
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:COPZ1-C19orf18 (FusionGDB2 ID:18666) |
Fusion Gene Summary for COPZ1-C19orf18 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: COPZ1-C19orf18 | Fusion gene ID: 18666 | Hgene | Tgene | Gene symbol | COPZ1 | C19orf18 | Gene ID | 22818 | 147685 |
| Gene name | COPI coat complex subunit zeta 1 | chromosome 19 open reading frame 18 | |
| Synonyms | CGI-120|COPZ|HSPC181|zeta-COP|zeta1-COP | - | |
| Cytomap | 12q13.13 | 19q13.43 | |
| Type of gene | protein-coding | protein-coding | |
| Description | coatomer subunit zeta-1coatomer protein complex subunit zeta 1coatomer protein complex, subunit zetazeta-1 COPzeta-1-coat protein | uncharacterized protein C19orf18 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P61923 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000548281, ENST00000262061, ENST00000416254, ENST00000455864, ENST00000548753, ENST00000549043, ENST00000549116, ENST00000551779, ENST00000552218, ENST00000552362, ENST00000553231, | ENST00000314391, | |
| Fusion gene scores | * DoF score | 12 X 4 X 10=480 | 4 X 4 X 4=64 |
| # samples | 13 | 5 | |
| ** MAII score | log2(13/480*10)=-1.88452278258006 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/64*10)=-0.356143810225275 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: COPZ1 [Title/Abstract] AND C19orf18 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | COPZ1(54741628)-C19orf18(58472919), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | COPZ1 | GO:0006891 | intra-Golgi vesicle-mediated transport | 11056392 |
 Fusion gene breakpoints across COPZ1 (5'-gene) Fusion gene breakpoints across COPZ1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
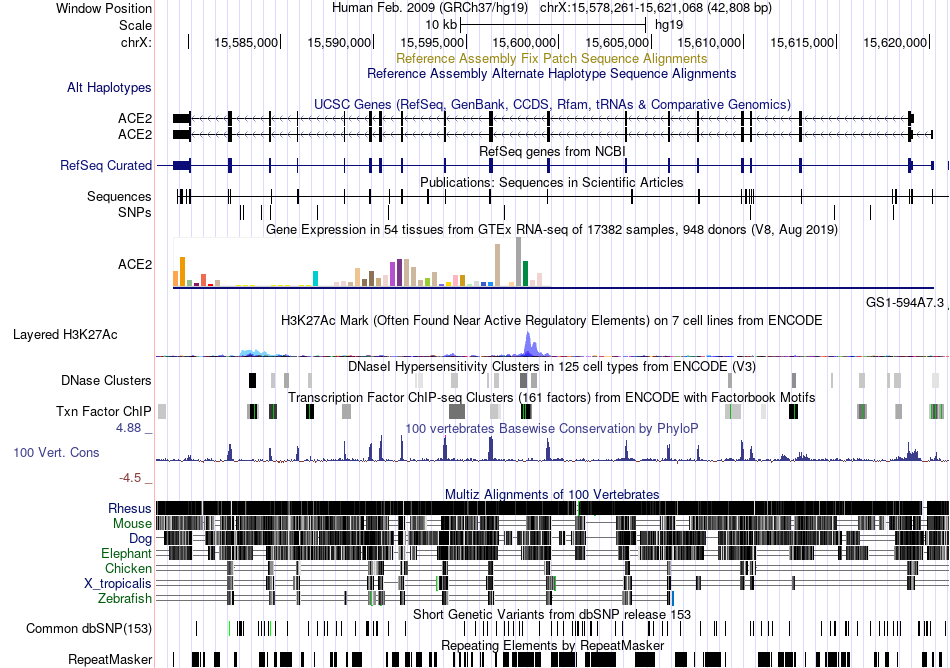 |
 Fusion gene breakpoints across C19orf18 (3'-gene) Fusion gene breakpoints across C19orf18 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
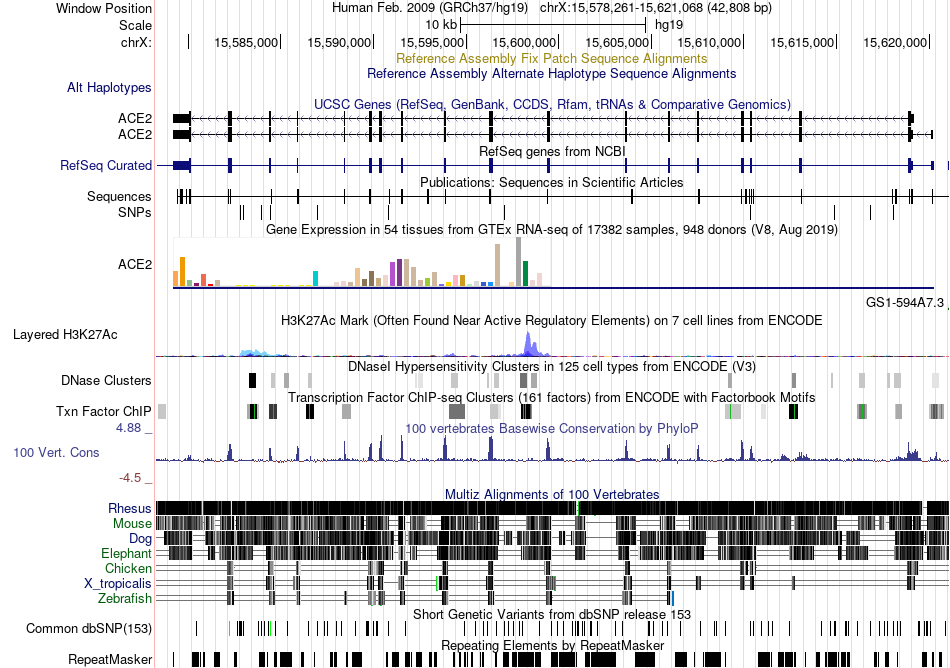 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-FE-A235-01A | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
Top |
Fusion Gene ORF analysis for COPZ1-C19orf18 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000548281 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000262061 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000416254 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000455864 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000548753 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000549043 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000549116 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000551779 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000552218 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000552362 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
| In-frame | ENST00000553231 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262061 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 874 | 432 | 25 | 708 | 227 |
| ENST00000549043 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 956 | 514 | 95 | 790 | 231 |
| ENST00000552218 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 933 | 491 | 21 | 767 | 248 |
| ENST00000553231 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 793 | 351 | 13 | 627 | 204 |
| ENST00000552362 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 862 | 420 | 13 | 696 | 227 |
| ENST00000455864 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 793 | 351 | 13 | 627 | 204 |
| ENST00000416254 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 770 | 328 | 86 | 604 | 172 |
| ENST00000549116 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 683 | 241 | 8 | 517 | 169 |
| ENST00000551779 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 850 | 408 | 1 | 684 | 227 |
| ENST00000548753 | COPZ1 | chr12 | 54741628 | + | ENST00000314391 | C19orf18 | chr19 | 58472919 | - | 918 | 476 | 108 | 752 | 214 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262061 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.001052941 | 0.9989471 |
| ENST00000549043 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.002118667 | 0.99788135 |
| ENST00000552218 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.003066138 | 0.9969338 |
| ENST00000553231 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.002193907 | 0.99780613 |
| ENST00000552362 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.001155024 | 0.998845 |
| ENST00000455864 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.002193907 | 0.99780613 |
| ENST00000416254 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.007384641 | 0.99261534 |
| ENST00000549116 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.003378084 | 0.99662197 |
| ENST00000551779 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.001005289 | 0.99899477 |
| ENST00000548753 | ENST00000314391 | COPZ1 | chr12 | 54741628 | + | C19orf18 | chr19 | 58472919 | - | 0.005706738 | 0.99429333 |
Top |
Fusion Genomic Features for COPZ1-C19orf18 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
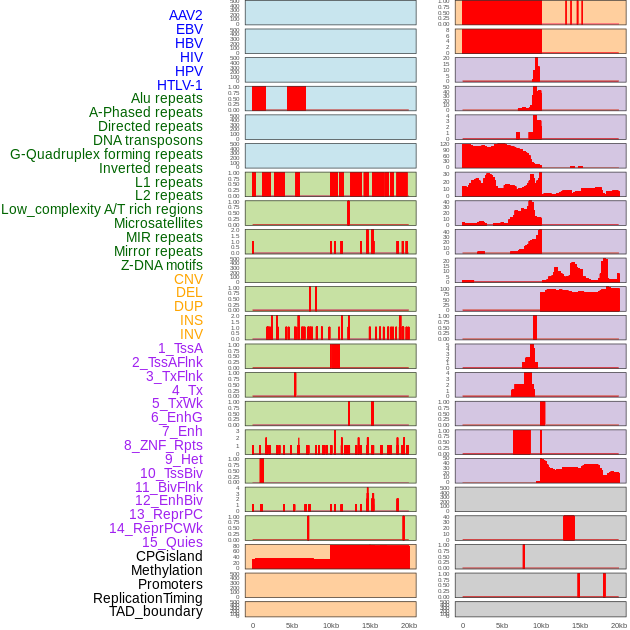 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for COPZ1-C19orf18 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:54741628/chr19:58472919) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| COPZ1 | . |
| FUNCTION: The coatomer is a cytosolic protein complex that binds to dilysine motifs and reversibly associates with Golgi non-clathrin-coated vesicles, which further mediate biosynthetic protein transport from the ER, via the Golgi up to the trans Golgi network. Coatomer complex is required for budding from Golgi membranes, and is essential for the retrograde Golgi-to-ER transport of dilysine-tagged proteins (By similarity). The zeta subunit may be involved in regulating the coat assembly and, hence, the rate of biosynthetic protein transport due to its association-dissociation properties with the coatomer complex (By similarity). {ECO:0000250|UniProtKB:P53600}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | C19orf18 | chr12:54741628 | chr19:58472919 | ENST00000314391 | 3 | 6 | 122_215 | 123 | 216.0 | Topological domain | Cytoplasmic |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | C19orf18 | chr12:54741628 | chr19:58472919 | ENST00000314391 | 3 | 6 | 30_100 | 123 | 216.0 | Topological domain | Extracellular | |
| Tgene | C19orf18 | chr12:54741628 | chr19:58472919 | ENST00000314391 | 3 | 6 | 101_121 | 123 | 216.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for COPZ1-C19orf18 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >18666_18666_1_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000262061_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=874nt_BP=432nt TTTTGCGGCTCCACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGAT TCTGGACAATGATGGAGATCGACTTTTTGCCAAGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACAT TTTCAACAAGACCCATCGGACTGACAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCTCTATTTCTA TGTGATTGGCAGCTCCTATGAAAATGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAA TGTAGAAAAGCGAGCACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGA GGAAAGACAACAGCTCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTC CACGCACCTACTTCCAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAA ACTGAAGGAAGAGCAAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGAC AGAGGTGCCGGCTGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAA >18666_18666_1_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000262061_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=227AA_BP=136 MRGKMEALILEPSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYEN ELMLMAVLNCLFDSLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLLPENE -------------------------------------------------------------- >18666_18666_2_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000416254_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=770nt_BP=328nt ACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTGGACAATGA TGGAGATCGACTTTTTGCCAAGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACATTTTCAACAAGAC CCATCGGACTGACACTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAATGTAGAAAAGCGAG CACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGACAACAGC TCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACCTACTTC CAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGGAAGAGC AAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGCCGGCTG AGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACATCAAAGA >18666_18666_2_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000416254_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=172AA_BP=81 MMEIDFLPSTMTTPTPVSRSKRPLRRTFSTRPIGLTLMLMAVLNCLFDSLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQ -------------------------------------------------------------- >18666_18666_3_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000455864_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=793nt_BP=351nt ACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGA GAAGAACATTTTCAACAAGACCCATCGGACTGACAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCT CTATTTCTATGTGATTGGCAGCTCCTATGAAAATGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCT GAGGAAAAATGTAGAAAAGCGAGCACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGC ACAGGCTGAGGAAAGACAACAGCTCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGA GGGTGAGTCCACGCACCTACTTCCAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATAT TAAGAAGAAACTGAAGGAAGAGCAAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAAC GCAGACGACAGAGGTGCCGGCTGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAAC >18666_18666_3_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000455864_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=204AA_BP=113 MRGKMEALILYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYENELMLMAVLNCLFDSLSQMLRKNV EKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLLPENENELEKFIHSVIISKRSKNIKKKL -------------------------------------------------------------- >18666_18666_4_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000548753_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=918nt_BP=476nt TGATGGTTTCCCAAGTTTGACTCTCCATTGCCGATTCCCTGCCACATTCCTTCAGGAAACTGTTTTGTCTTCGGTTTCCTGTTAATCCCT ATTATATAGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTGGACAATGATGGAGATCGACTTTTTGCCAAGTACTATGACGAC ACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACATTTTCAACAAGACCCATCGGACTGACAGTGAAATTGCCCTCTTGGAA GGCCTGACAGTGGTATACAAAAGCAGTATAGATCTCTATTTCTATGTGATTGGCAGCTCCTATGAAAATGAGCTGATGCTTATGGCTGTT CTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAATGTAGAAAAGCGAGCACTGCTGGAGAACATGGAGGGGCTGTTCTTG GCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGACAACAGCTCGAGTCACTTTATAAGAACCTCAGGATACCG TTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACCTACTTCCAGAGAACGAAAATGAGCTGGAAAAGTTCATC CACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGGAAGAGCAAAACTCAGTAACAGAAAACAAAACAAAGAAT GCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGCCGGCTGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAG ACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACATCAAAGAAAGAAATGCCAGACCTGTATCCCAGAAAATAA >18666_18666_4_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000548753_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=214AA_BP=123 MYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYENELMLMAVLNCLFD SLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLLPENENELEKFIHSVIIS -------------------------------------------------------------- >18666_18666_5_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000549043_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=956nt_BP=514nt TTGCGGCTCCACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGCCCCAGTGGGACTAGGGGAAGGTATCTGGTTTACCAG TGGTGATGGGAGGCTTCAGGACTGAAGGTATGTTCGTATCTCTTCAGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTGGACA ATGATGGAGATCGACTTTTTGCCAAGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACATTTTCAACA AGACCCATCGGACTGACAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCTCTATTTCTATGTGATTG GCAGCTCCTATGAAAATGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAATGTAGAAA AGCGAGCACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGAC AACAGCTCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACC TACTTCCAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGG AAGAGCAAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGC CGGCTGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACAT >18666_18666_5_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000549043_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=231AA_BP=140 MGGFRTEGMFVSLQEPSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGS SYENELMLMAVLNCLFDSLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLL -------------------------------------------------------------- >18666_18666_6_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000549116_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=683nt_BP=241nt GGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTGGACAATGATGGAG ATCGACTTTTTGCCAAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAATGTAGAAAAGC GAGCACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGACAAC AGCTCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACCTAC TTCCAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGGAAG AGCAAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGCCGG CTGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACATCAA >18666_18666_6_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000549116_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=169AA_BP=78 MRGKMEALILEPSLYTVKAILILDNDGDRLFAKLMLMAVLNCLFDSLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLE -------------------------------------------------------------- >18666_18666_7_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000551779_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=850nt_BP=408nt GCTGCGGGGCAAGATGGAGGCGCTGATTTTGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTGGACAATGATGGAGATCGACT TTTTGCCAAGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACATTTTCAACAAGACCCATCGGACTGA CAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCTCTATTTCTATGTGATTGGCAGCTCCTATGAAAA TGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAATGTAGAAAAGCGAGCACTGCTGGA GAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGACAACAGCTCGAGTCACT TTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACCTACTTCCAGAGAACGA AAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGGAAGAGCAAAACTCAGT AACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGCCGGCTGAGGCAGAGGA GAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACATCAAAGAAAGAAATGCC >18666_18666_7_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000551779_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=227AA_BP=136 LRGKMEALILEPSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYEN ELMLMAVLNCLFDSLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLLPENE -------------------------------------------------------------- >18666_18666_8_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000552218_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=933nt_BP=491nt GCGGCTCCACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTG GACAATGATGGAGATCGACTTTTTGCCAAGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACATTTTC AACAAGACCCATCGGACTGACAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCTCTATTTCTATGTG ATTGGCAGCTCCTATGAAAATGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGAACAGGAAGC ACCCAGGCAGGAGGACTGCATGAGTCGAGTTCAAGACAAGCTTGGGCAACACAGAAAAATGTAGAAAAGCGAGCACTGCTGGAGAACATG GAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGACAACAGCTCGAGTCACTTTATAAG AACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACCTACTTCCAGAGAACGAAAATGAG CTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGGAAGAGCAAAACTCAGTAACAGAA AACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGCCGGCTGAGGCAGAGGAGAAACTA TGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACATCAAAGAAAGAAATGCCAGACCTG >18666_18666_8_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000552218_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=248AA_BP=122 MRGKMEALILEPSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYEN ELMLMAVLNCLFDSLSQMLRTGSTQAGGLHESSSRQAWATQKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLL -------------------------------------------------------------- >18666_18666_9_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000552362_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=862nt_BP=420nt ACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGGAACCTTCCCTGTATACTGTCAAAGCCATCCTGATTCTGGACAATGA TGGAGATCGACTTTTTGCCAAGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGAGAAGAACATTTTCAACAAGAC CCATCGGACTGACAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCTCTATTTCTATGTGATTGGCAG CTCCTATGAAAATGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCTGAGGAAAAATGTAGAAAAGCG AGCACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGCACAGGCTGAGGAAAGACAACA GCTCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGAGGGTGAGTCCACGCACCTACT TCCAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATATTAAGAAGAAACTGAAGGAAGA GCAAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAACGCAGACGACAGAGGTGCCGGC TGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAACCTTATGGAAGAGGACATCAAA >18666_18666_9_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000552362_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=227AA_BP=136 MRGKMEALILEPSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYEN ELMLMAVLNCLFDSLSQMLRKNVEKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLLPENE -------------------------------------------------------------- >18666_18666_10_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000553231_C19orf18_chr19_58472919_ENST00000314391_length(transcript)=793nt_BP=351nt ACGTCGGCACCAGCTGCGGGGCAAGATGGAGGCGCTGATTTTGTACTATGACGACACCTACCCCAGTGTCAAGGAGCAAAAGGCCTTTGA GAAGAACATTTTCAACAAGACCCATCGGACTGACAGTGAAATTGCCCTCTTGGAAGGCCTGACAGTGGTATACAAAAGCAGTATAGATCT CTATTTCTATGTGATTGGCAGCTCCTATGAAAATGAGCTGATGCTTATGGCTGTTCTGAACTGTCTCTTCGACTCATTGAGCCAGATGCT GAGGAAAAATGTAGAAAAGCGAGCACTGCTGGAGAACATGGAGGGGCTGTTCTTGGCTGTGGATGAAATTGTAGATGGAGGTCGACTGGC ACAGGCTGAGGAAAGACAACAGCTCGAGTCACTTTATAAGAACCTCAGGATACCGTTATTAGGAGATGAAGAAGAGGGCTCAGAGGACGA GGGTGAGTCCACGCACCTACTTCCAGAGAACGAAAATGAGCTGGAAAAGTTCATCCACTCAGTTATTATATCAAAAAGAAGCAAAAATAT TAAGAAGAAACTGAAGGAAGAGCAAAACTCAGTAACAGAAAACAAAACAAAGAATGCGTCACATAATGGAAAAATGGAAGACTTGTGAAC GCAGACGACAGAGGTGCCGGCTGAGGCAGAGGAGAAACTATGGGGGTGCTGGGAGACTGAGCCTGTGGGCGTGGCTTGCTCCCAGAGAAC >18666_18666_10_COPZ1-C19orf18_COPZ1_chr12_54741628_ENST00000553231_C19orf18_chr19_58472919_ENST00000314391_length(amino acids)=204AA_BP=113 MRGKMEALILYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIALLEGLTVVYKSSIDLYFYVIGSSYENELMLMAVLNCLFDSLSQMLRKNV EKRALLENMEGLFLAVDEIVDGGRLAQAEERQQLESLYKNLRIPLLGDEEEGSEDEGESTHLLPENENELEKFIHSVIISKRSKNIKKKL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for COPZ1-C19orf18 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for COPZ1-C19orf18 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for COPZ1-C19orf18 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
