
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CLIP2-NCF1 (FusionGDB2 ID:17179) |
Fusion Gene Summary for CLIP2-NCF1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CLIP2-NCF1 | Fusion gene ID: 17179 | Hgene | Tgene | Gene symbol | CLIP2 | NCF1 | Gene ID | 7461 | 653361 |
| Gene name | CAP-Gly domain containing linker protein 2 | neutrophil cytosolic factor 1 | |
| Synonyms | CLIP|CLIP-115|CYLN2|WBSCR3|WBSCR4|WSCR3|WSCR4 | NCF1A|NOXO2|SH3PXD1A|p47phox | |
| Cytomap | 7q11.23 | 7q11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CAP-Gly domain-containing linker protein 2Williams-Beuren syndrome chromosome region 3Williams-Beuren syndrome chromosome region 4cytoplasmic linker 2cytoplasmic linker protein 115cytoplasmic linker protein 2testicular tissue protein Li 40williams- | neutrophil cytosol factor 147 kDa autosomal chronic granulomatous disease protein47 kDa neutrophil oxidase factorNADPH oxidase organizer 2NCF-1NCF-47KSH3 and PX domain-containing protein 1Aneutrophil NADPH oxidase factor 1neutrophil cytosolic fact | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9UDT6 | P14598 | |
| Ensembl transtripts involved in fusion gene | ENST00000223398, ENST00000361545, ENST00000395060, | ENST00000443956, ENST00000289473, | |
| Fusion gene scores | * DoF score | 9 X 8 X 5=360 | 2 X 2 X 2=8 |
| # samples | 11 | 2 | |
| ** MAII score | log2(11/360*10)=-1.71049338280502 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: CLIP2 [Title/Abstract] AND NCF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CLIP2(73778645)-NCF1(74193427), # samples:1 CLIP2(73787366)-NCF1(74193427), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NCF1 | GO:0006612 | protein targeting to membrane | 12356722 |
| Tgene | NCF1 | GO:0034614 | cellular response to reactive oxygen species | 26514923 |
| Tgene | NCF1 | GO:0071276 | cellular response to cadmium ion | 26514923 |
 Fusion gene breakpoints across CLIP2 (5'-gene) Fusion gene breakpoints across CLIP2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
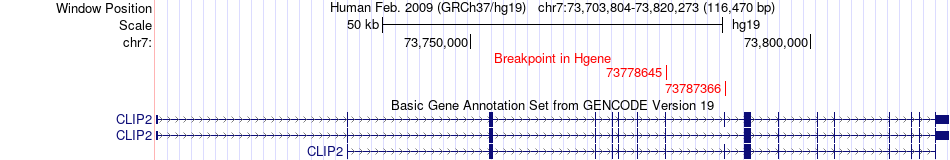 |
 Fusion gene breakpoints across NCF1 (3'-gene) Fusion gene breakpoints across NCF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
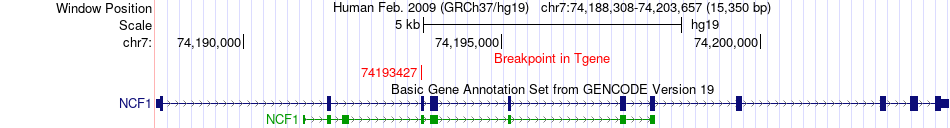 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-D1-A3DG | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| ChimerDB4 | UCEC | TCGA-D1-A3DG | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
Top |
Fusion Gene ORF analysis for CLIP2-NCF1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000223398 | ENST00000443956 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| 5CDS-3UTR | ENST00000223398 | ENST00000443956 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
| 5CDS-3UTR | ENST00000361545 | ENST00000443956 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| 5CDS-3UTR | ENST00000395060 | ENST00000443956 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| 5CDS-3UTR | ENST00000395060 | ENST00000443956 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
| In-frame | ENST00000223398 | ENST00000289473 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| In-frame | ENST00000223398 | ENST00000289473 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
| In-frame | ENST00000361545 | ENST00000289473 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| In-frame | ENST00000395060 | ENST00000289473 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + |
| In-frame | ENST00000395060 | ENST00000289473 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
| intron-3CDS | ENST00000361545 | ENST00000289473 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
| intron-3UTR | ENST00000361545 | ENST00000443956 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000361545 | CLIP2 | chr7 | 73778645 | + | ENST00000289473 | NCF1 | chr7 | 74193427 | + | 2880 | 1707 | 309 | 2726 | 805 |
| ENST00000223398 | CLIP2 | chr7 | 73778645 | + | ENST00000289473 | NCF1 | chr7 | 74193427 | + | 2880 | 1707 | 309 | 2726 | 805 |
| ENST00000395060 | CLIP2 | chr7 | 73778645 | + | ENST00000289473 | NCF1 | chr7 | 74193427 | + | 2553 | 1380 | 0 | 2399 | 799 |
| ENST00000223398 | CLIP2 | chr7 | 73787366 | + | ENST00000289473 | NCF1 | chr7 | 74193427 | + | 2985 | 1812 | 309 | 2831 | 840 |
| ENST00000395060 | CLIP2 | chr7 | 73787366 | + | ENST00000289473 | NCF1 | chr7 | 74193427 | + | 2658 | 1485 | 0 | 2504 | 834 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000361545 | ENST00000289473 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + | 0.016851611 | 0.98314834 |
| ENST00000223398 | ENST00000289473 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + | 0.016851611 | 0.98314834 |
| ENST00000395060 | ENST00000289473 | CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + | 0.017183028 | 0.98281693 |
| ENST00000223398 | ENST00000289473 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + | 0.016697062 | 0.983303 |
| ENST00000395060 | ENST00000289473 | CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + | 0.017098986 | 0.98290104 |
Top |
Fusion Genomic Features for CLIP2-NCF1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + | 6.26E-17 | 1 |
| CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + | 7.03E-11 | 1 |
| CLIP2 | chr7 | 73778645 | + | NCF1 | chr7 | 74193427 | + | 6.26E-17 | 1 |
| CLIP2 | chr7 | 73787366 | + | NCF1 | chr7 | 74193427 | + | 7.03E-11 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
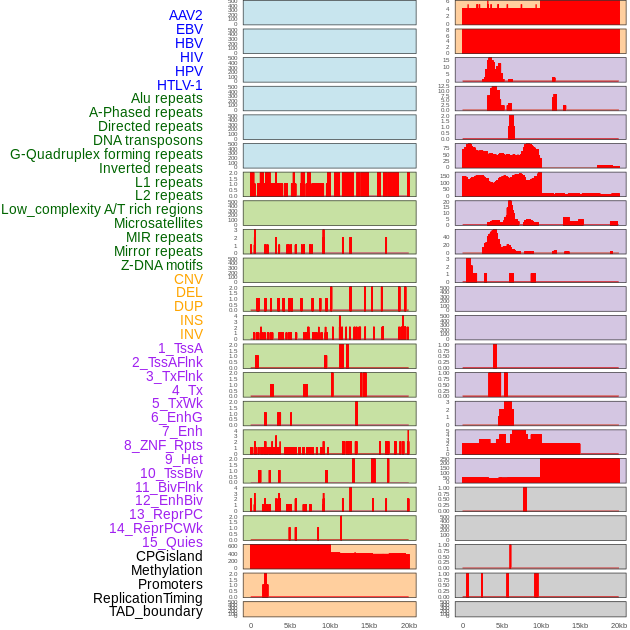 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
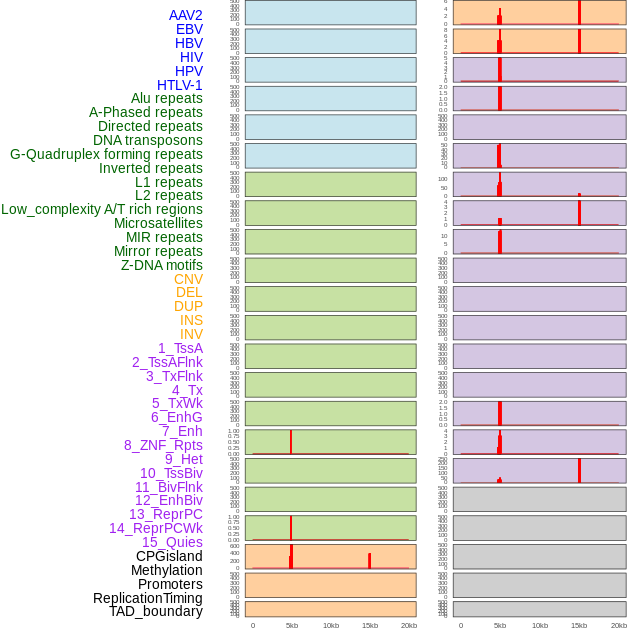 |
Top |
Fusion Protein Features for CLIP2-NCF1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:73778645/chr7:74193427) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CLIP2 | NCF1 |
| FUNCTION: Seems to link microtubules to dendritic lamellar body (DLB), a membranous organelle predominantly present in bulbous dendritic appendages of neurons linked by dendrodendritic gap junctions. May operate in the control of brain-specific organelle translocations (By similarity). {ECO:0000250}. | FUNCTION: NCF2, NCF1, and a membrane bound cytochrome b558 are required for activation of the latent NADPH oxidase (necessary for superoxide production). {ECO:0000269|PubMed:19801500, ECO:0000269|PubMed:2547247, ECO:0000269|PubMed:2550933}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000223398 | + | 8 | 17 | 314_345 | 460 | 1047.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000361545 | + | 8 | 16 | 314_345 | 460 | 1012.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000395060 | + | 7 | 16 | 314_345 | 460 | 1047.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000223398 | + | 9 | 17 | 314_345 | 495 | 1047.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000395060 | + | 8 | 16 | 314_345 | 495 | 1047.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000223398 | + | 8 | 17 | 239_281 | 460 | 1047.0 | Domain | CAP-Gly 2 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000223398 | + | 8 | 17 | 99_141 | 460 | 1047.0 | Domain | CAP-Gly 1 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000361545 | + | 8 | 16 | 239_281 | 460 | 1012.0 | Domain | CAP-Gly 2 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000361545 | + | 8 | 16 | 99_141 | 460 | 1012.0 | Domain | CAP-Gly 1 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000395060 | + | 7 | 16 | 239_281 | 460 | 1047.0 | Domain | CAP-Gly 2 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000395060 | + | 7 | 16 | 99_141 | 460 | 1047.0 | Domain | CAP-Gly 1 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000223398 | + | 9 | 17 | 239_281 | 495 | 1047.0 | Domain | CAP-Gly 2 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000223398 | + | 9 | 17 | 99_141 | 495 | 1047.0 | Domain | CAP-Gly 1 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000395060 | + | 8 | 16 | 239_281 | 495 | 1047.0 | Domain | CAP-Gly 2 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000395060 | + | 8 | 16 | 99_141 | 495 | 1047.0 | Domain | CAP-Gly 1 |
| Tgene | NCF1 | chr7:73778645 | chr7:74193427 | ENST00000289473 | 1 | 11 | 211_254 | 51 | 391.0 | Compositional bias | Note=Asp/Glu-rich (highly acidic) | |
| Tgene | NCF1 | chr7:73778645 | chr7:74193427 | ENST00000289473 | 1 | 11 | 292_390 | 51 | 391.0 | Compositional bias | Note=Arg/Lys-rich (highly basic) | |
| Tgene | NCF1 | chr7:73787366 | chr7:74193427 | ENST00000289473 | 1 | 11 | 211_254 | 51 | 391.0 | Compositional bias | Note=Asp/Glu-rich (highly acidic) | |
| Tgene | NCF1 | chr7:73787366 | chr7:74193427 | ENST00000289473 | 1 | 11 | 292_390 | 51 | 391.0 | Compositional bias | Note=Arg/Lys-rich (highly basic) | |
| Tgene | NCF1 | chr7:73778645 | chr7:74193427 | ENST00000289473 | 1 | 11 | 156_215 | 51 | 391.0 | Domain | SH3 1 | |
| Tgene | NCF1 | chr7:73778645 | chr7:74193427 | ENST00000289473 | 1 | 11 | 226_285 | 51 | 391.0 | Domain | SH3 2 | |
| Tgene | NCF1 | chr7:73787366 | chr7:74193427 | ENST00000289473 | 1 | 11 | 156_215 | 51 | 391.0 | Domain | SH3 1 | |
| Tgene | NCF1 | chr7:73787366 | chr7:74193427 | ENST00000289473 | 1 | 11 | 226_285 | 51 | 391.0 | Domain | SH3 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000223398 | + | 8 | 17 | 354_524 | 460 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000223398 | + | 8 | 17 | 561_636 | 460 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000223398 | + | 8 | 17 | 673_1016 | 460 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000361545 | + | 8 | 16 | 354_524 | 460 | 1012.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000361545 | + | 8 | 16 | 561_636 | 460 | 1012.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000361545 | + | 8 | 16 | 673_1016 | 460 | 1012.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000395060 | + | 7 | 16 | 354_524 | 460 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000395060 | + | 7 | 16 | 561_636 | 460 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73778645 | chr7:74193427 | ENST00000395060 | + | 7 | 16 | 673_1016 | 460 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000223398 | + | 9 | 17 | 354_524 | 495 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000223398 | + | 9 | 17 | 561_636 | 495 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000223398 | + | 9 | 17 | 673_1016 | 495 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000361545 | + | 1 | 16 | 354_524 | 0 | 1012.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000361545 | + | 1 | 16 | 561_636 | 0 | 1012.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000361545 | + | 1 | 16 | 673_1016 | 0 | 1012.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000395060 | + | 8 | 16 | 354_524 | 495 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000395060 | + | 8 | 16 | 561_636 | 495 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000395060 | + | 8 | 16 | 673_1016 | 495 | 1047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000361545 | + | 1 | 16 | 314_345 | 0 | 1012.0 | Compositional bias | Note=Ser-rich |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000361545 | + | 1 | 16 | 239_281 | 0 | 1012.0 | Domain | CAP-Gly 2 |
| Hgene | CLIP2 | chr7:73787366 | chr7:74193427 | ENST00000361545 | + | 1 | 16 | 99_141 | 0 | 1012.0 | Domain | CAP-Gly 1 |
| Tgene | NCF1 | chr7:73778645 | chr7:74193427 | ENST00000289473 | 1 | 11 | 4_125 | 51 | 391.0 | Domain | PX | |
| Tgene | NCF1 | chr7:73787366 | chr7:74193427 | ENST00000289473 | 1 | 11 | 4_125 | 51 | 391.0 | Domain | PX |
Top |
Fusion Gene Sequence for CLIP2-NCF1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >17179_17179_1_CLIP2-NCF1_CLIP2_chr7_73778645_ENST00000223398_NCF1_chr7_74193427_ENST00000289473_length(transcript)=2880nt_BP=1707nt CGGCTCGGCCGCGGGGCGCGCAGGCGGCTGCTGGGCGGCCTCGGTGCGCGCCTCCCGCCTTCCCAGAGACGTGGCGCGAGGCCCGGGCCC TGAGCACCTATCGCGGGGATCCCCGGCGCCAGGAGGGGGTGCAGCCGGTGGGCAGCGCCGCGCAGGGAGGGGCCGCAGCATCCTCGCCCC CCAGCGCGCCCGGGCCCGAGAGGAGGAGGCCGGGGCTCTCCGGGCCTCCCGCCGCTTAGCCTGATGCTGGAAGGACGAAGGTGAGTGAAG ATGGCAGAGAGGACGTGACCAGCACTCACCCTTGTCCACCTGCCCAGTGGCACCGCCATGCAGAAGCCCAGCGGCCTGAAGCCCCCCGGC CGTGGGGGGAAGCACTCCAGCCCCATGGGCCGGACATCTACTGGGTCAGCTTCATCCTCGGCGGCGGTGGCCGCTAGCTCCAAGGAAGGC TCCCCACTGCACAAACAGTCATCTGGACCCTCCTCCTCCCCGGCCGCAGCTGCTGCCCCCGAGAAGCCGGGCCCCAAGGCGGCGGAAGTG GGGGATGACTTCCTGGGGGACTTTGTGGTGGGCGAGCGGGTGTGGGTGAACGGCGTGAAGCCAGGCGTGGTGCAGTATCTGGGAGAGACG CAGTTCGCACCGGGCCAGTGGGCTGGCGTGGTGCTGGACGACCCGGTGGGCAAGAATGATGGCGCGGTGGGCGGCGTGCGCTACTTCGAG TGCCCGGCCCTCCAGGGTATCTTCACGCGGCCCTCCAAGCTGACCCGGCAGCCCACGGCCGAGGGCTCGGGGAGTGATGCCCACTCCGTG GAGTCGCTGACTGCCCAGAACCTGTCATTGCATTCGGGCACGGCCACGCCCCCGCTGACCAGCCGCGTCATCCCCCTGCGGGAGAGCGTC CTCAACAGCTCCGTGAAGACTGGCAACGAGTCGGGATCCAACCTCTCAGACAGCGGCTCTGTGAAGCGGGGCGAAAAGGACCTGCGCCTG GGGGACCGCGTGCTGGTTGGCGGGACGAAGACTGGCGTGGTGCGGTACGTGGGGGAGACAGACTTTGCCAAGGGCGAGTGGTGTGGCGTG GAGCTGGACGAGCCCCTTGGGAAGAATGATGGGGCGGTGGCGGGCACCAGGTACTTCCAGTGCCCACCCAAGTTTGGTCTCTTCGCGCCC ATCCACAAAGTGATCCGTATCGGCTTCCCATCTACCAGCCCAGCCAAGGCCAAGAAGACCAAGCGTATGGCCATGGGTGTGTCAGCACTG ACCCACAGTCCCAGCAGTTCCTCCATCAGCTCCGTCAGCTCTGTGGCCTCCTCCGTGGGGGGTCGGCCCAGCCGCAGTGGCCTGCTCACG GAGACCTCTTCACGCTACGCCCGCAAGATCTCGGGCACCACGGCCTTGCAGGAGGCACTGAAGGAGAAGCAGCAGCACATTGAGCAGCTG CTGGCTGAACGAGACCTGGAACGGGCTGAGGTGGCCAAGGCCACAAGCCACATCTGCGAGGTGGAGAAGGAGATTGCCCTGCTCAAGGCA CAGCATGAGCAGTATGTTGCAGAAGCCGAGGAGAAGCTGCAGCGAGCCCGGCTGCTCGTGGAGAGCGTGCGGAAAGAGAAGGTGGACCTG TCCAACCAGCTGGAGGAGGAGAGGAGGAAGGTGGAGGATCTGCAGTTCCGCGTGGAGGAGGAGTCCATCACCAAGGGAGACCTGGAGAAA ACCTTAAAAGAAATGTTCCCTATTGAGGCAGGGGCGATCAATCCAGAGAACAGGATCATCCCCCACCTCCCAGCTCCCAAGTGGTTTGAC GGGCAGCGGGCCGCCGAGAACCGCCAGGGCACACTTACCGAGTACTGCGGCACGCTCATGAGCCTGCCCACCAAGATCTCCCGCTGTCCC CACCTCCTCGACTTCTTCAAGGTGCGCCCTGATGACCTCAAGCTCCCCACGGACAACCAGACAAAAAAGCCAGAGACATACTTGATGCCC AAAGATGGCAAGAGTACCGCGACAGACATCACCGGCCCCATCATCCTGCAGACGTACCGCGCCATTGCCAACTACGAGAAGACCTCGGGC TCCGAGATGGCTCTGTCCACGGGGGACGTGGTGGAGGTCGTAGAGAAGAGCGAGAGCGGTTGGTGGTTCTGTCAGATGAAAGCAAAGCGA GGCTGGATCCCAGCGTCCTTCCTCGAGCCCCTGGACAGTCCTGACGAGACGGAAGACCCTGAGCCCAACTATGCAGGTGAGCCATACGTC GCCATCAAGGCCTACACTGCTGTGGAGGGGGACGAGGTGTCCCTGCTCGAGGGTGAAGCTGTTGAGGTCATTCACAAGCTCCTGGACGGC TGGTGGGTCATCAGGAAAGACGACGTCACAGGCTACTTCCCGTCCATGTACCTGCAAAAGTCAGGGCAAGACGTGTCCCAGGCCCAACGC CAGATCAAGCGGGGGGCGCCGCCCCGCAGGTCGTCCATCCGCAACGCGCACAGCATCCACCAGCGGTCGCGGAAGCGCCTCAGCCAGGAC GCCTATCGCCGCAACAGCGTCCGTTTTCTGCAGCAGCGACGCCGCCAGGCGCGGCCGGGACCGCAGAGCCCCGGGAGCCCGCTCGAGGAG GAGCGGCAGACGCAGCGCTCTAAACCGCAGCCGGCGGTGCCCCCGCGGCCGAGCGCCGACCTCATCCTGAACCGCTGCAGCGAGAGCACC AAGCGGAAGCTGGCGTCTGCCGTCTGAGGCTGGAGCGCAGTCCCCAGCTAGCGTCTCGGCCCTTGCCGCCCCGTGCCTGTACATACGTGT TCTATAGAGCCTGGCGTCTGGACGCCGAGGGCAGCCCCGACCCCTGTCCAGCGCGGCTCCCGCCACCCTCAATAAATGTTGCTTGGAGTG >17179_17179_1_CLIP2-NCF1_CLIP2_chr7_73778645_ENST00000223398_NCF1_chr7_74193427_ENST00000289473_length(amino acids)=805AA_BP=1 MPSGTAMQKPSGLKPPGRGGKHSSPMGRTSTGSASSSAAVAASSKEGSPLHKQSSGPSSSPAAAAAPEKPGPKAAEVGDDFLGDFVVGER VWVNGVKPGVVQYLGETQFAPGQWAGVVLDDPVGKNDGAVGGVRYFECPALQGIFTRPSKLTRQPTAEGSGSDAHSVESLTAQNLSLHSG TATPPLTSRVIPLRESVLNSSVKTGNESGSNLSDSGSVKRGEKDLRLGDRVLVGGTKTGVVRYVGETDFAKGEWCGVELDEPLGKNDGAV AGTRYFQCPPKFGLFAPIHKVIRIGFPSTSPAKAKKTKRMAMGVSALTHSPSSSSISSVSSVASSVGGRPSRSGLLTETSSRYARKISGT TALQEALKEKQQHIEQLLAERDLERAEVAKATSHICEVEKEIALLKAQHEQYVAEAEEKLQRARLLVESVRKEKVDLSNQLEEERRKVED LQFRVEEESITKGDLEKTLKEMFPIEAGAINPENRIIPHLPAPKWFDGQRAAENRQGTLTEYCGTLMSLPTKISRCPHLLDFFKVRPDDL KLPTDNQTKKPETYLMPKDGKSTATDITGPIILQTYRAIANYEKTSGSEMALSTGDVVEVVEKSESGWWFCQMKAKRGWIPASFLEPLDS PDETEDPEPNYAGEPYVAIKAYTAVEGDEVSLLEGEAVEVIHKLLDGWWVIRKDDVTGYFPSMYLQKSGQDVSQAQRQIKRGAPPRRSSI -------------------------------------------------------------- >17179_17179_2_CLIP2-NCF1_CLIP2_chr7_73778645_ENST00000361545_NCF1_chr7_74193427_ENST00000289473_length(transcript)=2880nt_BP=1707nt CGGCTCGGCCGCGGGGCGCGCAGGCGGCTGCTGGGCGGCCTCGGTGCGCGCCTCCCGCCTTCCCAGAGACGTGGCGCGAGGCCCGGGCCC TGAGCACCTATCGCGGGGATCCCCGGCGCCAGGAGGGGGTGCAGCCGGTGGGCAGCGCCGCGCAGGGAGGGGCCGCAGCATCCTCGCCCC CCAGCGCGCCCGGGCCCGAGAGGAGGAGGCCGGGGCTCTCCGGGCCTCCCGCCGCTTAGCCTGATGCTGGAAGGACGAAGGTGAGTGAAG ATGGCAGAGAGGACGTGACCAGCACTCACCCTTGTCCACCTGCCCAGTGGCACCGCCATGCAGAAGCCCAGCGGCCTGAAGCCCCCCGGC CGTGGGGGGAAGCACTCCAGCCCCATGGGCCGGACATCTACTGGGTCAGCTTCATCCTCGGCGGCGGTGGCCGCTAGCTCCAAGGAAGGC TCCCCACTGCACAAACAGTCATCTGGACCCTCCTCCTCCCCGGCCGCAGCTGCTGCCCCCGAGAAGCCGGGCCCCAAGGCGGCGGAAGTG GGGGATGACTTCCTGGGGGACTTTGTGGTGGGCGAGCGGGTGTGGGTGAACGGCGTGAAGCCAGGCGTGGTGCAGTATCTGGGAGAGACG CAGTTCGCACCGGGCCAGTGGGCTGGCGTGGTGCTGGACGACCCGGTGGGCAAGAATGATGGCGCGGTGGGCGGCGTGCGCTACTTCGAG TGCCCGGCCCTCCAGGGTATCTTCACGCGGCCCTCCAAGCTGACCCGGCAGCCCACGGCCGAGGGCTCGGGGAGTGATGCCCACTCCGTG GAGTCGCTGACTGCCCAGAACCTGTCATTGCATTCGGGCACGGCCACGCCCCCGCTGACCAGCCGCGTCATCCCCCTGCGGGAGAGCGTC CTCAACAGCTCCGTGAAGACTGGCAACGAGTCGGGATCCAACCTCTCAGACAGCGGCTCTGTGAAGCGGGGCGAAAAGGACCTGCGCCTG GGGGACCGCGTGCTGGTTGGCGGGACGAAGACTGGCGTGGTGCGGTACGTGGGGGAGACAGACTTTGCCAAGGGCGAGTGGTGTGGCGTG GAGCTGGACGAGCCCCTTGGGAAGAATGATGGGGCGGTGGCGGGCACCAGGTACTTCCAGTGCCCACCCAAGTTTGGTCTCTTCGCGCCC ATCCACAAAGTGATCCGTATCGGCTTCCCATCTACCAGCCCAGCCAAGGCCAAGAAGACCAAGCGTATGGCCATGGGTGTGTCAGCACTG ACCCACAGTCCCAGCAGTTCCTCCATCAGCTCCGTCAGCTCTGTGGCCTCCTCCGTGGGGGGTCGGCCCAGCCGCAGTGGCCTGCTCACG GAGACCTCTTCACGCTACGCCCGCAAGATCTCGGGCACCACGGCCTTGCAGGAGGCACTGAAGGAGAAGCAGCAGCACATTGAGCAGCTG CTGGCTGAACGAGACCTGGAACGGGCTGAGGTGGCCAAGGCCACAAGCCACATCTGCGAGGTGGAGAAGGAGATTGCCCTGCTCAAGGCA CAGCATGAGCAGTATGTTGCAGAAGCCGAGGAGAAGCTGCAGCGAGCCCGGCTGCTCGTGGAGAGCGTGCGGAAAGAGAAGGTGGACCTG TCCAACCAGCTGGAGGAGGAGAGGAGGAAGGTGGAGGATCTGCAGTTCCGCGTGGAGGAGGAGTCCATCACCAAGGGAGACCTGGAGAAA ACCTTAAAAGAAATGTTCCCTATTGAGGCAGGGGCGATCAATCCAGAGAACAGGATCATCCCCCACCTCCCAGCTCCCAAGTGGTTTGAC GGGCAGCGGGCCGCCGAGAACCGCCAGGGCACACTTACCGAGTACTGCGGCACGCTCATGAGCCTGCCCACCAAGATCTCCCGCTGTCCC CACCTCCTCGACTTCTTCAAGGTGCGCCCTGATGACCTCAAGCTCCCCACGGACAACCAGACAAAAAAGCCAGAGACATACTTGATGCCC AAAGATGGCAAGAGTACCGCGACAGACATCACCGGCCCCATCATCCTGCAGACGTACCGCGCCATTGCCAACTACGAGAAGACCTCGGGC TCCGAGATGGCTCTGTCCACGGGGGACGTGGTGGAGGTCGTAGAGAAGAGCGAGAGCGGTTGGTGGTTCTGTCAGATGAAAGCAAAGCGA GGCTGGATCCCAGCGTCCTTCCTCGAGCCCCTGGACAGTCCTGACGAGACGGAAGACCCTGAGCCCAACTATGCAGGTGAGCCATACGTC GCCATCAAGGCCTACACTGCTGTGGAGGGGGACGAGGTGTCCCTGCTCGAGGGTGAAGCTGTTGAGGTCATTCACAAGCTCCTGGACGGC TGGTGGGTCATCAGGAAAGACGACGTCACAGGCTACTTCCCGTCCATGTACCTGCAAAAGTCAGGGCAAGACGTGTCCCAGGCCCAACGC CAGATCAAGCGGGGGGCGCCGCCCCGCAGGTCGTCCATCCGCAACGCGCACAGCATCCACCAGCGGTCGCGGAAGCGCCTCAGCCAGGAC GCCTATCGCCGCAACAGCGTCCGTTTTCTGCAGCAGCGACGCCGCCAGGCGCGGCCGGGACCGCAGAGCCCCGGGAGCCCGCTCGAGGAG GAGCGGCAGACGCAGCGCTCTAAACCGCAGCCGGCGGTGCCCCCGCGGCCGAGCGCCGACCTCATCCTGAACCGCTGCAGCGAGAGCACC AAGCGGAAGCTGGCGTCTGCCGTCTGAGGCTGGAGCGCAGTCCCCAGCTAGCGTCTCGGCCCTTGCCGCCCCGTGCCTGTACATACGTGT TCTATAGAGCCTGGCGTCTGGACGCCGAGGGCAGCCCCGACCCCTGTCCAGCGCGGCTCCCGCCACCCTCAATAAATGTTGCTTGGAGTG >17179_17179_2_CLIP2-NCF1_CLIP2_chr7_73778645_ENST00000361545_NCF1_chr7_74193427_ENST00000289473_length(amino acids)=805AA_BP=1 MPSGTAMQKPSGLKPPGRGGKHSSPMGRTSTGSASSSAAVAASSKEGSPLHKQSSGPSSSPAAAAAPEKPGPKAAEVGDDFLGDFVVGER VWVNGVKPGVVQYLGETQFAPGQWAGVVLDDPVGKNDGAVGGVRYFECPALQGIFTRPSKLTRQPTAEGSGSDAHSVESLTAQNLSLHSG TATPPLTSRVIPLRESVLNSSVKTGNESGSNLSDSGSVKRGEKDLRLGDRVLVGGTKTGVVRYVGETDFAKGEWCGVELDEPLGKNDGAV AGTRYFQCPPKFGLFAPIHKVIRIGFPSTSPAKAKKTKRMAMGVSALTHSPSSSSISSVSSVASSVGGRPSRSGLLTETSSRYARKISGT TALQEALKEKQQHIEQLLAERDLERAEVAKATSHICEVEKEIALLKAQHEQYVAEAEEKLQRARLLVESVRKEKVDLSNQLEEERRKVED LQFRVEEESITKGDLEKTLKEMFPIEAGAINPENRIIPHLPAPKWFDGQRAAENRQGTLTEYCGTLMSLPTKISRCPHLLDFFKVRPDDL KLPTDNQTKKPETYLMPKDGKSTATDITGPIILQTYRAIANYEKTSGSEMALSTGDVVEVVEKSESGWWFCQMKAKRGWIPASFLEPLDS PDETEDPEPNYAGEPYVAIKAYTAVEGDEVSLLEGEAVEVIHKLLDGWWVIRKDDVTGYFPSMYLQKSGQDVSQAQRQIKRGAPPRRSSI -------------------------------------------------------------- >17179_17179_3_CLIP2-NCF1_CLIP2_chr7_73778645_ENST00000395060_NCF1_chr7_74193427_ENST00000289473_length(transcript)=2553nt_BP=1380nt ATGCAGAAGCCCAGCGGCCTGAAGCCCCCCGGCCGTGGGGGGAAGCACTCCAGCCCCATGGGCCGGACATCTACTGGGTCAGCTTCATCC TCGGCGGCGGTGGCCGCTAGCTCCAAGGAAGGCTCCCCACTGCACAAACAGTCATCTGGACCCTCCTCCTCCCCGGCCGCAGCTGCTGCC CCCGAGAAGCCGGGCCCCAAGGCGGCGGAAGTGGGGGATGACTTCCTGGGGGACTTTGTGGTGGGCGAGCGGGTGTGGGTGAACGGCGTG AAGCCAGGCGTGGTGCAGTATCTGGGAGAGACGCAGTTCGCACCGGGCCAGTGGGCTGGCGTGGTGCTGGACGACCCGGTGGGCAAGAAT GATGGCGCGGTGGGCGGCGTGCGCTACTTCGAGTGCCCGGCCCTCCAGGGTATCTTCACGCGGCCCTCCAAGCTGACCCGGCAGCCCACG GCCGAGGGCTCGGGGAGTGATGCCCACTCCGTGGAGTCGCTGACTGCCCAGAACCTGTCATTGCATTCGGGCACGGCCACGCCCCCGCTG ACCAGCCGCGTCATCCCCCTGCGGGAGAGCGTCCTCAACAGCTCCGTGAAGACTGGCAACGAGTCGGGATCCAACCTCTCAGACAGCGGC TCTGTGAAGCGGGGCGAAAAGGACCTGCGCCTGGGGGACCGCGTGCTGGTTGGCGGGACGAAGACTGGCGTGGTGCGGTACGTGGGGGAG ACAGACTTTGCCAAGGGCGAGTGGTGTGGCGTGGAGCTGGACGAGCCCCTTGGGAAGAATGATGGGGCGGTGGCGGGCACCAGGTACTTC CAGTGCCCACCCAAGTTTGGTCTCTTCGCGCCCATCCACAAAGTGATCCGTATCGGCTTCCCATCTACCAGCCCAGCCAAGGCCAAGAAG ACCAAGCGTATGGCCATGGGTGTGTCAGCACTGACCCACAGTCCCAGCAGTTCCTCCATCAGCTCCGTCAGCTCTGTGGCCTCCTCCGTG GGGGGTCGGCCCAGCCGCAGTGGCCTGCTCACGGAGACCTCTTCACGCTACGCCCGCAAGATCTCGGGCACCACGGCCTTGCAGGAGGCA CTGAAGGAGAAGCAGCAGCACATTGAGCAGCTGCTGGCTGAACGAGACCTGGAACGGGCTGAGGTGGCCAAGGCCACAAGCCACATCTGC GAGGTGGAGAAGGAGATTGCCCTGCTCAAGGCACAGCATGAGCAGTATGTTGCAGAAGCCGAGGAGAAGCTGCAGCGAGCCCGGCTGCTC GTGGAGAGCGTGCGGAAAGAGAAGGTGGACCTGTCCAACCAGCTGGAGGAGGAGAGGAGGAAGGTGGAGGATCTGCAGTTCCGCGTGGAG GAGGAGTCCATCACCAAGGGAGACCTGGAGAAAACCTTAAAAGAAATGTTCCCTATTGAGGCAGGGGCGATCAATCCAGAGAACAGGATC ATCCCCCACCTCCCAGCTCCCAAGTGGTTTGACGGGCAGCGGGCCGCCGAGAACCGCCAGGGCACACTTACCGAGTACTGCGGCACGCTC ATGAGCCTGCCCACCAAGATCTCCCGCTGTCCCCACCTCCTCGACTTCTTCAAGGTGCGCCCTGATGACCTCAAGCTCCCCACGGACAAC CAGACAAAAAAGCCAGAGACATACTTGATGCCCAAAGATGGCAAGAGTACCGCGACAGACATCACCGGCCCCATCATCCTGCAGACGTAC CGCGCCATTGCCAACTACGAGAAGACCTCGGGCTCCGAGATGGCTCTGTCCACGGGGGACGTGGTGGAGGTCGTAGAGAAGAGCGAGAGC GGTTGGTGGTTCTGTCAGATGAAAGCAAAGCGAGGCTGGATCCCAGCGTCCTTCCTCGAGCCCCTGGACAGTCCTGACGAGACGGAAGAC CCTGAGCCCAACTATGCAGGTGAGCCATACGTCGCCATCAAGGCCTACACTGCTGTGGAGGGGGACGAGGTGTCCCTGCTCGAGGGTGAA GCTGTTGAGGTCATTCACAAGCTCCTGGACGGCTGGTGGGTCATCAGGAAAGACGACGTCACAGGCTACTTCCCGTCCATGTACCTGCAA AAGTCAGGGCAAGACGTGTCCCAGGCCCAACGCCAGATCAAGCGGGGGGCGCCGCCCCGCAGGTCGTCCATCCGCAACGCGCACAGCATC CACCAGCGGTCGCGGAAGCGCCTCAGCCAGGACGCCTATCGCCGCAACAGCGTCCGTTTTCTGCAGCAGCGACGCCGCCAGGCGCGGCCG GGACCGCAGAGCCCCGGGAGCCCGCTCGAGGAGGAGCGGCAGACGCAGCGCTCTAAACCGCAGCCGGCGGTGCCCCCGCGGCCGAGCGCC GACCTCATCCTGAACCGCTGCAGCGAGAGCACCAAGCGGAAGCTGGCGTCTGCCGTCTGAGGCTGGAGCGCAGTCCCCAGCTAGCGTCTC GGCCCTTGCCGCCCCGTGCCTGTACATACGTGTTCTATAGAGCCTGGCGTCTGGACGCCGAGGGCAGCCCCGACCCCTGTCCAGCGCGGC >17179_17179_3_CLIP2-NCF1_CLIP2_chr7_73778645_ENST00000395060_NCF1_chr7_74193427_ENST00000289473_length(amino acids)=799AA_BP=0 MQKPSGLKPPGRGGKHSSPMGRTSTGSASSSAAVAASSKEGSPLHKQSSGPSSSPAAAAAPEKPGPKAAEVGDDFLGDFVVGERVWVNGV KPGVVQYLGETQFAPGQWAGVVLDDPVGKNDGAVGGVRYFECPALQGIFTRPSKLTRQPTAEGSGSDAHSVESLTAQNLSLHSGTATPPL TSRVIPLRESVLNSSVKTGNESGSNLSDSGSVKRGEKDLRLGDRVLVGGTKTGVVRYVGETDFAKGEWCGVELDEPLGKNDGAVAGTRYF QCPPKFGLFAPIHKVIRIGFPSTSPAKAKKTKRMAMGVSALTHSPSSSSISSVSSVASSVGGRPSRSGLLTETSSRYARKISGTTALQEA LKEKQQHIEQLLAERDLERAEVAKATSHICEVEKEIALLKAQHEQYVAEAEEKLQRARLLVESVRKEKVDLSNQLEEERRKVEDLQFRVE EESITKGDLEKTLKEMFPIEAGAINPENRIIPHLPAPKWFDGQRAAENRQGTLTEYCGTLMSLPTKISRCPHLLDFFKVRPDDLKLPTDN QTKKPETYLMPKDGKSTATDITGPIILQTYRAIANYEKTSGSEMALSTGDVVEVVEKSESGWWFCQMKAKRGWIPASFLEPLDSPDETED PEPNYAGEPYVAIKAYTAVEGDEVSLLEGEAVEVIHKLLDGWWVIRKDDVTGYFPSMYLQKSGQDVSQAQRQIKRGAPPRRSSIRNAHSI -------------------------------------------------------------- >17179_17179_4_CLIP2-NCF1_CLIP2_chr7_73787366_ENST00000223398_NCF1_chr7_74193427_ENST00000289473_length(transcript)=2985nt_BP=1812nt CGGCTCGGCCGCGGGGCGCGCAGGCGGCTGCTGGGCGGCCTCGGTGCGCGCCTCCCGCCTTCCCAGAGACGTGGCGCGAGGCCCGGGCCC TGAGCACCTATCGCGGGGATCCCCGGCGCCAGGAGGGGGTGCAGCCGGTGGGCAGCGCCGCGCAGGGAGGGGCCGCAGCATCCTCGCCCC CCAGCGCGCCCGGGCCCGAGAGGAGGAGGCCGGGGCTCTCCGGGCCTCCCGCCGCTTAGCCTGATGCTGGAAGGACGAAGGTGAGTGAAG ATGGCAGAGAGGACGTGACCAGCACTCACCCTTGTCCACCTGCCCAGTGGCACCGCCATGCAGAAGCCCAGCGGCCTGAAGCCCCCCGGC CGTGGGGGGAAGCACTCCAGCCCCATGGGCCGGACATCTACTGGGTCAGCTTCATCCTCGGCGGCGGTGGCCGCTAGCTCCAAGGAAGGC TCCCCACTGCACAAACAGTCATCTGGACCCTCCTCCTCCCCGGCCGCAGCTGCTGCCCCCGAGAAGCCGGGCCCCAAGGCGGCGGAAGTG GGGGATGACTTCCTGGGGGACTTTGTGGTGGGCGAGCGGGTGTGGGTGAACGGCGTGAAGCCAGGCGTGGTGCAGTATCTGGGAGAGACG CAGTTCGCACCGGGCCAGTGGGCTGGCGTGGTGCTGGACGACCCGGTGGGCAAGAATGATGGCGCGGTGGGCGGCGTGCGCTACTTCGAG TGCCCGGCCCTCCAGGGTATCTTCACGCGGCCCTCCAAGCTGACCCGGCAGCCCACGGCCGAGGGCTCGGGGAGTGATGCCCACTCCGTG GAGTCGCTGACTGCCCAGAACCTGTCATTGCATTCGGGCACGGCCACGCCCCCGCTGACCAGCCGCGTCATCCCCCTGCGGGAGAGCGTC CTCAACAGCTCCGTGAAGACTGGCAACGAGTCGGGATCCAACCTCTCAGACAGCGGCTCTGTGAAGCGGGGCGAAAAGGACCTGCGCCTG GGGGACCGCGTGCTGGTTGGCGGGACGAAGACTGGCGTGGTGCGGTACGTGGGGGAGACAGACTTTGCCAAGGGCGAGTGGTGTGGCGTG GAGCTGGACGAGCCCCTTGGGAAGAATGATGGGGCGGTGGCGGGCACCAGGTACTTCCAGTGCCCACCCAAGTTTGGTCTCTTCGCGCCC ATCCACAAAGTGATCCGTATCGGCTTCCCATCTACCAGCCCAGCCAAGGCCAAGAAGACCAAGCGTATGGCCATGGGTGTGTCAGCACTG ACCCACAGTCCCAGCAGTTCCTCCATCAGCTCCGTCAGCTCTGTGGCCTCCTCCGTGGGGGGTCGGCCCAGCCGCAGTGGCCTGCTCACG GAGACCTCTTCACGCTACGCCCGCAAGATCTCGGGCACCACGGCCTTGCAGGAGGCACTGAAGGAGAAGCAGCAGCACATTGAGCAGCTG CTGGCTGAACGAGACCTGGAACGGGCTGAGGTGGCCAAGGCCACAAGCCACATCTGCGAGGTGGAGAAGGAGATTGCCCTGCTCAAGGCA CAGCATGAGCAGTATGTTGCAGAAGCCGAGGAGAAGCTGCAGCGAGCCCGGCTGCTCGTGGAGAGCGTGCGGAAAGAGAAGGTGGACCTG TCCAACCAGCTGGAGGAGGAGAGGAGGAAGGTGGAGGATCTGCAGTTCCGCGTGGAGGAGGAGTCCATCACCAAGGGAGACCTGGAGACC CAGACGCAGCTGGAGCACGCGCGCATTGGGGAGCTGGAACAGAGCCTGCTACTGGAGAAGGCGCAGGCCGAGCGGCTGCTCCGAGAATTA GCGGACAACAGGAAAACCTTAAAAGAAATGTTCCCTATTGAGGCAGGGGCGATCAATCCAGAGAACAGGATCATCCCCCACCTCCCAGCT CCCAAGTGGTTTGACGGGCAGCGGGCCGCCGAGAACCGCCAGGGCACACTTACCGAGTACTGCGGCACGCTCATGAGCCTGCCCACCAAG ATCTCCCGCTGTCCCCACCTCCTCGACTTCTTCAAGGTGCGCCCTGATGACCTCAAGCTCCCCACGGACAACCAGACAAAAAAGCCAGAG ACATACTTGATGCCCAAAGATGGCAAGAGTACCGCGACAGACATCACCGGCCCCATCATCCTGCAGACGTACCGCGCCATTGCCAACTAC GAGAAGACCTCGGGCTCCGAGATGGCTCTGTCCACGGGGGACGTGGTGGAGGTCGTAGAGAAGAGCGAGAGCGGTTGGTGGTTCTGTCAG ATGAAAGCAAAGCGAGGCTGGATCCCAGCGTCCTTCCTCGAGCCCCTGGACAGTCCTGACGAGACGGAAGACCCTGAGCCCAACTATGCA GGTGAGCCATACGTCGCCATCAAGGCCTACACTGCTGTGGAGGGGGACGAGGTGTCCCTGCTCGAGGGTGAAGCTGTTGAGGTCATTCAC AAGCTCCTGGACGGCTGGTGGGTCATCAGGAAAGACGACGTCACAGGCTACTTCCCGTCCATGTACCTGCAAAAGTCAGGGCAAGACGTG TCCCAGGCCCAACGCCAGATCAAGCGGGGGGCGCCGCCCCGCAGGTCGTCCATCCGCAACGCGCACAGCATCCACCAGCGGTCGCGGAAG CGCCTCAGCCAGGACGCCTATCGCCGCAACAGCGTCCGTTTTCTGCAGCAGCGACGCCGCCAGGCGCGGCCGGGACCGCAGAGCCCCGGG AGCCCGCTCGAGGAGGAGCGGCAGACGCAGCGCTCTAAACCGCAGCCGGCGGTGCCCCCGCGGCCGAGCGCCGACCTCATCCTGAACCGC TGCAGCGAGAGCACCAAGCGGAAGCTGGCGTCTGCCGTCTGAGGCTGGAGCGCAGTCCCCAGCTAGCGTCTCGGCCCTTGCCGCCCCGTG CCTGTACATACGTGTTCTATAGAGCCTGGCGTCTGGACGCCGAGGGCAGCCCCGACCCCTGTCCAGCGCGGCTCCCGCCACCCTCAATAA >17179_17179_4_CLIP2-NCF1_CLIP2_chr7_73787366_ENST00000223398_NCF1_chr7_74193427_ENST00000289473_length(amino acids)=840AA_BP=1 MPSGTAMQKPSGLKPPGRGGKHSSPMGRTSTGSASSSAAVAASSKEGSPLHKQSSGPSSSPAAAAAPEKPGPKAAEVGDDFLGDFVVGER VWVNGVKPGVVQYLGETQFAPGQWAGVVLDDPVGKNDGAVGGVRYFECPALQGIFTRPSKLTRQPTAEGSGSDAHSVESLTAQNLSLHSG TATPPLTSRVIPLRESVLNSSVKTGNESGSNLSDSGSVKRGEKDLRLGDRVLVGGTKTGVVRYVGETDFAKGEWCGVELDEPLGKNDGAV AGTRYFQCPPKFGLFAPIHKVIRIGFPSTSPAKAKKTKRMAMGVSALTHSPSSSSISSVSSVASSVGGRPSRSGLLTETSSRYARKISGT TALQEALKEKQQHIEQLLAERDLERAEVAKATSHICEVEKEIALLKAQHEQYVAEAEEKLQRARLLVESVRKEKVDLSNQLEEERRKVED LQFRVEEESITKGDLETQTQLEHARIGELEQSLLLEKAQAERLLRELADNRKTLKEMFPIEAGAINPENRIIPHLPAPKWFDGQRAAENR QGTLTEYCGTLMSLPTKISRCPHLLDFFKVRPDDLKLPTDNQTKKPETYLMPKDGKSTATDITGPIILQTYRAIANYEKTSGSEMALSTG DVVEVVEKSESGWWFCQMKAKRGWIPASFLEPLDSPDETEDPEPNYAGEPYVAIKAYTAVEGDEVSLLEGEAVEVIHKLLDGWWVIRKDD VTGYFPSMYLQKSGQDVSQAQRQIKRGAPPRRSSIRNAHSIHQRSRKRLSQDAYRRNSVRFLQQRRRQARPGPQSPGSPLEEERQTQRSK -------------------------------------------------------------- >17179_17179_5_CLIP2-NCF1_CLIP2_chr7_73787366_ENST00000395060_NCF1_chr7_74193427_ENST00000289473_length(transcript)=2658nt_BP=1485nt ATGCAGAAGCCCAGCGGCCTGAAGCCCCCCGGCCGTGGGGGGAAGCACTCCAGCCCCATGGGCCGGACATCTACTGGGTCAGCTTCATCC TCGGCGGCGGTGGCCGCTAGCTCCAAGGAAGGCTCCCCACTGCACAAACAGTCATCTGGACCCTCCTCCTCCCCGGCCGCAGCTGCTGCC CCCGAGAAGCCGGGCCCCAAGGCGGCGGAAGTGGGGGATGACTTCCTGGGGGACTTTGTGGTGGGCGAGCGGGTGTGGGTGAACGGCGTG AAGCCAGGCGTGGTGCAGTATCTGGGAGAGACGCAGTTCGCACCGGGCCAGTGGGCTGGCGTGGTGCTGGACGACCCGGTGGGCAAGAAT GATGGCGCGGTGGGCGGCGTGCGCTACTTCGAGTGCCCGGCCCTCCAGGGTATCTTCACGCGGCCCTCCAAGCTGACCCGGCAGCCCACG GCCGAGGGCTCGGGGAGTGATGCCCACTCCGTGGAGTCGCTGACTGCCCAGAACCTGTCATTGCATTCGGGCACGGCCACGCCCCCGCTG ACCAGCCGCGTCATCCCCCTGCGGGAGAGCGTCCTCAACAGCTCCGTGAAGACTGGCAACGAGTCGGGATCCAACCTCTCAGACAGCGGC TCTGTGAAGCGGGGCGAAAAGGACCTGCGCCTGGGGGACCGCGTGCTGGTTGGCGGGACGAAGACTGGCGTGGTGCGGTACGTGGGGGAG ACAGACTTTGCCAAGGGCGAGTGGTGTGGCGTGGAGCTGGACGAGCCCCTTGGGAAGAATGATGGGGCGGTGGCGGGCACCAGGTACTTC CAGTGCCCACCCAAGTTTGGTCTCTTCGCGCCCATCCACAAAGTGATCCGTATCGGCTTCCCATCTACCAGCCCAGCCAAGGCCAAGAAG ACCAAGCGTATGGCCATGGGTGTGTCAGCACTGACCCACAGTCCCAGCAGTTCCTCCATCAGCTCCGTCAGCTCTGTGGCCTCCTCCGTG GGGGGTCGGCCCAGCCGCAGTGGCCTGCTCACGGAGACCTCTTCACGCTACGCCCGCAAGATCTCGGGCACCACGGCCTTGCAGGAGGCA CTGAAGGAGAAGCAGCAGCACATTGAGCAGCTGCTGGCTGAACGAGACCTGGAACGGGCTGAGGTGGCCAAGGCCACAAGCCACATCTGC GAGGTGGAGAAGGAGATTGCCCTGCTCAAGGCACAGCATGAGCAGTATGTTGCAGAAGCCGAGGAGAAGCTGCAGCGAGCCCGGCTGCTC GTGGAGAGCGTGCGGAAAGAGAAGGTGGACCTGTCCAACCAGCTGGAGGAGGAGAGGAGGAAGGTGGAGGATCTGCAGTTCCGCGTGGAG GAGGAGTCCATCACCAAGGGAGACCTGGAGACCCAGACGCAGCTGGAGCACGCGCGCATTGGGGAGCTGGAACAGAGCCTGCTACTGGAG AAGGCGCAGGCCGAGCGGCTGCTCCGAGAATTAGCGGACAACAGGAAAACCTTAAAAGAAATGTTCCCTATTGAGGCAGGGGCGATCAAT CCAGAGAACAGGATCATCCCCCACCTCCCAGCTCCCAAGTGGTTTGACGGGCAGCGGGCCGCCGAGAACCGCCAGGGCACACTTACCGAG TACTGCGGCACGCTCATGAGCCTGCCCACCAAGATCTCCCGCTGTCCCCACCTCCTCGACTTCTTCAAGGTGCGCCCTGATGACCTCAAG CTCCCCACGGACAACCAGACAAAAAAGCCAGAGACATACTTGATGCCCAAAGATGGCAAGAGTACCGCGACAGACATCACCGGCCCCATC ATCCTGCAGACGTACCGCGCCATTGCCAACTACGAGAAGACCTCGGGCTCCGAGATGGCTCTGTCCACGGGGGACGTGGTGGAGGTCGTA GAGAAGAGCGAGAGCGGTTGGTGGTTCTGTCAGATGAAAGCAAAGCGAGGCTGGATCCCAGCGTCCTTCCTCGAGCCCCTGGACAGTCCT GACGAGACGGAAGACCCTGAGCCCAACTATGCAGGTGAGCCATACGTCGCCATCAAGGCCTACACTGCTGTGGAGGGGGACGAGGTGTCC CTGCTCGAGGGTGAAGCTGTTGAGGTCATTCACAAGCTCCTGGACGGCTGGTGGGTCATCAGGAAAGACGACGTCACAGGCTACTTCCCG TCCATGTACCTGCAAAAGTCAGGGCAAGACGTGTCCCAGGCCCAACGCCAGATCAAGCGGGGGGCGCCGCCCCGCAGGTCGTCCATCCGC AACGCGCACAGCATCCACCAGCGGTCGCGGAAGCGCCTCAGCCAGGACGCCTATCGCCGCAACAGCGTCCGTTTTCTGCAGCAGCGACGC CGCCAGGCGCGGCCGGGACCGCAGAGCCCCGGGAGCCCGCTCGAGGAGGAGCGGCAGACGCAGCGCTCTAAACCGCAGCCGGCGGTGCCC CCGCGGCCGAGCGCCGACCTCATCCTGAACCGCTGCAGCGAGAGCACCAAGCGGAAGCTGGCGTCTGCCGTCTGAGGCTGGAGCGCAGTC CCCAGCTAGCGTCTCGGCCCTTGCCGCCCCGTGCCTGTACATACGTGTTCTATAGAGCCTGGCGTCTGGACGCCGAGGGCAGCCCCGACC >17179_17179_5_CLIP2-NCF1_CLIP2_chr7_73787366_ENST00000395060_NCF1_chr7_74193427_ENST00000289473_length(amino acids)=834AA_BP=0 MQKPSGLKPPGRGGKHSSPMGRTSTGSASSSAAVAASSKEGSPLHKQSSGPSSSPAAAAAPEKPGPKAAEVGDDFLGDFVVGERVWVNGV KPGVVQYLGETQFAPGQWAGVVLDDPVGKNDGAVGGVRYFECPALQGIFTRPSKLTRQPTAEGSGSDAHSVESLTAQNLSLHSGTATPPL TSRVIPLRESVLNSSVKTGNESGSNLSDSGSVKRGEKDLRLGDRVLVGGTKTGVVRYVGETDFAKGEWCGVELDEPLGKNDGAVAGTRYF QCPPKFGLFAPIHKVIRIGFPSTSPAKAKKTKRMAMGVSALTHSPSSSSISSVSSVASSVGGRPSRSGLLTETSSRYARKISGTTALQEA LKEKQQHIEQLLAERDLERAEVAKATSHICEVEKEIALLKAQHEQYVAEAEEKLQRARLLVESVRKEKVDLSNQLEEERRKVEDLQFRVE EESITKGDLETQTQLEHARIGELEQSLLLEKAQAERLLRELADNRKTLKEMFPIEAGAINPENRIIPHLPAPKWFDGQRAAENRQGTLTE YCGTLMSLPTKISRCPHLLDFFKVRPDDLKLPTDNQTKKPETYLMPKDGKSTATDITGPIILQTYRAIANYEKTSGSEMALSTGDVVEVV EKSESGWWFCQMKAKRGWIPASFLEPLDSPDETEDPEPNYAGEPYVAIKAYTAVEGDEVSLLEGEAVEVIHKLLDGWWVIRKDDVTGYFP SMYLQKSGQDVSQAQRQIKRGAPPRRSSIRNAHSIHQRSRKRLSQDAYRRNSVRFLQQRRRQARPGPQSPGSPLEEERQTQRSKPQPAVP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CLIP2-NCF1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CLIP2-NCF1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CLIP2-NCF1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
