14 | 495_531 | 143 | 822.0 | Repeat | Note=HEAT 1 | |
| Tgene | BRAT1 | chr3:33557515 | chr7:2583596 | ENST00000340611 | 3 | 14 | 544_576 | 143 | 822.0 | Repeat | Note=HEAT 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 1104_1108 | 0 | 432.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 310_587 | 0 | 432.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 1017_1294 | 0 | 432.0 | Region | Note=Required for cortical localization |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 60_311 | 0 | 432.0 | Region | TOG 1 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 649_881 | 0 | 432.0 | Region | TOG 2 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 1054_1091 | 0 | 432.0 | Repeat | HEAT 6 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 1098_1135 | 0 | 432.0 | Repeat | HEAT 7 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 1216_1253 | 0 | 432.0 | Repeat | HEAT 8 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 173_208 | 0 | 432.0 | Repeat | HEAT 1 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 209_245 | 0 | 432.0 | Repeat | HEAT 2 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 250_287 | 0 | 432.0 | Repeat | HEAT 3 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 710_747 | 0 | 432.0 | Repeat | HEAT 4 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 772_809 | 0 | 432.0 | Repeat | HEAT 5 |
Top |
Fusion Gene Sequence for CLASP2-BRAT1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16941_16941_1_CLASP2-BRAT1_CLASP2_chr3_33557515_ENST00000307312_BRAT1_chr7_2583596_ENST00000340611_length(transcript)=6692nt_BP=4464nt AGAGGTCCTATCGCTCCCAGCGGTTTCCGCAGCCACCTCCACCACCTCCGCAGCAAAACGCTAGCCGGACTGGAGGGCCCTCGCCGGCGT CGTGCTGACGTCACGCGCGTGCTGACGTCGCCCGCGGCCGCGGCCTCTGAAGCGGGCTGGGGATCGGGGGGCGCCGAGTTTGACTAGTTT GGGGGCGGCTGGGCGCTTGGCGTTCCTCCCGCCGCCCGCTGCGCCCCGCAAGCCGCGCCCCTGGCGGGCTAAGTGAGTCCCGCCCGCTCC CGCGGGGACCCGCACTGGAGGCTGGGCGGCTCTCGGCGAAAGTTGGCCGCTCACAGACTGGCAGGCGGGCGGGCGGCCGCAGCCATGGAG CCCCGCAGCATGGAGTACTTCTGCGCCCAGGTGCAGCAGAAGGACGTCGGCGGCCGGCTGCAGGTCGGCCAGGAGCTCCTGCTCTACCTT GGCGCCCCCGGCGCCATCTCGGACCTGGAGGAGGACCTGGGCCGCCTAGGCAAGACAGTCGACGCGCTCACCGGCTGGGTGGGTTCGAGC AACTACCGGGTATCATTAATGGGATTGGAAATTTTAAGTGCCTTTGTGGACAGATTATCAACACGCTTTAAATCCTATGTAGCAATGGTT ATTGTAGCTTTAATAGACAGAATGGGAGATGCCAAAGACAAGGTTCGAGATGAAGCTCAGACTCTGATATTGAAGTTAATGGATCAAGTA GCACCACCTATGTACATTTGGGAGCAGTTGGCTTCTGGTTTTAAACACAAGAATTTTCGATCTCGAGAAGGCGTGTGTCTGTGTCTTATT GAAACCTTAAACATTTTTGGGGCTCAGCCACTAGTCATCAGCAAATTGATACCACATTTGTGTATCCTGTTTGGAGACTCCAACAGTCAG GTGAGAGATGCTGCAATATTGGCTATAGTGGAGATTTATAGACATGTGGGAGAAAAAGTGAGGATGGATCTTTATAAGAGAGGAATTCCC CCTGCTAGATTAGAAATGATATTTGCCAAATTTGATGAAGTGCAAAGTTCAGGCGGTATGATTTTGAGTGTCTGCAAAGATAAAAGCTTC GATGATGAAGAATCAGTGGATGGAAATAGGCCATCATCAGCTGCATCAGCCTTCAAGGTTCCTGCACCTAAAACATCCGGAAATCCTGCC AACAGTGCAAGGAAGCCTGGTTCAGCAGGTGGCCCTAAGGTTGGAGGTGCTTCTAAGGAAGGAGGTGCTGGAGCAGTTGATGAAGATGAT TTTATAAAAGCTTTTACAGATGTCCCTTCTATTCAGATTTATTCTAGTCGAGAACTCGAAGAAACATTAAATAAAATCAGGGAAATTTTG TCAGATGATAAACATGACTGGGATCAGCGTGCCAATGCACTGAAGAAAATTCGATCACTGCTTGTTGCTGGAGCTGCACAGTATGATTGC TTTTTTCAACATTTACGATTGTTGGATGGAGCACTTAAACTTTCAGCTAAGGATCTTAGATCCCAGGTGGTTAGAGAAGCTTGTATTACT GTAGCCCACCTTTCAACAGTTTTGGGAAACAAGTTTGATCATGGCGCTGAAGCCATTGTACCTACACTTTTTAATCTCGTCCCCAATAGT GCAAAAGTCATGGCAACTTCTGGATGTGCAGCAATCAGATTTATCATTCGGCATACTCATGTACCCAGACTTATACCTTTAATAACAAGC AATTGCACATCAAAATCAGTTCCCGTGAGGAGACGTTCATTTGAATTTTTAGATTTATTGTTGCAAGAGTGGCAGACTCATTCATTGGAA AGACATGCAGCCGTCTTGGTTGAAACTATTAAAAAGGGAATTCATGATGCTGACGCTGAGGCCAGAGTGGAGGCAAGAAAGACATACATG GGTCTTAGAAACCACTTTCCTGGTGAAGCTGAAACATTATATAATTCCCTTGAGCCATCTTATCAGAAGAGTCTTCAAACTTACTTAAAG AGTTCTGGCAGTGTAGCATCTCTTCCACAATCAGACAGGTCCTCATCCAGCTCACAGGAAAGTCTCAATCGCCCTTTTTCTTCCAAATGG TCTACAGCAAATCCATCAACTGTGGCTGGAAGAGTATCAGCAGGCAGCAGCAAAGCCAGTTCCCTTCCAGGAAGCCTGCAGCGTTCACGA AGTGACATTGATGTGAATGCTGCTGCAGGTGCCAAGGCACATCATGCTGCTGGACAGTCTGTGCGAAGCGGGCGCTTAGGTGCAGGTGCC CTGAATGCAGGTTCCTATGCGTCACTAGAGGATACTTCTGACAAGCTGGATGGAACAGCATCTGAAGATGGCCGGGTGAGAGCAAAACTT TCAGCACCACTTGCTGGCATGGGAAATGCCAAGGCAGATTCTAGAGGAAGAAGTCGAACAAAAATGGTGTCTCAATCACAGCCTGGTAGC CGGTCTGGGTCTCCAGGAAGAGTTCTGACCACAACAGCCCTGTCCACTGTGAGCTCTGGTGTTCAAAGAGTCCTGGTCAATTCAGCCTCA GCACAAAAAAGAAGCAAGATACCACGGAGCCAGGGCTGTAGCAGAGAGGCTAGTCCATCTAGGCTTTCAGTGGCCCGAAGCAGTCGTATT CCTCGACCAAGTGTGAGTCAAGGATGCAGCCGGGAAGCTAGTCGGGAGAGCAGCAGAGACACAAGTCCTGTTCGCTCTTTTCAGCCCCTC GGTCCAGGTTATGGGATCAGCCAATCAAGTCGACTGTCGTCTTCTGTTAGTGCCATGCGAGTCCTGAACACAGGTTCTGATGTGGAGGAG GCGGTGGCAGATGCCTTGAAAAAACCAGCTCGAAGAAGATATGAATCATATGGAATGCATTCAGATGATGACGCCAACAGCGATGCATCT AGTGCTTGTTCAGAACGCTCCTATAGTTCTCGAAATGGTAGTATTCCTACATATATGAGGCAGACGGAAGATGTGGCAGAAGTCCTCAAT AGATGTGCTAGTTCCAATTGGTCAGAAAGGAAAGAAGGCCTCCTAGGTCTGCAGAACTTATTAAAAAATCAGAGAACACTAAGTCGAGTT GAACTGAAAAGATTATGTGAAATTTTCACAAGAATGTTTGCTGACCCTCATGGCAAGAGGGTATTCAGCATGTTTTTGGAGACTCTAGTG GATTTCATACAAGTCCACAAAGATGATCTTCAAGATTGGTTGTTTGTACTGCTGACACAACTACTAAAAAAAATGGGTGCTGATTTGCTT GGATCTGTTCAGGCAAAAGTTCAGAAAGCCCTTGATGTTACAAGAGAGTCTTTTCCAAATGATCTTCAGTTCAATATTCTAATGAGATTT ACAGTTGATCAGACCCAGACACCAAGCTTAAAGGTGAAGGTTGCTATCCTTAAATACATAGAAACTCTGGCCAAACAGATGGATCCAGGA GATTTTATAAATTCCAGTGAAACTCGCCTAGCAGTGTCTCGGGTCATCACTTGGACAACAGAACCCAAAAGTTCTGATGTTCGGAAGGCA GCACAGTCAGTGCTGATTTCATTATTTGAACTCAATACCCCAGAGTTTACAATGTTATTAGGAGCTTTACCAAAAACTTTTCAGGATGGT GCTACCAAGCTTCTTCATAATCACCTTCGAAACACTGGCAATGGAACCCAGAGTTCCATGGGGAGTCCTTTGACAAGACCAACACCACGA TCACCAGCTAACTGGTCCAGTCCTCTTACTTCTCCTACCAATACATCACAGAATACTTTATCTCCAAGTGCATTTGATTATGACACAGAA AATATGAACTCTGAAGATATTTATAGCTCTCTTAGAGGTGTCACTGAAGCAATCCAGAATTTCAGCTTCCGTAGCCAAGAAGATATGAAT GAGCCATTGAAAAGGGATTCTAAAAAAGATGATGGCGATTCAATGTGTGGTGGTCCTGGGATGTCTGACCCAAGAGCAGGAGGTGATGCT ACTGACTCAAGTCAAACAGCTCTTGATAATAAAGCTTCATTGCTCCATTCAATGCCTACTCACTCCTCTCCACGCTCTCGAGACTATAAT CCATATAACTATTCAGATAGCATCAGTCCCTTCAACAAGTCTGCCCTCAAGGAAGCCATGTTTGATGATGATGCTGACCAGTTTCCTGAC GATCTTTCCCTAGATCATTCTGACCTAGTTGCAGAGTTGTTGAAGGAGCTGTCTAACCATAATGAGCGTGTAGAAGAAAGAAAAATTGCC CTCTATGAACTTATGAAACTGACACAGGAAGAATCTTTTAGTGTTTGGGATGAACACTTCAAAACAATATTGCTTTTATTGCTTGAAACG CTTGGAGATAAAGAGCCTACAATCAGGGCTTTGGCATTAAAGGTTTTAAGAGAAATCCTAAGGCATCAACCAGCAAGATTTAAAAACTAT GCAGAATTGACTGTCATGAAAACATTGGAAGCACATAAAGATCCTCATAAGGAGGTGCGGTCGACACCATCTTCTCCCTGCAGGGAGACT CCAGCCTGTTTGTGGCCTCGGCGGCCAGTCAGCTCCTGGTGCACGTCCTGGCTTTGTCCATGCGAGGTGGAGCCGAGGGGCAGCCCTGCC TGCCGGGGGGTGACTGGCCCGCGTGTGCCCAGAAGATCATGGATCACGTTGAAGAGTCCTTGTGCTCCGCGGCCACCCCCAAGGTCACTC AGGCCCTGAACGTCCTGACCACGACCTTCGGGCGCTGCCAGAGCCCCTGGACGGAAGCCCTGTGGGTGCGGCTGAGTCCCCGCGTGGCCT GTCTGCTGGAGAGAGACCCCATCCCCGCCGCACACTCGTTCGTGGACCTGCTTCTCTGTGTGGCTCGTTCTCCCGTGTTCAGTTCTTCCG ACGGCAGCCTGTGGGAGACAGTGGCGCGGGCTCTGAGCTGCCTGGGTCCCACCCACATGGGACCCCTGGCTTTGGGGATCCTGAAGCTCG AGCACTGTCCACAGGCACTGAGGACCCAGGCCTTCCAGGTCCTTCTCCAGCCCCTGGCCTGTGTCCTGAAGGCCACGGTTCAGGCCCCCG GACCCCCAGGCTTGCTGGACGGGACGGCAGACGATGCCACGACGGTGGACACACTCCTGGCCTCCAAGTCGTCCTGCGCCGGCCTCCTGT GCCGCACCCTGGCTCACCTGGAGGAGCTGCAGCCGCTGCCCCAGCGCCCTTCACCGTGGCCCCAGGCGTCTCTACTGGGGGCTACAGTGA CTGTCCTGCGGCTCTGTGACGGCTCGGCTGCCCCTGCCTCCAGTGTGGGGGGCCACCTCTGTGGGACCCTGGCGGGCTGCGTCCGGGTCC AGCGAGCAGCCCTCGACTTCCTGGGGACGCTGTCACAGGGGACAGGCCCCCAGGAGCTGGTGACGCAGGCGCTTGCTGTCCTCCTGGAGT GCCTCGAGAGCCCCGGCTCCAGCCCCACGGTTCTGAAGAAGGCCTTCCAGGCCACGCTCAGGTGGCTCCTGAGCTCACCCAAGACCCCCG GCTGCTCTGATCTCGGCCCCCTCATCCCGCAGTTCCTCAGAGAGCTGTTCCCTGTGCTGCAGAAACGCCTGTGCCACCCCTGCTGGGAGG TGAGGGACTCCGCCCTCGAGTTCCTGACCCAGCTGAGCAGGCACTGGGGAGGACAGGCTGACTTCAGATGCGCACTCTTGGCTTCAGAGG TGCCTCAGCTGGCCCTGCAGCTCCTCCAGGACCCTGAGAGTTATGTCCGAGCGAGTGCAGTGACCGCCATGGGGCAGCTGTCCAGCCAGG GCCTGCACGCCCCCACCAGCCCTGAGCATGCAGAGGCCCGGCAGAGCCTGTTCCTGGAGCTCCTGCACATCCTCTCCGTAGACTCGGAGG GCTTCCCACGGCGGGCGGTCATGCAAGTCTTCACTGAGTGGCTGCGGGACGGCCACGCCGACGCGGCCCAGGACACGGAGCAGTTCGTGG CCACTGTGCTGCAGGCGGCGAGCCGAGACCTGGACTGGGAGGTCCGCGCCCAGGGCCTGGAGCTGGCCCTCGTGTTCCTGGGCCAGACTT TGGGGCCGCCGCGTACCCACTGCCCCTATGCCGTGGCCCTACCCGAGGTGGCCCCAGCCCAGCCACTCACCGAGGCACTGAGGGCTCTCT GCCACGTGGGGCTCTTTGACTTCGCCTTTTGTGCCTTGTTTGACTGCGACCGCCCTGTGGCGCAGAAGTCTTGTGACCTCCTTCTCTTCC TGAGGGACAAGATTGCTTCCTACAGCAGCCTGCGGGAGGCCAGGGGCAGCCCCAACACTGCCTCCGCAGAGGCCACCCTGCCGAGGTGGC GGGCGGGTGAGCAGGCCCAGCCCCCAGGGGACCAGGAGCCTGAGGCTGTGCTGGCCATGCTCAGGTCCCTAGACCTGGAGGGCCTGCGGA GCACGCTGGCCGAGAGCAGCGACCACGTGGAAAAGAGTCCCCAGTCCCTCCTGCAGGACATGCTGGCCACGGGAGGCTTCCTGCAGGGGG ACGAGGCCGACTGCTACTGAGCAGAACCAGAGTCTGCCACTGGGGCTCAGGACCAAGGGAGGCAGCACCATGTCCTTCTGTGGGACACTG CCAGCCCCAGGGCTCCAGCCCAGCCCGGTGGATCCTCTGGGGAAGCCAGGACCAGGAGAGAAGCAAGGTCAAGAAATCCCACAGTTTGAT >16941_16941_1_CLASP2-BRAT1_CLASP2_chr3_33557515_ENST00000307312_BRAT1_chr7_2583596_ENST00000340611_length(amino acids)=1465AA_BP=1391 MGGSRRKLAAHRLAGGRAAAAMEPRSMEYFCAQVQQKDVGGRLQVGQELLLYLGAPGAISDLEEDLGRLGKTVDALTGWVGSSNYRVSLM GLEILSAFVDRLSTRFKSYVAMVIVALIDRMGDAKDKVRDEAQTLILKLMDQVAPPMYIWEQLASGFKHKNFRSREGVCLCLIETLNIFG AQPLVISKLIPHLCILFGDSNSQVRDAAILAIVEIYRHVGEKVRMDLYKRGIPPARLEMIFAKFDEVQSSGGMILSVCKDKSFDDEESVD GNRPSSAASAFKVPAPKTSGNPANSARKPGSAGGPKVGGASKEGGAGAVDEDDFIKAFTDVPSIQIYSSRELEETLNKIREILSDDKHDW DQRANALKKIRSLLVAGAAQYDCFFQHLRLLDGALKLSAKDLRSQVVREACITVAHLSTVLGNKFDHGAEAIVPTLFNLVPNSAKVMATS GCAAIRFIIRHTHVPRLIPLITSNCTSKSVPVRRRSFEFLDLLLQEWQTHSLERHAAVLVETIKKGIHDADAEARVEARKTYMGLRNHFP GEAETLYNSLEPSYQKSLQTYLKSSGSVASLPQSDRSSSSSQESLNRPFSSKWSTANPSTVAGRVSAGSSKASSLPGSLQRSRSDIDVNA AAGAKAHHAAGQSVRSGRLGAGALNAGSYASLEDTSDKLDGTASEDGRVRAKLSAPLAGMGNAKADSRGRSRTKMVSQSQPGSRSGSPGR VLTTTALSTVSSGVQRVLVNSASAQKRSKIPRSQGCSREASPSRLSVARSSRIPRPSVSQGCSREASRESSRDTSPVRSFQPLGPGYGIS QSSRLSSSVSAMRVLNTGSDVEEAVADALKKPARRRYESYGMHSDDDANSDASSACSERSYSSRNGSIPTYMRQTEDVAEVLNRCASSNW SERKEGLLGLQNLLKNQRTLSRVELKRLCEIFTRMFADPHGKRVFSMFLETLVDFIQVHKDDLQDWLFVLLTQLLKKMGADLLGSVQAKV QKALDVTRESFPNDLQFNILMRFTVDQTQTPSLKVKVAILKYIETLAKQMDPGDFINSSETRLAVSRVITWTTEPKSSDVRKAAQSVLIS LFELNTPEFTMLLGALPKTFQDGATKLLHNHLRNTGNGTQSSMGSPLTRPTPRSPANWSSPLTSPTNTSQNTLSPSAFDYDTENMNSEDI YSSLRGVTEAIQNFSFRSQEDMNEPLKRDSKKDDGDSMCGGPGMSDPRAGGDATDSSQTALDNKASLLHSMPTHSSPRSRDYNPYNYSDS ISPFNKSALKEAMFDDDADQFPDDLSLDHSDLVAELLKELSNHNERVEERKIALYELMKLTQEESFSVWDEHFKTILLLLLETLGDKEPT IRALALKVLREILRHQPARFKNYAELTVMKTLEAHKDPHKEVRSTPSSPCRETPACLWPRRPVSSWCTSWLCPCEVEPRGSPACRGVTGP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CLASP2-BRAT1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 444_580 | 0 | 432.0 | microtubules%2C MAPRE1 and MAPRE3 |
| Hgene | CLASP2 | chr3:33557515 | chr7:2583596 | ENST00000313350 | - | 1 | 13 | 872_1294 | 0 | 432.0 | RSN and localization to the Golgi and kinetochores |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CLASP2-BRAT1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CLASP2-BRAT1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
