
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CREB3L3-PKN3 (FusionGDB2 ID:19332) |
Fusion Gene Summary for CREB3L3-PKN3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CREB3L3-PKN3 | Fusion gene ID: 19332 | Hgene | Tgene | Gene symbol | CREB3L3 | PKN3 | Gene ID | 84699 | 29941 |
| Gene name | cAMP responsive element binding protein 3 like 3 | protein kinase N3 | |
| Synonyms | CREB-H|CREBH|HYST1481 | UTDP4-1 | |
| Cytomap | 19p13.3 | 9q34.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cyclic AMP-responsive element-binding protein 3-like protein 3CREB/ATF family transcription factorcAMP-responsive element-binding protein, hepatic-specific | serine/threonine-protein kinase N3protein kinase PKN-betaprotein-kinase C-related kinase 3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q68CJ9 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000078445, ENST00000595923, ENST00000602147, ENST00000602257, ENST00000252587, | ENST00000485301, ENST00000291906, | |
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 3 X 3 X 2=18 |
| # samples | 2 | 3 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: CREB3L3 [Title/Abstract] AND PKN3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CREB3L3(4171172)-PKN3(131467582), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CREB3L3-PKN3 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. CREB3L3-PKN3 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CREB3L3-PKN3 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CREB3L3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 16469704 |
| Tgene | PKN3 | GO:0010631 | epithelial cell migration | 21754995 |
 Fusion gene breakpoints across CREB3L3 (5'-gene) Fusion gene breakpoints across CREB3L3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
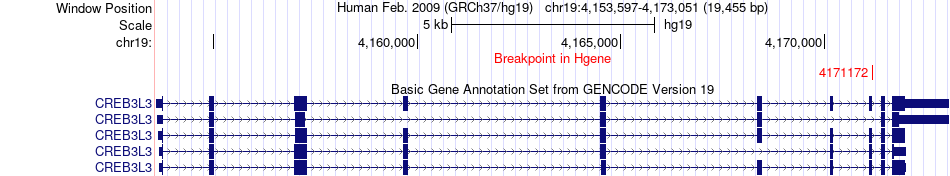 |
 Fusion gene breakpoints across PKN3 (3'-gene) Fusion gene breakpoints across PKN3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
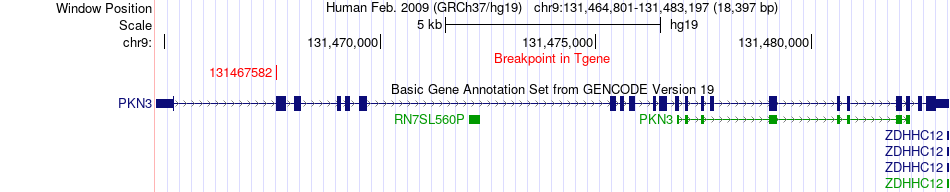 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-G3-A7M7-01A | CREB3L3 | chr19 | 4171172 | - | PKN3 | chr9 | 131467582 | + |
| ChimerDB4 | LIHC | TCGA-G3-A7M7-01A | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
Top |
Fusion Gene ORF analysis for CREB3L3-PKN3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000078445 | ENST00000485301 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| 5CDS-intron | ENST00000595923 | ENST00000485301 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| 5CDS-intron | ENST00000602147 | ENST00000485301 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| 5CDS-intron | ENST00000602257 | ENST00000485301 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| Frame-shift | ENST00000602147 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| In-frame | ENST00000078445 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| In-frame | ENST00000595923 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| In-frame | ENST00000602257 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| intron-3CDS | ENST00000252587 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
| intron-intron | ENST00000252587 | ENST00000485301 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000078445 | CREB3L3 | chr19 | 4171172 | + | ENST00000291906 | PKN3 | chr9 | 131467582 | + | 4080 | 1122 | 75 | 3767 | 1230 |
| ENST00000595923 | CREB3L3 | chr19 | 4171172 | + | ENST00000291906 | PKN3 | chr9 | 131467582 | + | 4038 | 1080 | 36 | 3725 | 1229 |
| ENST00000602257 | CREB3L3 | chr19 | 4171172 | + | ENST00000291906 | PKN3 | chr9 | 131467582 | + | 4007 | 1049 | 8 | 3694 | 1228 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000078445 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + | 0.004451758 | 0.99554825 |
| ENST00000595923 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + | 0.00435012 | 0.9956499 |
| ENST00000602257 | ENST00000291906 | CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467582 | + | 0.00439611 | 0.9956039 |
Top |
Fusion Genomic Features for CREB3L3-PKN3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467581 | + | 1.48E-07 | 0.9999999 |
| CREB3L3 | chr19 | 4171172 | + | PKN3 | chr9 | 131467581 | + | 1.48E-07 | 0.9999999 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
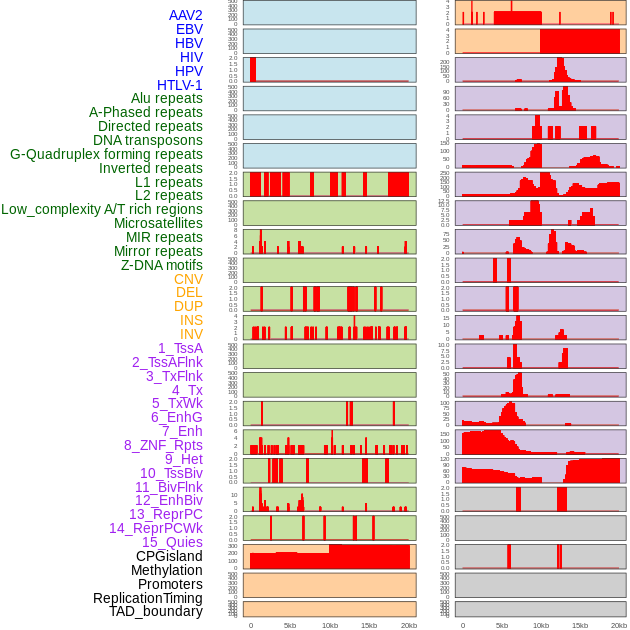 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
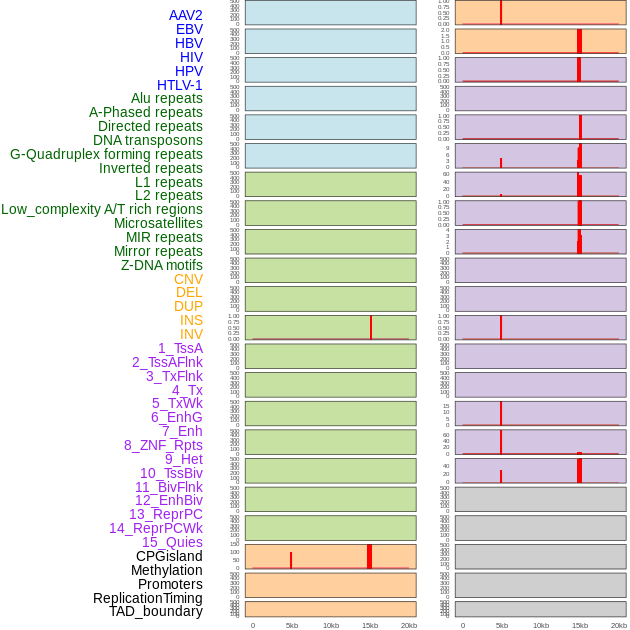 |
Top |
Fusion Protein Features for CREB3L3-PKN3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:4171172/chr9:131467582) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CREB3L3 | . |
| FUNCTION: Transcription factor that may act during endoplasmic reticulum stress by activating unfolded protein response target genes. Activated in response to cAMP stimulation. In vitro, binds to the cAMP response element (CRE) and box-B element. Activates transcription through box-B element. Activates transcription through CRE (By similarity). May function synergistically with ATF6. In acute inflammatory response, may activate expression of acute phase response (APR) genes. May be involved in growth suppression. Regulates FGF21 transcription (By similarity). {ECO:0000250, ECO:0000250|UniProtKB:Q91XE9, ECO:0000269|PubMed:11353085, ECO:0000269|PubMed:15800215, ECO:0000269|PubMed:16469704}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000078445 | + | 8 | 10 | 243_306 | 325 | 462.0 | Domain | bZIP |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000595923 | + | 8 | 10 | 243_306 | 324 | 461.0 | Domain | bZIP |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000078445 | + | 8 | 10 | 245_274 | 325 | 462.0 | Region | Basic motif |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000078445 | + | 8 | 10 | 285_306 | 325 | 462.0 | Region | Leucine-zipper |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000595923 | + | 8 | 10 | 245_274 | 324 | 461.0 | Region | Basic motif |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000595923 | + | 8 | 10 | 285_306 | 324 | 461.0 | Region | Leucine-zipper |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000078445 | + | 8 | 10 | 1_322 | 325 | 462.0 | Topological domain | Cytoplasmic |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000595923 | + | 8 | 10 | 1_322 | 324 | 461.0 | Topological domain | Cytoplasmic |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 463_554 | 8 | 890.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 165_245 | 8 | 890.0 | Domain | REM-1 3 | |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 559_818 | 8 | 890.0 | Domain | Protein kinase | |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 5_80 | 8 | 890.0 | Domain | REM-1 1 | |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 819_889 | 8 | 890.0 | Domain | AGC-kinase C-terminal | |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 93_163 | 8 | 890.0 | Domain | REM-1 2 | |
| Tgene | PKN3 | chr19:4171172 | chr9:131467582 | ENST00000291906 | 0 | 22 | 565_573 | 8 | 890.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000078445 | + | 8 | 10 | 344_461 | 325 | 462.0 | Topological domain | Lumenal |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000595923 | + | 8 | 10 | 344_461 | 324 | 461.0 | Topological domain | Lumenal |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000078445 | + | 8 | 10 | 323_343 | 325 | 462.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | CREB3L3 | chr19:4171172 | chr9:131467582 | ENST00000595923 | + | 8 | 10 | 323_343 | 324 | 461.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for CREB3L3-PKN3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19332_19332_1_CREB3L3-PKN3_CREB3L3_chr19_4171172_ENST00000078445_PKN3_chr9_131467582_ENST00000291906_length(transcript)=4080nt_BP=1122nt GTGACCGCTTCAGCATCTTTTGGGAGTGGTGACAGAGCCACAGAGGGCTGTGAGCTTGCCCGGCCCCAGGTAACGCTGGCGGTGGGTGGG CCTCCAGCTTGGAGCAGAGACCCCCCGAGGCATCTGCAGACAGAACTGGATGGACCCATGAATACGGATTTAGCTGCTGGAAAGATGGCT TCTGCTGCCTGCTCCATGGACCCCATCGACAGCTTTGAGCTCCTGGATCTCCTGTTTGACCGGCAGGACGGCATCCTGAGACACGTGGAG CTGGGCGAGGGCTGGGGTCACGTCAAGGACCAGCAGGTCCTGCCAAACCCCGACTCTGACGACTTCCTCAGCTCCATCCTGGGCTCTGGA GACTCACTGCCCAGCTCCCCACTCTGGTCCCCCGAAGGCAGTGATAGTGGCATCTCCGAAGACCTCCCCTCCGACCCCCAGGACACCCCT CCACGCAGCGGACCAGCCACCTCCCCCGCCGGCTGCCATCCTGCCCAGCCTGGCAAGGGGCCCTGCCTCTCCTATCATCCTGGCAACTCT TGCTCCACCACAACCCCAGGGCCAGTGATCCAAGTACCTGAAGCCTCTGTGACCATAGACCTGGAAATGTGGAGCCCAGGAGGAAGGATC TGTGCTGAGAAGCCGGCTGATCCGGTGGACCTGTCCCCACGATGCAATCTCACCGTGAAAGACCTCCTCCTTTCGGGCAGCAGTGGGGAC CTGCAACAGCATCACCTGGGGGCCTCCTACCTCCTGCGACCTGGGGCTGGGCACTGTCAGGAGCTGGTGCTCACCGAGGATGAGAAGAAG CTGCTGGCTAAAGAAGGCATCACCCTGCCCACTCAGCTGCCCCTCACTAAGTACGAGGAGCGAGTGCTGAAAAAAATCCGCCGGAAAATC CGGAACAAGCAGTCGGCGCAAGAAAGCAGGAAGAAGAAGAAGGAATATATCGATGGCCTGGAGACTCGGATGTCAGCTTGCACTGCTCAG AATCAGGAGTTACAGAGGAAAGTCTTGCATCTCGAGAAGCAAAACCTGTCCCTCTTGGAGCAACTGAAGAAACTCCAGGCCATTGTGGTG CAGTCCACCAGCAAGTCAGCCCAGACAGGCACCTGTGTCGCACCTGGGCCGAGCCAGTGGCCCCCAGAGGATGAGAAGGAGGTGATCCGC CGGGCCATCCAGAAAGAGCTGAAGATCAAGGAGGGGGTGGAGAACCTGCGGCGCGTGGCCACAGACCGCCGCCACTTGGGCCATGTGCAG CAGCTGCTGCGGTCCTCCAACCGCCGCCTGGAGCAGCTGCATGGCGAGCTGCGGGAGCTGCACGCCCGAATCCTGCTGCCCGGCCCTGGG CCTGGCCCAGCTGAGCCTGTGGCCTCAGGACCCCGGCCGTGGGCAGAGCAGCTCAGGGCTCGGCACCTAGAGGCTCTCCGGAGGCAGCTG CATGTGGAGCTGAAGGTGAAGCAGGGGGCTGAGAACATGACCCACACGTGCGCCAGTGGCACCCCCAAGGAGAGGAAGCTCCTGGCAGCT GCCCAGCAGATGCTGCGGGACAGCCAGCTGAAGGTGGCCCTGCTGCGGATGAAGATCAGCAGCCTGGAGGCCAGTGGGTCCCCGGAGCCA GGGCCTGAGCTGCTGGCGGAGGAGCTACAGCATCGACTGCACGTTGAGGCAGCTGTGGCTGAGGGCGCCAAGAACGTGGTGAAACTGCTT AGTAGCCGGAGAACACAGGACCGCAAGGCACTGGCTGAGGCCCAGGCCCAGCTACAGGAGTCCTCTCAGAAACTGGACCTCCTGCGCCTG GCCTTGGAGCAGCTGCTGGAGCAACTGCCTCCTGCCCACCCTTTGCGCAGCAGAGTGACCCGAGAGTTGCGGGCTGCGGTGCCTGGATAC CCCCAGCCTTCAGGGACACCTGTGAAGCCCACCGCCCTAACAGGGACACTGCAGGTCCGCCTCCTGGGCTGTGAACAGTTGCTGACAGCC GTGCCTGGGCGCTCCCCAGCGGCCGCACTGGCCAGCAGCCCCTCCGAGGGCTGGCTTCGGACCAAGGCCAAGCACCAGCGTGGCCGAGGC GAGCTTGCCAGCGAGGTGCTGGCTGTGCTAAAGGTGGACAACCGTGTTGTGGGGCAGACGGGCTGGGGGCAGGTGGCCGAACAGTCCTGG GACCAGACCTTTGTCATCCCACTGGAGCGAGCCCGTGAGCTGGAGATTGGGGTACACTGGCGGGACTGGCGGCAGCTATGTGGCGTGGCC TTCCTGAGACTTGAGGACTTCCTGGACAATGCCTGTCACCAACTGTCCCTCAGCCTGGTACCGCAGGGACTGCTTTTTGCCCAGGTGACC TTCTGCGATCCTGTCATTGAGAGGCGGCCCCGGCTGCAGAGGCAGGAACGCATCTTCTCTAAACGCAGAGGCCAGGACTTCCTGAGGGCT TCGCAGATGAACCTCGGCATGGCGGCCTGGGGGCGCCTCGTCATGAACCTGCTGCCCCCCTGCAGCTCCCCGAGCACAATCAGCCCCCCT AAAGGATGCCCTCGGACCCCAACAACACTGCGAGAGGCCTCTGACCCTGCCACTCCCAGTAATTTCCTGCCCAAGAAGACCCCCTTGGGT GAAGAGATGACACCCCCACCCAAGCCCCCACGCCTCTACCTCCCCCAGGAGCCAACATCCGAGGAGACTCCGCGCACCAAACGTCCCCAT ATGGAGCCTAGGACTCGACGTGGGCCATCTCCACCAGCCTCCCCCACCAGGAAACCCCCTCGGCTTCAGGACTTCCGCTGCTTAGCTGTG CTGGGCCGGGGACACTTTGGGAAGGTCCTCCTGGTCCAGTTCAAGGGGACAGGGAAATACTACGCCATCAAAGCACTGAAGAAGCAGGAG GTGCTCAGCCGGGACGAGATAGAGAGCCTGTACTGCGAGAAGCGGATCCTGGAGGCTGTGGGCTGCACAGGGCACCCTTTCCTGCTCTCC CTCCTTGCCTGCTTCCAGACCTCCAGCCATGCCTGCTTTGTGACTGAGTTTGTGCCTGGTGGTGACCTCATGATGCAGATCCACGAGGAT GTCTTCCCCGAGCCCCAGGCCCGCTTCTACGTGGCTTGTGTTGTCCTGGGGCTGCAGTTCTTACACGAGAAGAAGATCATTTACAGGGAC CTGAAGTTGGATAACCTTCTGCTGGATGCCCAGGGATTCCTGAAGATCGCAGACTTTGGACTCTGCAAGGAAGGGATCGGCTTCGGGGAC CGGACTAGCACCTTCTGTGGCACCCCGGAGTTCCTGGCTCCCGAGGTGCTGACCCAGGAGGCATACACACGGGCTGTGGACTGGTGGGGG CTGGGTGTGCTGCTCTACGAGATGCTGGTGGGTGAGTGCCCGTTCCCAGGGGACACAGAGGAAGAGGTGTTTGACTGCATCGTCAACATG GACGCCCCCTACCCCGGCTTTCTGTCGGTGCAAGGGCTTGAGTTCATTCAGAAGCTCCTCCAGAAGTGCCCGGAGAAGCGCCTCGGGGCA GGTGAGCAGGATGCCGAGGAGATCAAGGTCCAGCCATTCTTCAGGACCACCAACTGGCAAGCCCTGCTCGCCCGCACCATCCAGCCCCCC TTCGTGCCTACCCTGTGTGGCCCTGCGGACCTGCGCTACTTTGAGGGCGAGTTCACAGGGCTGCCGCCTGCCCTGACCCCACCTGCACCC CACAGCCTCCTCACTGCCCGCCAACAGGCCGCCTTCCGGGACTTCGACTTTGTGTCAGAGCGATTCCTGGAACCCTGAGGGCATCTCCTG GCACCTCTGTCCCCTTCCCCCACAGACTGTTAGAGCCTCTGCTCGTTCACCCGTGCGCCCTGCCTGGAGGTCCAGGCCTTGCTGGGTACT TCTGAGCCCTTGGGATTCAAAGTGGCAGCCATGGGGCCACTGTTGTGGGCTTTGCTCAGTGTCACTGGGCAAAGTGTGTCCCTTCCCCCT CCAGCTCGCCCTCTTCTACCTCCCAGCGAGACCTGGCCCAGAAAGGGTGCCGCAGCAAGGAGTGATATGGTTTGTCTTTTTAAGACTGGA >19332_19332_1_CREB3L3-PKN3_CREB3L3_chr19_4171172_ENST00000078445_PKN3_chr9_131467582_ENST00000291906_length(amino acids)=1230AA_BP=349 MAVGGPPAWSRDPPRHLQTELDGPMNTDLAAGKMASAACSMDPIDSFELLDLLFDRQDGILRHVELGEGWGHVKDQQVLPNPDSDDFLSS ILGSGDSLPSSPLWSPEGSDSGISEDLPSDPQDTPPRSGPATSPAGCHPAQPGKGPCLSYHPGNSCSTTTPGPVIQVPEASVTIDLEMWS PGGRICAEKPADPVDLSPRCNLTVKDLLLSGSSGDLQQHHLGASYLLRPGAGHCQELVLTEDEKKLLAKEGITLPTQLPLTKYEERVLKK IRRKIRNKQSAQESRKKKKEYIDGLETRMSACTAQNQELQRKVLHLEKQNLSLLEQLKKLQAIVVQSTSKSAQTGTCVAPGPSQWPPEDE KEVIRRAIQKELKIKEGVENLRRVATDRRHLGHVQQLLRSSNRRLEQLHGELRELHARILLPGPGPGPAEPVASGPRPWAEQLRARHLEA LRRQLHVELKVKQGAENMTHTCASGTPKERKLLAAAQQMLRDSQLKVALLRMKISSLEASGSPEPGPELLAEELQHRLHVEAAVAEGAKN VVKLLSSRRTQDRKALAEAQAQLQESSQKLDLLRLALEQLLEQLPPAHPLRSRVTRELRAAVPGYPQPSGTPVKPTALTGTLQVRLLGCE QLLTAVPGRSPAAALASSPSEGWLRTKAKHQRGRGELASEVLAVLKVDNRVVGQTGWGQVAEQSWDQTFVIPLERARELEIGVHWRDWRQ LCGVAFLRLEDFLDNACHQLSLSLVPQGLLFAQVTFCDPVIERRPRLQRQERIFSKRRGQDFLRASQMNLGMAAWGRLVMNLLPPCSSPS TISPPKGCPRTPTTLREASDPATPSNFLPKKTPLGEEMTPPPKPPRLYLPQEPTSEETPRTKRPHMEPRTRRGPSPPASPTRKPPRLQDF RCLAVLGRGHFGKVLLVQFKGTGKYYAIKALKKQEVLSRDEIESLYCEKRILEAVGCTGHPFLLSLLACFQTSSHACFVTEFVPGGDLMM QIHEDVFPEPQARFYVACVVLGLQFLHEKKIIYRDLKLDNLLLDAQGFLKIADFGLCKEGIGFGDRTSTFCGTPEFLAPEVLTQEAYTRA VDWWGLGVLLYEMLVGECPFPGDTEEEVFDCIVNMDAPYPGFLSVQGLEFIQKLLQKCPEKRLGAGEQDAEEIKVQPFFRTTNWQALLAR -------------------------------------------------------------- >19332_19332_2_CREB3L3-PKN3_CREB3L3_chr19_4171172_ENST00000595923_PKN3_chr9_131467582_ENST00000291906_length(transcript)=4038nt_BP=1080nt ACAGAGGGCTGTGAGCTTGCCCGGCCCCAGGTAACGCTGGCGGTGGGTGGGCCTCCAGCTTGGAGCAGAGACCCCCCGAGGCATCTGCAG ACAGAACTGGATGGACCCATGAATACGGATTTAGCTGCTGGAAAGATGGCTTCTGCTGCCTGCTCCATGGACCCCATCGACAGCTTTGAG CTCCTGGATCTCCTGTTTGACCGGCAGGACGGCATCCTGAGACACGTGGAGCTGGGCGAGGGCTGGGGTCACGTCAAGGACCAGGTCCTG CCAAACCCCGACTCTGACGACTTCCTCAGCTCCATCCTGGGCTCTGGAGACTCACTGCCCAGCTCCCCACTCTGGTCCCCCGAAGGCAGT GATAGTGGCATCTCCGAAGACCTCCCCTCCGACCCCCAGGACACCCCTCCACGCAGCGGACCAGCCACCTCCCCCGCCGGCTGCCATCCT GCCCAGCCTGGCAAGGGGCCCTGCCTCTCCTATCATCCTGGCAACTCTTGCTCCACCACAACCCCAGGGCCAGTGATCCAAGTACCTGAA GCCTCTGTGACCATAGACCTGGAAATGTGGAGCCCAGGAGGAAGGATCTGTGCTGAGAAGCCGGCTGATCCGGTGGACCTGTCCCCACGA TGCAATCTCACCGTGAAAGACCTCCTCCTTTCGGGCAGCAGTGGGGACCTGCAACAGCATCACCTGGGGGCCTCCTACCTCCTGCGACCT GGGGCTGGGCACTGTCAGGAGCTGGTGCTCACCGAGGATGAGAAGAAGCTGCTGGCTAAAGAAGGCATCACCCTGCCCACTCAGCTGCCC CTCACTAAGTACGAGGAGCGAGTGCTGAAAAAAATCCGCCGGAAAATCCGGAACAAGCAGTCGGCGCAAGAAAGCAGGAAGAAGAAGAAG GAATATATCGATGGCCTGGAGACTCGGATGTCAGCTTGCACTGCTCAGAATCAGGAGTTACAGAGGAAAGTCTTGCATCTCGAGAAGCAA AACCTGTCCCTCTTGGAGCAACTGAAGAAACTCCAGGCCATTGTGGTGCAGTCCACCAGCAAGTCAGCCCAGACAGGCACCTGTGTCGCA CCTGGGCCGAGCCAGTGGCCCCCAGAGGATGAGAAGGAGGTGATCCGCCGGGCCATCCAGAAAGAGCTGAAGATCAAGGAGGGGGTGGAG AACCTGCGGCGCGTGGCCACAGACCGCCGCCACTTGGGCCATGTGCAGCAGCTGCTGCGGTCCTCCAACCGCCGCCTGGAGCAGCTGCAT GGCGAGCTGCGGGAGCTGCACGCCCGAATCCTGCTGCCCGGCCCTGGGCCTGGCCCAGCTGAGCCTGTGGCCTCAGGACCCCGGCCGTGG GCAGAGCAGCTCAGGGCTCGGCACCTAGAGGCTCTCCGGAGGCAGCTGCATGTGGAGCTGAAGGTGAAGCAGGGGGCTGAGAACATGACC CACACGTGCGCCAGTGGCACCCCCAAGGAGAGGAAGCTCCTGGCAGCTGCCCAGCAGATGCTGCGGGACAGCCAGCTGAAGGTGGCCCTG CTGCGGATGAAGATCAGCAGCCTGGAGGCCAGTGGGTCCCCGGAGCCAGGGCCTGAGCTGCTGGCGGAGGAGCTACAGCATCGACTGCAC GTTGAGGCAGCTGTGGCTGAGGGCGCCAAGAACGTGGTGAAACTGCTTAGTAGCCGGAGAACACAGGACCGCAAGGCACTGGCTGAGGCC CAGGCCCAGCTACAGGAGTCCTCTCAGAAACTGGACCTCCTGCGCCTGGCCTTGGAGCAGCTGCTGGAGCAACTGCCTCCTGCCCACCCT TTGCGCAGCAGAGTGACCCGAGAGTTGCGGGCTGCGGTGCCTGGATACCCCCAGCCTTCAGGGACACCTGTGAAGCCCACCGCCCTAACA GGGACACTGCAGGTCCGCCTCCTGGGCTGTGAACAGTTGCTGACAGCCGTGCCTGGGCGCTCCCCAGCGGCCGCACTGGCCAGCAGCCCC TCCGAGGGCTGGCTTCGGACCAAGGCCAAGCACCAGCGTGGCCGAGGCGAGCTTGCCAGCGAGGTGCTGGCTGTGCTAAAGGTGGACAAC CGTGTTGTGGGGCAGACGGGCTGGGGGCAGGTGGCCGAACAGTCCTGGGACCAGACCTTTGTCATCCCACTGGAGCGAGCCCGTGAGCTG GAGATTGGGGTACACTGGCGGGACTGGCGGCAGCTATGTGGCGTGGCCTTCCTGAGACTTGAGGACTTCCTGGACAATGCCTGTCACCAA CTGTCCCTCAGCCTGGTACCGCAGGGACTGCTTTTTGCCCAGGTGACCTTCTGCGATCCTGTCATTGAGAGGCGGCCCCGGCTGCAGAGG CAGGAACGCATCTTCTCTAAACGCAGAGGCCAGGACTTCCTGAGGGCTTCGCAGATGAACCTCGGCATGGCGGCCTGGGGGCGCCTCGTC ATGAACCTGCTGCCCCCCTGCAGCTCCCCGAGCACAATCAGCCCCCCTAAAGGATGCCCTCGGACCCCAACAACACTGCGAGAGGCCTCT GACCCTGCCACTCCCAGTAATTTCCTGCCCAAGAAGACCCCCTTGGGTGAAGAGATGACACCCCCACCCAAGCCCCCACGCCTCTACCTC CCCCAGGAGCCAACATCCGAGGAGACTCCGCGCACCAAACGTCCCCATATGGAGCCTAGGACTCGACGTGGGCCATCTCCACCAGCCTCC CCCACCAGGAAACCCCCTCGGCTTCAGGACTTCCGCTGCTTAGCTGTGCTGGGCCGGGGACACTTTGGGAAGGTCCTCCTGGTCCAGTTC AAGGGGACAGGGAAATACTACGCCATCAAAGCACTGAAGAAGCAGGAGGTGCTCAGCCGGGACGAGATAGAGAGCCTGTACTGCGAGAAG CGGATCCTGGAGGCTGTGGGCTGCACAGGGCACCCTTTCCTGCTCTCCCTCCTTGCCTGCTTCCAGACCTCCAGCCATGCCTGCTTTGTG ACTGAGTTTGTGCCTGGTGGTGACCTCATGATGCAGATCCACGAGGATGTCTTCCCCGAGCCCCAGGCCCGCTTCTACGTGGCTTGTGTT GTCCTGGGGCTGCAGTTCTTACACGAGAAGAAGATCATTTACAGGGACCTGAAGTTGGATAACCTTCTGCTGGATGCCCAGGGATTCCTG AAGATCGCAGACTTTGGACTCTGCAAGGAAGGGATCGGCTTCGGGGACCGGACTAGCACCTTCTGTGGCACCCCGGAGTTCCTGGCTCCC GAGGTGCTGACCCAGGAGGCATACACACGGGCTGTGGACTGGTGGGGGCTGGGTGTGCTGCTCTACGAGATGCTGGTGGGTGAGTGCCCG TTCCCAGGGGACACAGAGGAAGAGGTGTTTGACTGCATCGTCAACATGGACGCCCCCTACCCCGGCTTTCTGTCGGTGCAAGGGCTTGAG TTCATTCAGAAGCTCCTCCAGAAGTGCCCGGAGAAGCGCCTCGGGGCAGGTGAGCAGGATGCCGAGGAGATCAAGGTCCAGCCATTCTTC AGGACCACCAACTGGCAAGCCCTGCTCGCCCGCACCATCCAGCCCCCCTTCGTGCCTACCCTGTGTGGCCCTGCGGACCTGCGCTACTTT GAGGGCGAGTTCACAGGGCTGCCGCCTGCCCTGACCCCACCTGCACCCCACAGCCTCCTCACTGCCCGCCAACAGGCCGCCTTCCGGGAC TTCGACTTTGTGTCAGAGCGATTCCTGGAACCCTGAGGGCATCTCCTGGCACCTCTGTCCCCTTCCCCCACAGACTGTTAGAGCCTCTGC TCGTTCACCCGTGCGCCCTGCCTGGAGGTCCAGGCCTTGCTGGGTACTTCTGAGCCCTTGGGATTCAAAGTGGCAGCCATGGGGCCACTG TTGTGGGCTTTGCTCAGTGTCACTGGGCAAAGTGTGTCCCTTCCCCCTCCAGCTCGCCCTCTTCTACCTCCCAGCGAGACCTGGCCCAGA >19332_19332_2_CREB3L3-PKN3_CREB3L3_chr19_4171172_ENST00000595923_PKN3_chr9_131467582_ENST00000291906_length(amino acids)=1229AA_BP=348 MAVGGPPAWSRDPPRHLQTELDGPMNTDLAAGKMASAACSMDPIDSFELLDLLFDRQDGILRHVELGEGWGHVKDQVLPNPDSDDFLSSI LGSGDSLPSSPLWSPEGSDSGISEDLPSDPQDTPPRSGPATSPAGCHPAQPGKGPCLSYHPGNSCSTTTPGPVIQVPEASVTIDLEMWSP GGRICAEKPADPVDLSPRCNLTVKDLLLSGSSGDLQQHHLGASYLLRPGAGHCQELVLTEDEKKLLAKEGITLPTQLPLTKYEERVLKKI RRKIRNKQSAQESRKKKKEYIDGLETRMSACTAQNQELQRKVLHLEKQNLSLLEQLKKLQAIVVQSTSKSAQTGTCVAPGPSQWPPEDEK EVIRRAIQKELKIKEGVENLRRVATDRRHLGHVQQLLRSSNRRLEQLHGELRELHARILLPGPGPGPAEPVASGPRPWAEQLRARHLEAL RRQLHVELKVKQGAENMTHTCASGTPKERKLLAAAQQMLRDSQLKVALLRMKISSLEASGSPEPGPELLAEELQHRLHVEAAVAEGAKNV VKLLSSRRTQDRKALAEAQAQLQESSQKLDLLRLALEQLLEQLPPAHPLRSRVTRELRAAVPGYPQPSGTPVKPTALTGTLQVRLLGCEQ LLTAVPGRSPAAALASSPSEGWLRTKAKHQRGRGELASEVLAVLKVDNRVVGQTGWGQVAEQSWDQTFVIPLERARELEIGVHWRDWRQL CGVAFLRLEDFLDNACHQLSLSLVPQGLLFAQVTFCDPVIERRPRLQRQERIFSKRRGQDFLRASQMNLGMAAWGRLVMNLLPPCSSPST ISPPKGCPRTPTTLREASDPATPSNFLPKKTPLGEEMTPPPKPPRLYLPQEPTSEETPRTKRPHMEPRTRRGPSPPASPTRKPPRLQDFR CLAVLGRGHFGKVLLVQFKGTGKYYAIKALKKQEVLSRDEIESLYCEKRILEAVGCTGHPFLLSLLACFQTSSHACFVTEFVPGGDLMMQ IHEDVFPEPQARFYVACVVLGLQFLHEKKIIYRDLKLDNLLLDAQGFLKIADFGLCKEGIGFGDRTSTFCGTPEFLAPEVLTQEAYTRAV DWWGLGVLLYEMLVGECPFPGDTEEEVFDCIVNMDAPYPGFLSVQGLEFIQKLLQKCPEKRLGAGEQDAEEIKVQPFFRTTNWQALLART -------------------------------------------------------------- >19332_19332_3_CREB3L3-PKN3_CREB3L3_chr19_4171172_ENST00000602257_PKN3_chr9_131467582_ENST00000291906_length(transcript)=4007nt_BP=1049nt AGGTAACGCTGGCGGTGGGTGGGCCTCCAGCTTGGAGCAGAGACCCCCCGAGGCATCTGCAGACAGAACTGGATGGACCCATGAATACGG ATTTAGCTGCTGGAAAGATGGCTTCTGCTGCCTGCTCCATGGACCCCATCGACAGCTTTGAGCTCCTGGATCTCCTGTTTGACCGGCAGG ACGGCATCCTGAGACACGTGGAGCTGGGCGAGGGCTGGGGTCACGTCAAGGACCAGCAGGTCCTGCCAAACCCCGACTCTGACGACTTCC TCAGCTCCATCCTGGGCTCTGGAGACTCACTGCCCAGCTCCCCACTCTGGTCCCCCGAAGGCAGTGATAGTGGCATCTCCGAAGACCTCC CCTCCGACCCCCAGGACACCCCTCCACGCAGCGGACCAGCCACCTCCCCCGCCGGCTGCCATCCTGCCCAGCCTGGCAAGGGGCCCTGCC TCTCCTATCATCCTGGCAACTCTTGCTCCACCACAACCCCAGGGCCAGTGATCCAAGTACCTGAAGCCTCTGTGACCATAGACCTGGAAA TGTGGAGCCCAGGAGGAAGGATCTGTGCTGAGAAGCCGGCTGATCCGGTGGACCTGTCCCCACGATGCAATCTCACCGTGAAAGACCTCC TCCTTTCGGGCAGCAGTGGGGACCTGCATCACCTGGGGGCCTCCTACCTCCTGCGACCTGGGGCTGGGCACTGTCAGGAGCTGGTGCTCA CCGAGGATGAGAAGAAGCTGCTGGCTAAAGAAGGCATCACCCTGCCCACTCAGCTGCCCCTCACTAAGTACGAGGAGCGAGTGCTGAAAA AAATCCGCCGGAAAATCCGGAACAAGCAGTCGGCGCAAGAAAGCAGGAAGAAGAAGAAGGAATATATCGATGGCCTGGAGACTCGGATGT CAGCTTGCACTGCTCAGAATCAGGAGTTACAGAGGAAAGTCTTGCATCTCGAGAAGCAAAACCTGTCCCTCTTGGAGCAACTGAAGAAAC TCCAGGCCATTGTGGTGCAGTCCACCAGCAAGTCAGCCCAGACAGGCACCTGTGTCGCACCTGGGCCGAGCCAGTGGCCCCCAGAGGATG AGAAGGAGGTGATCCGCCGGGCCATCCAGAAAGAGCTGAAGATCAAGGAGGGGGTGGAGAACCTGCGGCGCGTGGCCACAGACCGCCGCC ACTTGGGCCATGTGCAGCAGCTGCTGCGGTCCTCCAACCGCCGCCTGGAGCAGCTGCATGGCGAGCTGCGGGAGCTGCACGCCCGAATCC TGCTGCCCGGCCCTGGGCCTGGCCCAGCTGAGCCTGTGGCCTCAGGACCCCGGCCGTGGGCAGAGCAGCTCAGGGCTCGGCACCTAGAGG CTCTCCGGAGGCAGCTGCATGTGGAGCTGAAGGTGAAGCAGGGGGCTGAGAACATGACCCACACGTGCGCCAGTGGCACCCCCAAGGAGA GGAAGCTCCTGGCAGCTGCCCAGCAGATGCTGCGGGACAGCCAGCTGAAGGTGGCCCTGCTGCGGATGAAGATCAGCAGCCTGGAGGCCA GTGGGTCCCCGGAGCCAGGGCCTGAGCTGCTGGCGGAGGAGCTACAGCATCGACTGCACGTTGAGGCAGCTGTGGCTGAGGGCGCCAAGA ACGTGGTGAAACTGCTTAGTAGCCGGAGAACACAGGACCGCAAGGCACTGGCTGAGGCCCAGGCCCAGCTACAGGAGTCCTCTCAGAAAC TGGACCTCCTGCGCCTGGCCTTGGAGCAGCTGCTGGAGCAACTGCCTCCTGCCCACCCTTTGCGCAGCAGAGTGACCCGAGAGTTGCGGG CTGCGGTGCCTGGATACCCCCAGCCTTCAGGGACACCTGTGAAGCCCACCGCCCTAACAGGGACACTGCAGGTCCGCCTCCTGGGCTGTG AACAGTTGCTGACAGCCGTGCCTGGGCGCTCCCCAGCGGCCGCACTGGCCAGCAGCCCCTCCGAGGGCTGGCTTCGGACCAAGGCCAAGC ACCAGCGTGGCCGAGGCGAGCTTGCCAGCGAGGTGCTGGCTGTGCTAAAGGTGGACAACCGTGTTGTGGGGCAGACGGGCTGGGGGCAGG TGGCCGAACAGTCCTGGGACCAGACCTTTGTCATCCCACTGGAGCGAGCCCGTGAGCTGGAGATTGGGGTACACTGGCGGGACTGGCGGC AGCTATGTGGCGTGGCCTTCCTGAGACTTGAGGACTTCCTGGACAATGCCTGTCACCAACTGTCCCTCAGCCTGGTACCGCAGGGACTGC TTTTTGCCCAGGTGACCTTCTGCGATCCTGTCATTGAGAGGCGGCCCCGGCTGCAGAGGCAGGAACGCATCTTCTCTAAACGCAGAGGCC AGGACTTCCTGAGGGCTTCGCAGATGAACCTCGGCATGGCGGCCTGGGGGCGCCTCGTCATGAACCTGCTGCCCCCCTGCAGCTCCCCGA GCACAATCAGCCCCCCTAAAGGATGCCCTCGGACCCCAACAACACTGCGAGAGGCCTCTGACCCTGCCACTCCCAGTAATTTCCTGCCCA AGAAGACCCCCTTGGGTGAAGAGATGACACCCCCACCCAAGCCCCCACGCCTCTACCTCCCCCAGGAGCCAACATCCGAGGAGACTCCGC GCACCAAACGTCCCCATATGGAGCCTAGGACTCGACGTGGGCCATCTCCACCAGCCTCCCCCACCAGGAAACCCCCTCGGCTTCAGGACT TCCGCTGCTTAGCTGTGCTGGGCCGGGGACACTTTGGGAAGGTCCTCCTGGTCCAGTTCAAGGGGACAGGGAAATACTACGCCATCAAAG CACTGAAGAAGCAGGAGGTGCTCAGCCGGGACGAGATAGAGAGCCTGTACTGCGAGAAGCGGATCCTGGAGGCTGTGGGCTGCACAGGGC ACCCTTTCCTGCTCTCCCTCCTTGCCTGCTTCCAGACCTCCAGCCATGCCTGCTTTGTGACTGAGTTTGTGCCTGGTGGTGACCTCATGA TGCAGATCCACGAGGATGTCTTCCCCGAGCCCCAGGCCCGCTTCTACGTGGCTTGTGTTGTCCTGGGGCTGCAGTTCTTACACGAGAAGA AGATCATTTACAGGGACCTGAAGTTGGATAACCTTCTGCTGGATGCCCAGGGATTCCTGAAGATCGCAGACTTTGGACTCTGCAAGGAAG GGATCGGCTTCGGGGACCGGACTAGCACCTTCTGTGGCACCCCGGAGTTCCTGGCTCCCGAGGTGCTGACCCAGGAGGCATACACACGGG CTGTGGACTGGTGGGGGCTGGGTGTGCTGCTCTACGAGATGCTGGTGGGTGAGTGCCCGTTCCCAGGGGACACAGAGGAAGAGGTGTTTG ACTGCATCGTCAACATGGACGCCCCCTACCCCGGCTTTCTGTCGGTGCAAGGGCTTGAGTTCATTCAGAAGCTCCTCCAGAAGTGCCCGG AGAAGCGCCTCGGGGCAGGTGAGCAGGATGCCGAGGAGATCAAGGTCCAGCCATTCTTCAGGACCACCAACTGGCAAGCCCTGCTCGCCC GCACCATCCAGCCCCCCTTCGTGCCTACCCTGTGTGGCCCTGCGGACCTGCGCTACTTTGAGGGCGAGTTCACAGGGCTGCCGCCTGCCC TGACCCCACCTGCACCCCACAGCCTCCTCACTGCCCGCCAACAGGCCGCCTTCCGGGACTTCGACTTTGTGTCAGAGCGATTCCTGGAAC CCTGAGGGCATCTCCTGGCACCTCTGTCCCCTTCCCCCACAGACTGTTAGAGCCTCTGCTCGTTCACCCGTGCGCCCTGCCTGGAGGTCC AGGCCTTGCTGGGTACTTCTGAGCCCTTGGGATTCAAAGTGGCAGCCATGGGGCCACTGTTGTGGGCTTTGCTCAGTGTCACTGGGCAAA GTGTGTCCCTTCCCCCTCCAGCTCGCCCTCTTCTACCTCCCAGCGAGACCTGGCCCAGAAAGGGTGCCGCAGCAAGGAGTGATATGGTTT >19332_19332_3_CREB3L3-PKN3_CREB3L3_chr19_4171172_ENST00000602257_PKN3_chr9_131467582_ENST00000291906_length(amino acids)=1228AA_BP=347 MAVGGPPAWSRDPPRHLQTELDGPMNTDLAAGKMASAACSMDPIDSFELLDLLFDRQDGILRHVELGEGWGHVKDQQVLPNPDSDDFLSS ILGSGDSLPSSPLWSPEGSDSGISEDLPSDPQDTPPRSGPATSPAGCHPAQPGKGPCLSYHPGNSCSTTTPGPVIQVPEASVTIDLEMWS PGGRICAEKPADPVDLSPRCNLTVKDLLLSGSSGDLHHLGASYLLRPGAGHCQELVLTEDEKKLLAKEGITLPTQLPLTKYEERVLKKIR RKIRNKQSAQESRKKKKEYIDGLETRMSACTAQNQELQRKVLHLEKQNLSLLEQLKKLQAIVVQSTSKSAQTGTCVAPGPSQWPPEDEKE VIRRAIQKELKIKEGVENLRRVATDRRHLGHVQQLLRSSNRRLEQLHGELRELHARILLPGPGPGPAEPVASGPRPWAEQLRARHLEALR RQLHVELKVKQGAENMTHTCASGTPKERKLLAAAQQMLRDSQLKVALLRMKISSLEASGSPEPGPELLAEELQHRLHVEAAVAEGAKNVV KLLSSRRTQDRKALAEAQAQLQESSQKLDLLRLALEQLLEQLPPAHPLRSRVTRELRAAVPGYPQPSGTPVKPTALTGTLQVRLLGCEQL LTAVPGRSPAAALASSPSEGWLRTKAKHQRGRGELASEVLAVLKVDNRVVGQTGWGQVAEQSWDQTFVIPLERARELEIGVHWRDWRQLC GVAFLRLEDFLDNACHQLSLSLVPQGLLFAQVTFCDPVIERRPRLQRQERIFSKRRGQDFLRASQMNLGMAAWGRLVMNLLPPCSSPSTI SPPKGCPRTPTTLREASDPATPSNFLPKKTPLGEEMTPPPKPPRLYLPQEPTSEETPRTKRPHMEPRTRRGPSPPASPTRKPPRLQDFRC LAVLGRGHFGKVLLVQFKGTGKYYAIKALKKQEVLSRDEIESLYCEKRILEAVGCTGHPFLLSLLACFQTSSHACFVTEFVPGGDLMMQI HEDVFPEPQARFYVACVVLGLQFLHEKKIIYRDLKLDNLLLDAQGFLKIADFGLCKEGIGFGDRTSTFCGTPEFLAPEVLTQEAYTRAVD WWGLGVLLYEMLVGECPFPGDTEEEVFDCIVNMDAPYPGFLSVQGLEFIQKLLQKCPEKRLGAGEQDAEEIKVQPFFRTTNWQALLARTI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CREB3L3-PKN3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CREB3L3-PKN3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CREB3L3-PKN3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
