
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CNOT2-NUP107 (FusionGDB2 ID:17743) |
Fusion Gene Summary for CNOT2-NUP107 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CNOT2-NUP107 | Fusion gene ID: 17743 | Hgene | Tgene | Gene symbol | CNOT2 | NUP107 | Gene ID | 4848 | 57122 |
| Gene name | CCR4-NOT transcription complex subunit 2 | nucleoporin 107 | |
| Synonyms | CDC36|HSPC131|IDNADFS|NOT2|NOT2H | NPHS11|NUP84|ODG6|ODG6; GAMOS7 | |
| Cytomap | 12q15 | 12q15 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CCR4-NOT transcription complex subunit 2CCR4-associated factor 2negative regulator of transcription 2 | nuclear pore complex protein Nup107nucleoporin 107kDa | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NZN8 | P57740 | |
| Ensembl transtripts involved in fusion gene | ENST00000548230, ENST00000229195, ENST00000418359, ENST00000551483, | ENST00000229179, ENST00000378905, ENST00000539906, ENST00000401003, | |
| Fusion gene scores | * DoF score | 41 X 13 X 11=5863 | 27 X 22 X 9=5346 |
| # samples | 46 | 37 | |
| ** MAII score | log2(46/5863*10)=-3.6719332904521 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(37/5346*10)=-3.85286266172677 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CNOT2 [Title/Abstract] AND NUP107 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CNOT2(70637260)-NUP107(69090599), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CNOT2 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 14707134|16712523 |
| Tgene | NUP107 | GO:0006406 | mRNA export from nucleus | 11684705 |
 Fusion gene breakpoints across CNOT2 (5'-gene) Fusion gene breakpoints across CNOT2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
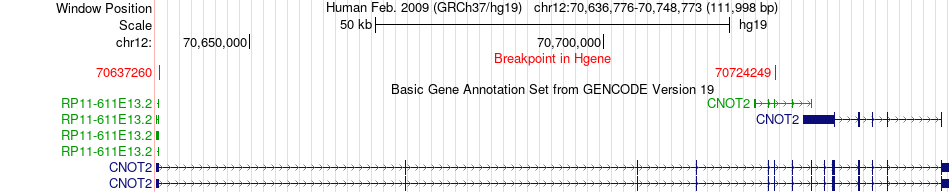 |
 Fusion gene breakpoints across NUP107 (3'-gene) Fusion gene breakpoints across NUP107 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
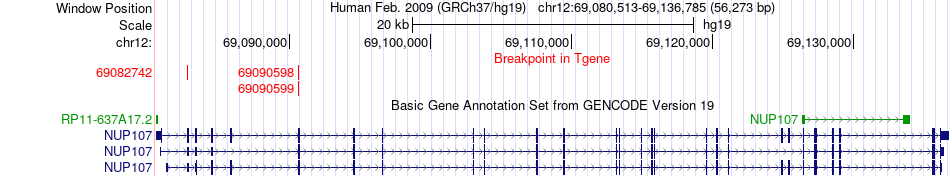 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A8-A08F-01A | CNOT2 | chr12 | 70637260 | - | NUP107 | chr12 | 69090599 | + |
| ChimerDB4 | BRCA | TCGA-A8-A08F-01A | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| ChimerDB4 | BRCA | TCGA-A8-A08F | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| ChimerDB4 | STAD | TCGA-FP-7998-01A | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
Top |
Fusion Gene ORF analysis for CNOT2-NUP107 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000548230 | ENST00000229179 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 3UTR-5UTR | ENST00000548230 | ENST00000378905 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 3UTR-5UTR | ENST00000548230 | ENST00000539906 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 3UTR-intron | ENST00000548230 | ENST00000401003 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5CDS-5UTR | ENST00000229195 | ENST00000378905 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5CDS-5UTR | ENST00000229195 | ENST00000539906 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5CDS-5UTR | ENST00000418359 | ENST00000378905 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5CDS-5UTR | ENST00000418359 | ENST00000539906 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5CDS-intron | ENST00000229195 | ENST00000401003 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5CDS-intron | ENST00000418359 | ENST00000401003 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| 5UTR-3CDS | ENST00000229195 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-3CDS | ENST00000229195 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-3CDS | ENST00000418359 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-3CDS | ENST00000418359 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-5UTR | ENST00000229195 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-5UTR | ENST00000229195 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-5UTR | ENST00000229195 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-5UTR | ENST00000229195 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-5UTR | ENST00000418359 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-5UTR | ENST00000418359 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-5UTR | ENST00000418359 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-5UTR | ENST00000418359 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-intron | ENST00000229195 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-intron | ENST00000229195 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| 5UTR-intron | ENST00000418359 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| 5UTR-intron | ENST00000418359 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| In-frame | ENST00000229195 | ENST00000229179 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| In-frame | ENST00000418359 | ENST00000229179 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| intron-3CDS | ENST00000548230 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-3CDS | ENST00000548230 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-3CDS | ENST00000551483 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-3CDS | ENST00000551483 | ENST00000229179 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-3CDS | ENST00000551483 | ENST00000229179 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| intron-5UTR | ENST00000548230 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-5UTR | ENST00000548230 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-5UTR | ENST00000548230 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-5UTR | ENST00000548230 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-5UTR | ENST00000551483 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-5UTR | ENST00000551483 | ENST00000378905 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-5UTR | ENST00000551483 | ENST00000378905 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| intron-5UTR | ENST00000551483 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-5UTR | ENST00000551483 | ENST00000539906 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-5UTR | ENST00000551483 | ENST00000539906 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
| intron-intron | ENST00000548230 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-intron | ENST00000548230 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-intron | ENST00000551483 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090599 | + |
| intron-intron | ENST00000551483 | ENST00000401003 | CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + |
| intron-intron | ENST00000551483 | ENST00000401003 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000229195 | CNOT2 | chr12 | 70724249 | - | ENST00000229179 | NUP107 | chr12 | 69082742 | + | 4461 | 1148 | 540 | 3917 | 1125 |
| ENST00000418359 | CNOT2 | chr12 | 70724249 | - | ENST00000229179 | NUP107 | chr12 | 69082742 | + | 4333 | 1020 | 412 | 3789 | 1125 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000229195 | ENST00000229179 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + | 0.000348003 | 0.999652 |
| ENST00000418359 | ENST00000229179 | CNOT2 | chr12 | 70724249 | - | NUP107 | chr12 | 69082742 | + | 0.000313095 | 0.99968696 |
Top |
Fusion Genomic Features for CNOT2-NUP107 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + | 9.72E-08 | 0.9999999 |
| CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + | 9.72E-08 | 0.9999999 |
| CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + | 9.72E-08 | 0.9999999 |
| CNOT2 | chr12 | 70637260 | + | NUP107 | chr12 | 69090598 | + | 9.72E-08 | 0.9999999 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
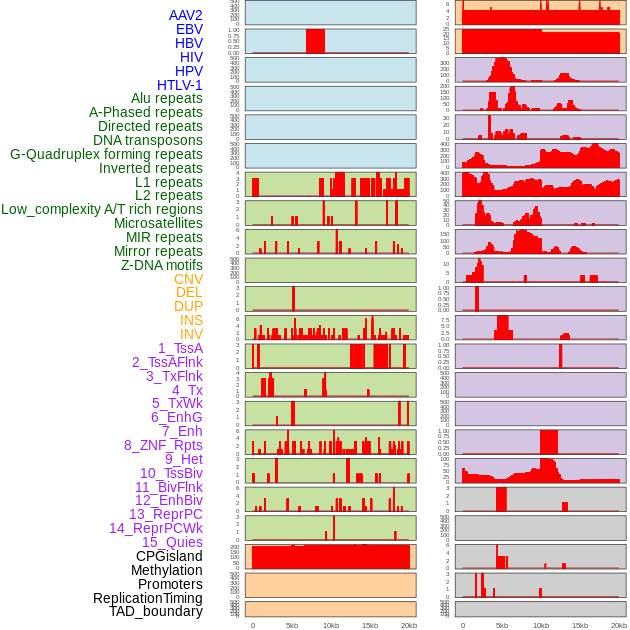 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
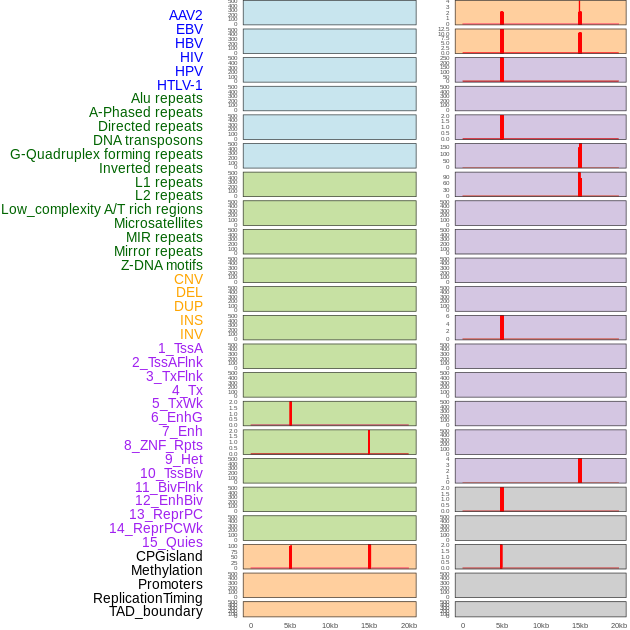 |
Top |
Fusion Protein Features for CNOT2-NUP107 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:70637260/chr12:69090599) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CNOT2 | NUP107 |
| FUNCTION: Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. Required for the CCR4-NOT complex structural integrity. Can repress transcription and may link the CCR4-NOT complex to transcriptional regulation; the repressive function may specifically involve the N-Cor repressor complex containing HDAC3, NCOR1 and NCOR2. Involved in the maintenance of embryonic stem (ES) cell identity. {ECO:0000269|PubMed:14707134, ECO:0000269|PubMed:16712523, ECO:0000269|PubMed:21299754, ECO:0000269|PubMed:22367759}. | FUNCTION: Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance (PubMed:12552102, PubMed:15229283, PubMed:30179222). Required for the assembly of peripheral proteins into the NPC (PubMed:15229283, PubMed:12552102). May anchor NUP62 to the NPC (PubMed:15229283). Involved in nephrogenesis (PubMed:30179222). {ECO:0000269|PubMed:12552102, ECO:0000269|PubMed:15229283, ECO:0000269|PubMed:30179222}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CNOT2 | chr12:70724249 | chr12:69082742 | ENST00000229195 | - | 6 | 16 | 437_540 | 189 | 541.0 | Region | Note=Repressor domain |
| Hgene | CNOT2 | chr12:70724249 | chr12:69082742 | ENST00000418359 | - | 7 | 17 | 437_540 | 189 | 541.0 | Region | Note=Repressor domain |
| Hgene | CNOT2 | chr12:70724249 | chr12:69082742 | ENST00000551483 | - | 1 | 7 | 437_540 | 0 | 192.0 | Region | Note=Repressor domain |
Top |
Fusion Gene Sequence for CNOT2-NUP107 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >17743_17743_1_CNOT2-NUP107_CNOT2_chr12_70724249_ENST00000229195_NUP107_chr12_69082742_ENST00000229179_length(transcript)=4461nt_BP=1148nt AACACAAACACAAACACGAACACGAACACGCTCAGCGTCCCCGGGGAGGGCGGACTGAAGCCACCTCACCATTCATCGCCCCTGGACCCC GCAGTCCCCCTCGGACTCAGGGCCAGAGCCTTCTGCGAAGCAACCTAGCCCCAGGGAAACGGTAGCGGCCGCAGAAGCCCCGCTCTCTGT AACGGCCGCGGAGGAGCAGCAAGCCTCCCTGTGAGTCGGTGGGAGACAAGACGGAGCGGCGGTAAGGGCGGTAGGGAAGTGAAGCGCCTC TCTCCACCTTGTTAGAAGCGCGCTGAAAGCGGCCTGCCTACCCACCCCATCGCTGCCGTTTTGCAGTCGTCGCTCCCACCTTCCGCTGCC GCCTGGAGGGAAGCCGGAGCGACGGGGGTCACGGCGGCGGTCAGAGGGTAAAGGTCTTGCTCCCAGCAGCCTCCGCGGTGGATACGTCGC CATCTTGGATCCGCGGGACAAGAAAATTCATGCGAGGGAGACGTGGTGGGCGGTCCTTCCTGTGACACGACCCTTGAGTGACAGTTCTAT TTGATTGCCTCCGGTACTGTGAGGAAAGGACACGACTCTATGGTGAGGACTGATGGACATACATTATCTGAGAAAAGAAACTACCAGGTG ACAAACAGCATGTTTGGTGCTTCAAGAAAGAAGTTTGTAGAGGGGGTCGACAGTGACTACCATGACGAAAACATGTACTACAGCCAGTCT TCTATGTTTCCACATCGGTCAGAAAAAGATATGCTGGCATCACCATCTACATCAGGTCAGCTGTCTCAGTTTGGGGCAAGTTTATACGGG CAACAAAGTGCACTAGGCCTTCCAATGAGGGGGATGAGCAACAATACCCCTCAGTTAAATCGCAGCTTATCACAAGGCACTCAGTTACCG AGCCACGTCACGCCAACAACAGGGGTACCAACAATGTCACTTCACACGCCTCCATCTCCAAGCAGGGGTATTTTGCCTATGAATCCTAGG AATATGATGAACCACTCCCAGGTTGGTCAGGGCATTGGAATTCCTAGCAGGACAAATAGCATGAGCAGTTCAGGGTTAGGTAGCCCCAAC AGAAGCTCGCCAAGCATAATATGTATGCCAAAGCAGCAGCCTTCTCGACAGCCTTTTACTGTGAACAGGAGTGGCTTTGGAGAGATATCA TCCCCTGTAATCCGGGAGGCAGAGGTGACACGGACTGCACGGAAACAGAGTGCTCAGAAAAGAGTTTTACTTCAGGCATCTCAAGATGAA AATTTTGGTAATACTACACCAAGAAACCAGGTTATCCCTCGAACTCCTAGCTCATTTCGACAGCCTTTTACCCCAACAAGCCGAAGCTTA CTAAGGCAGCCAGATATTTCCTGCATTCTTGGAACAGGAGGGAAGTCGCCCCGACTTACGCAGTCTTCAGGGTTCTTTGGAAATCTCTCC ATGGTTACTAATCTGGATGACAGTAACTGGGCAGCTGCATTTTCATCACAGCGTTCCGGGCTGTTCACAAACACAGAGCCCCACAGTATA ACAGAAGATGTAACTATCAGTGCTGTTATGTTACGTGAGGATGATCCTGGAGAAGCTGCATCCATGAGTATGTTTTCTGATTTCCTGCAG TCTTTTCTGAAGCACTCTTCGAGTACAGTTTTTGATCTTGTGGAAGAGTATGAAAACATCTGTGGTAGTCAGGTGAATATACTGAGTAAA ATAGTGAGTCGAGCAACACCTGGACTTCAAAAATTTTCAAAAACAGCCAGTATGCTCTGGCTTCTTCAACAGGAGATGGTCACATGGAGG CTGCTGGCTTCTTTGTATAGAGACAGAATACAGTCTGCATTAGAAGAGGAAAGTGTATTCGCAGTTACTGCTGTTAATGCCAGTGAAAAA ACAGTTGTGGAAGCGTTATTTCAGAGGGATTCACTTGTTCGACAAAGTCAGCTGGTGGTAGATTGGTTAGAGAGTATTGCCAAAGATGAA ATTGGAGAATTTTCTGATAATATTGAGTTTTATGCAAAATCAGTATATTGGGAAAATACTCTGCATACCTTAAAACAACGGCAGCTGACT TCTTACGTTGGAAGTGTTCGTCCGCTTGTCACTGAATTGGACCCTGATGCTCCCATAAGACAGAAAATGCCCCTTGATGATCTGGATAGA GAAGATGAAGTTAGATTACTCAAATATCTCTTTACTCTAATCCGTGCTGGAATGACAGAAGAGGCACAACGACTCTGTAAACGCTGTGGT CAAGCATGGAGAGCTGCAACACTTGAAGGCTGGAAACTGTACCATGACCCTAATGTTAATGGAGGAACAGAATTAGAACCTGTTGAAGGG AATCCATATAGACGCATTTGGAAAATAAGTTGCTGGAGAATGGCAGAAGATGAGCTTTTTAATAGATACGAGAGAGCAATTTATGCAGCT TTAAGTGGGAATCTTAAGCAGCTGCTTCCTGTCTGTGACACCTGGGAAGACACAGTTTGGGCCTACTTCCGGGTGATGGTGGACAGTCTG GTAGAACAGGAGATCCAGACATCAGTAGCAACTCTGGATGAAACTGAAGAACTCCCTAGAGAATATCTGGGAGCAAACTGGACGTTAGAA AAGGTTTTTGAGGAACTTCAAGCTACTGACAAAAAGAGAGTTCTGGAAGAGAATCAAGAACATTATCATATAGTTCAAAAGTTTCTTATC CTGGGAGACATTGATGGTTTGATGGATGAGTTTAGCAAATGGCTTTCCAAAAGCAGAAACAATCTACCTGGACACCTGCTTCGCTTTATG ACTCACCTTATTTTGTTTTTCCGTACTCTGGGACTACAGACCAAGGAGGAAGTTTCTATTGAAGTTTTAAAGACATACATACAGCTTTTA ATAAGAGAGAAACATACAAATCTTATAGCATTTTATACCTGTCATTTGCCTCAAGACCTAGCTGTTGCCCAGTATGCATTATTTTTGGAA AGTGTTACAGAATTTGAACAGCGCCACCATTGCCTGGAGTTGGCTAAAGAAGCAGATTTGGATGTTGCAACAATAACAAAAACTGTAGTT GAGAATATTCGAAAGAAAGATAATGGTGAATTTAGTCATCATGACCTGGCCCCAGCCCTAGATACTGGCACTACTGAGGAGGATCGTTTA AAAATTGATGTAATTGACTGGTTGGTATTTGACCCAGCGCAGAGGGCAGAAGCACTGAAACAAGGCAATGCAATTATGAGAAAATTCTTG GCATCAAAAAAGCACGAAGCTGCAAAAGAAGTATTTGTGAAAATTCCTCAGGATTCTATAGCAGAAATCTATAATCAGTGCGAGGAACAA GGAATGGAAAGTCCACTTCCTGCTGAAGATGATAATGCTATCCGAGAACATTTGTGCATCAGAGCTTATTTGGAAGCCCATGAAACCTTT AATGAGTGGTTTAAGCATATGAATTCAGTTCCACAAAAACCTGCTTTGATACCTCAACCAACTTTTACTGAGAAAGTGGCTCATGAACAC AAAGAAAAGAAATATGAAATGGATTTTGGTATTTGGAAAGGGCATTTGGATGCCCTAACTGCTGATGTGAAGGAGAAAATGTATAACGTC TTGTTGTTTGTTGATGGAGGGTGGATGGTGGATGTTAGAGAGGATGCCAAAGAAGACCATGAAAGAACACATCAAATGGTCTTACTGAGA AAGCTTTGTCTGCCAATGTTGTGTTTTCTGCTTCATACGATATTGCACAGTACTGGTCAGTATCAGGAATGCCTACAGTTAGCAGATATG GTATCCTCTGAGCGCCACAAACTGTACCTGGTATTTTCTAAGGAAGAGCTAAGGAAGTTGCTGCAGAAGCTCAGAGAGTCCTCTCTAATG CTCCTAGACCAGGGACTTGACCCATTAGGGTATGAAATTCAGTTATAGTTTAATCTTTGTAATCTCACTAATTTTCATGATAAATGAAGT TTTTAATAAAATATACTTGTTATTAGTAATTTTTTCTTTTGCATTACCATGTAAAATTTAGACATTTGAATTTTGTACTTTTCAGAATAT TATCGTGACACTTTCAACATGTAGGGATATCAGCGTTTCTCTGTGTGCTTGTTAATAAAACTATATAAAAATTAAAATTTTCTGTTTTTA CAAATTCCAGGGTTGTCTCTTTTTCATGTAATAAGAAAATTAAATTTTCTGTTCTAGTTTTTTCTGTAATCATAAGCATTCTGATATTTT AAAATCATAAAGATGATGGAAAGTAGATTTATTTTTAAGCCAGGTACGTAATTTTTTATTTATCATAAAATTTAGTATTTTTGAATATTA AAATTAAGAAGATGACTATAACAAGAATTTTTCTAACATAACATGCTATTATTATTTTAGTTATTTTGAAGACATTTCTAGTTTAACTTT >17743_17743_1_CNOT2-NUP107_CNOT2_chr12_70724249_ENST00000229195_NUP107_chr12_69082742_ENST00000229179_length(amino acids)=1125AA_BP=202 MIASGTVRKGHDSMVRTDGHTLSEKRNYQVTNSMFGASRKKFVEGVDSDYHDENMYYSQSSMFPHRSEKDMLASPSTSGQLSQFGASLYG QQSALGLPMRGMSNNTPQLNRSLSQGTQLPSHVTPTTGVPTMSLHTPPSPSRGILPMNPRNMMNHSQVGQGIGIPSRTNSMSSSGLGSPN RSSPSIICMPKQQPSRQPFTVNRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSL LRQPDISCILGTGGKSPRLTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQ SFLKHSSSTVFDLVEEYENICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEK TVVEALFQRDSLVRQSQLVVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDR EDEVRLLKYLFTLIRAGMTEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAA LSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLI LGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLE SVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFL ASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEH KEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADM -------------------------------------------------------------- >17743_17743_2_CNOT2-NUP107_CNOT2_chr12_70724249_ENST00000418359_NUP107_chr12_69082742_ENST00000229179_length(transcript)=4333nt_BP=1020nt AGCGTCCCCGGGGAGGGCGGACTGAAGCCACCTCACCATTCATCGCCCCTGGACCCCGCAGTCCCCCTCGGACTCAGGGCCAGAGCCTTC TGCGAAGCAACCTAGCCCCAGGGAAACGGTAGCGGCCGCAGAAGCCCCGCTCTCTGTAACGGCCGCGGAGGAGCAGCAAGCCTCCCTGTG AGTCGGTGGGAGACAAGACGGAGCGGCGTCGTCGCTCCCACCTTCCGCTGCCGCCTGGAGGGAAGCCGGAGCGACGGGGGTCACGGCGGC GGTCAGAGGGTAAAGGTCTTGCTCCCAGCAGCCTCCGCGGTGGATACGTCGCCATCTTGGATCCGCGGGACAAGAAAATTCATGCGAGGG AGACGTGGTGGGCGGTCCTTCCTGTGACACGACCCTTGAGTGACAGTTCTATTTGATTGCCTCCGGTACTGTGAGGAAAGGACACGACTC TATGGTGAGGACTGATGGACATACATTATCTGAGAAAAGAAACTACCAGGTGACAAACAGCATGTTTGGTGCTTCAAGAAAGAAGTTTGT AGAGGGGGTCGACAGTGACTACCATGACGAAAACATGTACTACAGCCAGTCTTCTATGTTTCCACATCGGTCAGAAAAAGATATGCTGGC ATCACCATCTACATCAGGTCAGCTGTCTCAGTTTGGGGCAAGTTTATACGGGCAACAAAGTGCACTAGGCCTTCCAATGAGGGGGATGAG CAACAATACCCCTCAGTTAAATCGCAGCTTATCACAAGGCACTCAGTTACCGAGCCACGTCACGCCAACAACAGGGGTACCAACAATGTC ACTTCACACGCCTCCATCTCCAAGCAGGGGTATTTTGCCTATGAATCCTAGGAATATGATGAACCACTCCCAGGTTGGTCAGGGCATTGG AATTCCTAGCAGGACAAATAGCATGAGCAGTTCAGGGTTAGGTAGCCCCAACAGAAGCTCGCCAAGCATAATATGTATGCCAAAGCAGCA GCCTTCTCGACAGCCTTTTACTGTGAACAGGAGTGGCTTTGGAGAGATATCATCCCCTGTAATCCGGGAGGCAGAGGTGACACGGACTGC ACGGAAACAGAGTGCTCAGAAAAGAGTTTTACTTCAGGCATCTCAAGATGAAAATTTTGGTAATACTACACCAAGAAACCAGGTTATCCC TCGAACTCCTAGCTCATTTCGACAGCCTTTTACCCCAACAAGCCGAAGCTTACTAAGGCAGCCAGATATTTCCTGCATTCTTGGAACAGG AGGGAAGTCGCCCCGACTTACGCAGTCTTCAGGGTTCTTTGGAAATCTCTCCATGGTTACTAATCTGGATGACAGTAACTGGGCAGCTGC ATTTTCATCACAGCGTTCCGGGCTGTTCACAAACACAGAGCCCCACAGTATAACAGAAGATGTAACTATCAGTGCTGTTATGTTACGTGA GGATGATCCTGGAGAAGCTGCATCCATGAGTATGTTTTCTGATTTCCTGCAGTCTTTTCTGAAGCACTCTTCGAGTACAGTTTTTGATCT TGTGGAAGAGTATGAAAACATCTGTGGTAGTCAGGTGAATATACTGAGTAAAATAGTGAGTCGAGCAACACCTGGACTTCAAAAATTTTC AAAAACAGCCAGTATGCTCTGGCTTCTTCAACAGGAGATGGTCACATGGAGGCTGCTGGCTTCTTTGTATAGAGACAGAATACAGTCTGC ATTAGAAGAGGAAAGTGTATTCGCAGTTACTGCTGTTAATGCCAGTGAAAAAACAGTTGTGGAAGCGTTATTTCAGAGGGATTCACTTGT TCGACAAAGTCAGCTGGTGGTAGATTGGTTAGAGAGTATTGCCAAAGATGAAATTGGAGAATTTTCTGATAATATTGAGTTTTATGCAAA ATCAGTATATTGGGAAAATACTCTGCATACCTTAAAACAACGGCAGCTGACTTCTTACGTTGGAAGTGTTCGTCCGCTTGTCACTGAATT GGACCCTGATGCTCCCATAAGACAGAAAATGCCCCTTGATGATCTGGATAGAGAAGATGAAGTTAGATTACTCAAATATCTCTTTACTCT AATCCGTGCTGGAATGACAGAAGAGGCACAACGACTCTGTAAACGCTGTGGTCAAGCATGGAGAGCTGCAACACTTGAAGGCTGGAAACT GTACCATGACCCTAATGTTAATGGAGGAACAGAATTAGAACCTGTTGAAGGGAATCCATATAGACGCATTTGGAAAATAAGTTGCTGGAG AATGGCAGAAGATGAGCTTTTTAATAGATACGAGAGAGCAATTTATGCAGCTTTAAGTGGGAATCTTAAGCAGCTGCTTCCTGTCTGTGA CACCTGGGAAGACACAGTTTGGGCCTACTTCCGGGTGATGGTGGACAGTCTGGTAGAACAGGAGATCCAGACATCAGTAGCAACTCTGGA TGAAACTGAAGAACTCCCTAGAGAATATCTGGGAGCAAACTGGACGTTAGAAAAGGTTTTTGAGGAACTTCAAGCTACTGACAAAAAGAG AGTTCTGGAAGAGAATCAAGAACATTATCATATAGTTCAAAAGTTTCTTATCCTGGGAGACATTGATGGTTTGATGGATGAGTTTAGCAA ATGGCTTTCCAAAAGCAGAAACAATCTACCTGGACACCTGCTTCGCTTTATGACTCACCTTATTTTGTTTTTCCGTACTCTGGGACTACA GACCAAGGAGGAAGTTTCTATTGAAGTTTTAAAGACATACATACAGCTTTTAATAAGAGAGAAACATACAAATCTTATAGCATTTTATAC CTGTCATTTGCCTCAAGACCTAGCTGTTGCCCAGTATGCATTATTTTTGGAAAGTGTTACAGAATTTGAACAGCGCCACCATTGCCTGGA GTTGGCTAAAGAAGCAGATTTGGATGTTGCAACAATAACAAAAACTGTAGTTGAGAATATTCGAAAGAAAGATAATGGTGAATTTAGTCA TCATGACCTGGCCCCAGCCCTAGATACTGGCACTACTGAGGAGGATCGTTTAAAAATTGATGTAATTGACTGGTTGGTATTTGACCCAGC GCAGAGGGCAGAAGCACTGAAACAAGGCAATGCAATTATGAGAAAATTCTTGGCATCAAAAAAGCACGAAGCTGCAAAAGAAGTATTTGT GAAAATTCCTCAGGATTCTATAGCAGAAATCTATAATCAGTGCGAGGAACAAGGAATGGAAAGTCCACTTCCTGCTGAAGATGATAATGC TATCCGAGAACATTTGTGCATCAGAGCTTATTTGGAAGCCCATGAAACCTTTAATGAGTGGTTTAAGCATATGAATTCAGTTCCACAAAA ACCTGCTTTGATACCTCAACCAACTTTTACTGAGAAAGTGGCTCATGAACACAAAGAAAAGAAATATGAAATGGATTTTGGTATTTGGAA AGGGCATTTGGATGCCCTAACTGCTGATGTGAAGGAGAAAATGTATAACGTCTTGTTGTTTGTTGATGGAGGGTGGATGGTGGATGTTAG AGAGGATGCCAAAGAAGACCATGAAAGAACACATCAAATGGTCTTACTGAGAAAGCTTTGTCTGCCAATGTTGTGTTTTCTGCTTCATAC GATATTGCACAGTACTGGTCAGTATCAGGAATGCCTACAGTTAGCAGATATGGTATCCTCTGAGCGCCACAAACTGTACCTGGTATTTTC TAAGGAAGAGCTAAGGAAGTTGCTGCAGAAGCTCAGAGAGTCCTCTCTAATGCTCCTAGACCAGGGACTTGACCCATTAGGGTATGAAAT TCAGTTATAGTTTAATCTTTGTAATCTCACTAATTTTCATGATAAATGAAGTTTTTAATAAAATATACTTGTTATTAGTAATTTTTTCTT TTGCATTACCATGTAAAATTTAGACATTTGAATTTTGTACTTTTCAGAATATTATCGTGACACTTTCAACATGTAGGGATATCAGCGTTT CTCTGTGTGCTTGTTAATAAAACTATATAAAAATTAAAATTTTCTGTTTTTACAAATTCCAGGGTTGTCTCTTTTTCATGTAATAAGAAA ATTAAATTTTCTGTTCTAGTTTTTTCTGTAATCATAAGCATTCTGATATTTTAAAATCATAAAGATGATGGAAAGTAGATTTATTTTTAA GCCAGGTACGTAATTTTTTATTTATCATAAAATTTAGTATTTTTGAATATTAAAATTAAGAAGATGACTATAACAAGAATTTTTCTAACA TAACATGCTATTATTATTTTAGTTATTTTGAAGACATTTCTAGTTTAACTTTCATTTAGCAAACTTCCTATCAAAATAAGAGCATATAAA >17743_17743_2_CNOT2-NUP107_CNOT2_chr12_70724249_ENST00000418359_NUP107_chr12_69082742_ENST00000229179_length(amino acids)=1125AA_BP=202 MIASGTVRKGHDSMVRTDGHTLSEKRNYQVTNSMFGASRKKFVEGVDSDYHDENMYYSQSSMFPHRSEKDMLASPSTSGQLSQFGASLYG QQSALGLPMRGMSNNTPQLNRSLSQGTQLPSHVTPTTGVPTMSLHTPPSPSRGILPMNPRNMMNHSQVGQGIGIPSRTNSMSSSGLGSPN RSSPSIICMPKQQPSRQPFTVNRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSL LRQPDISCILGTGGKSPRLTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQ SFLKHSSSTVFDLVEEYENICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEK TVVEALFQRDSLVRQSQLVVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDR EDEVRLLKYLFTLIRAGMTEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAA LSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLI LGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLE SVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFL ASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEH KEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CNOT2-NUP107 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CNOT2-NUP107 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CNOT2-NUP107 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
