
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CREBBP-ADCY9 (FusionGDB2 ID:19349) |
Fusion Gene Summary for CREBBP-ADCY9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CREBBP-ADCY9 | Fusion gene ID: 19349 | Hgene | Tgene | Gene symbol | CREBBP | ADCY9 | Gene ID | 1387 | 115 |
| Gene name | CREB binding protein | adenylate cyclase 9 | |
| Synonyms | CBP|KAT3A|MKHK1|RSTS|RSTS1 | AC9|ACIX | |
| Cytomap | 16p13.3 | 16p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CREB-binding proteinhistone lysine acetyltransferase CREBBPprotein-lysine acetyltransferase CREBBP | adenylate cyclase type 9ATP pyrophosphate-lyase 9adenylate cyclase type IXadenylyl cyclase 9type IX ATP pyrophosphate-lyase | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q92793 | O60503 | |
| Ensembl transtripts involved in fusion gene | ENST00000262367, ENST00000382070, | ENST00000571889, ENST00000294016, | |
| Fusion gene scores | * DoF score | 43 X 38 X 17=27778 | 8 X 7 X 6=336 |
| # samples | 61 | 9 | |
| ** MAII score | log2(61/27778*10)=-5.50898967694577 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/336*10)=-1.90046432644909 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CREBBP [Title/Abstract] AND ADCY9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ADCY9(4163751)-CREBBP(3901010), # samples:2 CREBBP(3929833)-ADCY9(4016967), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CREBBP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 21539536 |
| Hgene | CREBBP | GO:0006355 | regulation of transcription, DNA-templated | 12169688 |
| Hgene | CREBBP | GO:0006473 | protein acetylation | 15273251|24207024|24939902|28790157|30540930 |
| Hgene | CREBBP | GO:0016573 | histone acetylation | 11742995 |
| Hgene | CREBBP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 12435739 |
| Hgene | CREBBP | GO:0034644 | cellular response to UV | 24939902 |
| Hgene | CREBBP | GO:0045893 | positive regulation of transcription, DNA-templated | 11742995 |
| Hgene | CREBBP | GO:1990258 | histone glutamine methylation | 30540930 |
| Tgene | ADCY9 | GO:0006171 | cAMP biosynthetic process | 9628827|10987815 |
| Tgene | ADCY9 | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 9628827 |
 Fusion gene breakpoints across CREBBP (5'-gene) Fusion gene breakpoints across CREBBP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
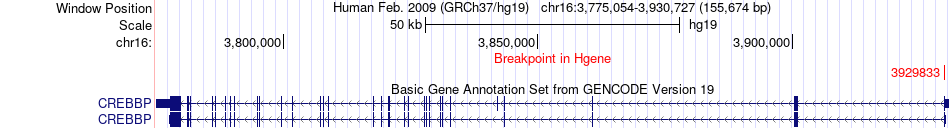 |
 Fusion gene breakpoints across ADCY9 (3'-gene) Fusion gene breakpoints across ADCY9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
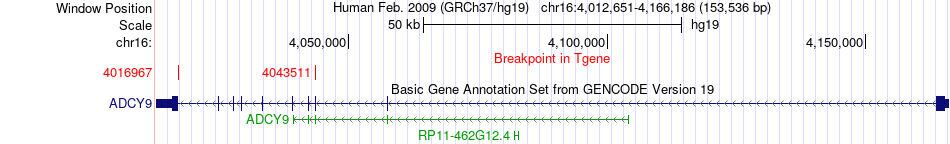 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-UY-A9PB-01A | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4016967 | - |
| ChimerDB4 | ESCA | TCGA-L5-A43M-01A | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - |
Top |
Fusion Gene ORF analysis for CREBBP-ADCY9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000262367 | ENST00000571889 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - |
| 5CDS-5UTR | ENST00000382070 | ENST00000571889 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - |
| 5CDS-intron | ENST00000262367 | ENST00000571889 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4016967 | - |
| 5CDS-intron | ENST00000382070 | ENST00000571889 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4016967 | - |
| In-frame | ENST00000262367 | ENST00000294016 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4016967 | - |
| In-frame | ENST00000262367 | ENST00000294016 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - |
| In-frame | ENST00000382070 | ENST00000294016 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4016967 | - |
| In-frame | ENST00000382070 | ENST00000294016 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262367 | CREBBP | chr16 | 3929833 | - | ENST00000294016 | ADCY9 | chr16 | 4043511 | - | 6197 | 895 | 814 | 3072 | 752 |
| ENST00000382070 | CREBBP | chr16 | 3929833 | - | ENST00000294016 | ADCY9 | chr16 | 4043511 | - | 5591 | 289 | 208 | 2466 | 752 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262367 | ENST00000294016 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - | 0.0017865 | 0.9982135 |
| ENST00000382070 | ENST00000294016 | CREBBP | chr16 | 3929833 | - | ADCY9 | chr16 | 4043511 | - | 0.001475505 | 0.9985245 |
Top |
Fusion Genomic Features for CREBBP-ADCY9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
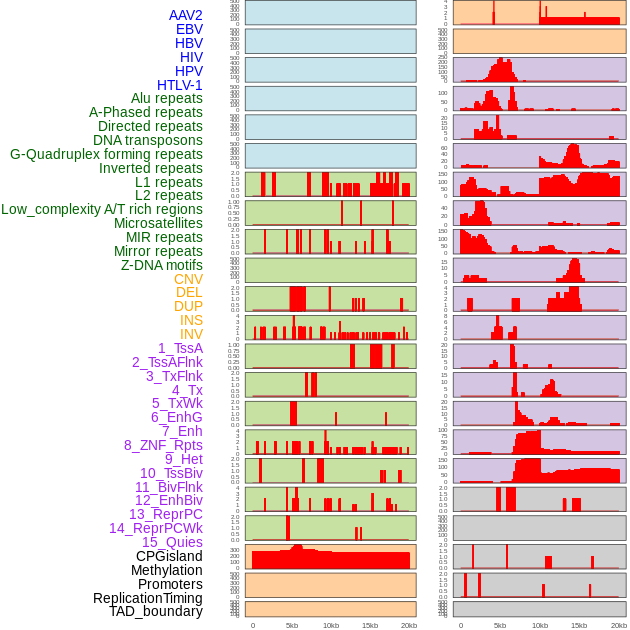 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CREBBP-ADCY9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:4163751/chr16:3901010) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CREBBP | ADCY9 |
| FUNCTION: Acetylates histones, giving a specific tag for transcriptional activation (PubMed:24616510). Also acetylates non-histone proteins, like DDX21, FBL, IRF2, MAFG, NCOA3, POLR1E/PAF53 and FOXO1 (PubMed:10490106, PubMed:11154691, PubMed:12738767, PubMed:12929931, PubMed:9707565, PubMed:24207024, PubMed:28790157, PubMed:30540930). Binds specifically to phosphorylated CREB and enhances its transcriptional activity toward cAMP-responsive genes. Acts as a coactivator of ALX1. Acts as a circadian transcriptional coactivator which enhances the activity of the circadian transcriptional activators: NPAS2-ARNTL/BMAL1 and CLOCK-ARNTL/BMAL1 heterodimers (PubMed:14645221). Acetylates PCNA; acetylation promotes removal of chromatin-bound PCNA and its degradation during nucleotide excision repair (NER) (PubMed:24939902). Acetylates POLR1E/PAF53, leading to decreased association of RNA polymerase I with the rDNA promoter region and coding region (PubMed:24207024). Acetylates DDX21, thereby inhibiting DDX21 helicase activity (PubMed:28790157). Acetylates FBL, preventing methylation of 'Gln-105' of histone H2A (H2AQ104me) (PubMed:30540930). Functions as a transcriptional coactivator for SMAD4 in the TGF-beta signaling pathway (PubMed:25514493). {ECO:0000269|PubMed:10490106, ECO:0000269|PubMed:11154691, ECO:0000269|PubMed:12738767, ECO:0000269|PubMed:12929931, ECO:0000269|PubMed:14645221, ECO:0000269|PubMed:24207024, ECO:0000269|PubMed:24616510, ECO:0000269|PubMed:24939902, ECO:0000269|PubMed:25514493, ECO:0000269|PubMed:28790157, ECO:0000269|PubMed:30540930, ECO:0000269|PubMed:9707565}. | FUNCTION: Adenylyl cyclase that catalyzes the formation of the signaling molecule cAMP in response to activation of G protein-coupled receptors (PubMed:9628827, PubMed:12972952, PubMed:15879435, PubMed:10987815). Contributes to signaling cascades activated by CRH (corticotropin-releasing factor), corticosteroids and beta-adrenergic receptors (PubMed:9628827). {ECO:0000269|PubMed:10987815, ECO:0000269|PubMed:12972952, ECO:0000269|PubMed:15879435, ECO:0000269|PubMed:9628827}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 1058_1198 | 956 | 1354.0 | Domain | Guanylate cyclase 2 | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 1058_1198 | 628 | 1354.0 | Domain | Guanylate cyclase 2 | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 1185_1187 | 956 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 1192_1196 | 956 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 1185_1187 | 628 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 1192_1196 | 628 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 997_1353 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 808_818 | 628 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 840_867 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 889_891 | 628 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 913_920 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 942_975 | 628 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 997_1353 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 976_996 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 787_807 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 819_839 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 868_888 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 892_912 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 921_941 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 976_996 | 628 | 1354.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1061_1064 | 28 | 2443.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1199_1487 | 28 | 2443.0 | Compositional bias | Note=Cys/His-rich |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1555_1562 | 28 | 2443.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1943_1948 | 28 | 2443.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1967_1970 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 2081_2085 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 2199_2216 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 2245_2248 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 2297_2300 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1061_1064 | 28 | 2405.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1199_1487 | 28 | 2405.0 | Compositional bias | Note=Cys/His-rich |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1555_1562 | 28 | 2405.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1943_1948 | 28 | 2405.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1967_1970 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 2081_2085 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 2199_2216 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 2245_2248 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 2297_2300 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1061_1064 | 28 | 2443.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1199_1487 | 28 | 2443.0 | Compositional bias | Note=Cys/His-rich |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1555_1562 | 28 | 2443.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1943_1948 | 28 | 2443.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1967_1970 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 2081_2085 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 2199_2216 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 2245_2248 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 2297_2300 | 28 | 2443.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1061_1064 | 28 | 2405.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1199_1487 | 28 | 2405.0 | Compositional bias | Note=Cys/His-rich |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1555_1562 | 28 | 2405.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1943_1948 | 28 | 2405.0 | Compositional bias | Note=Poly-Pro |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1967_1970 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 2081_2085 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 2199_2216 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 2245_2248 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 2297_2300 | 28 | 2405.0 | Compositional bias | Note=Poly-Gln |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1103_1175 | 28 | 2443.0 | Domain | Bromo |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1323_1700 | 28 | 2443.0 | Domain | CBP/p300-type HAT |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 587_666 | 28 | 2443.0 | Domain | KIX |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1103_1175 | 28 | 2405.0 | Domain | Bromo |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1323_1700 | 28 | 2405.0 | Domain | CBP/p300-type HAT |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 587_666 | 28 | 2405.0 | Domain | KIX |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1103_1175 | 28 | 2443.0 | Domain | Bromo |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1323_1700 | 28 | 2443.0 | Domain | CBP/p300-type HAT |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 587_666 | 28 | 2443.0 | Domain | KIX |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1103_1175 | 28 | 2405.0 | Domain | Bromo |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1323_1700 | 28 | 2405.0 | Domain | CBP/p300-type HAT |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 587_666 | 28 | 2405.0 | Domain | KIX |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1434_1436 | 28 | 2443.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1446_1447 | 28 | 2443.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1434_1436 | 28 | 2405.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1446_1447 | 28 | 2405.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1434_1436 | 28 | 2443.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1446_1447 | 28 | 2443.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1434_1436 | 28 | 2405.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1446_1447 | 28 | 2405.0 | Region | Acetyl-CoA binding |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1701_1744 | 28 | 2443.0 | Zinc finger | ZZ-type |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1765_1846 | 28 | 2443.0 | Zinc finger | TAZ-type 2 |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 347_433 | 28 | 2443.0 | Zinc finger | TAZ-type 1 |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1701_1744 | 28 | 2405.0 | Zinc finger | ZZ-type |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1765_1846 | 28 | 2405.0 | Zinc finger | TAZ-type 2 |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 347_433 | 28 | 2405.0 | Zinc finger | TAZ-type 1 |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1701_1744 | 28 | 2443.0 | Zinc finger | ZZ-type |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1765_1846 | 28 | 2443.0 | Zinc finger | TAZ-type 2 |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 347_433 | 28 | 2443.0 | Zinc finger | TAZ-type 1 |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1701_1744 | 28 | 2405.0 | Zinc finger | ZZ-type |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1765_1846 | 28 | 2405.0 | Zinc finger | TAZ-type 2 |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 347_433 | 28 | 2405.0 | Zinc finger | TAZ-type 1 |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 394_521 | 956 | 1354.0 | Domain | Guanylate cyclase 1 | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 394_521 | 628 | 1354.0 | Domain | Guanylate cyclase 1 | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 399_404 | 956 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 441_443 | 956 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 399_404 | 628 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 441_443 | 628 | 1354.0 | Nucleotide binding | ATP | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 139_141 | 956 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 163_171 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 193_215 | 956 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 1_117 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 236_241 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 260_280 | 956 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 302_786 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 808_818 | 956 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 840_867 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 889_891 | 956 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 913_920 | 956 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 942_975 | 956 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 139_141 | 628 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 163_171 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 193_215 | 628 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 1_117 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 236_241 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 260_280 | 628 | 1354.0 | Topological domain | Extracellular | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 302_786 | 628 | 1354.0 | Topological domain | Cytoplasmic | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 118_138 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 142_162 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 172_192 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 216_235 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 242_259 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 281_301 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 787_807 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 819_839 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 868_888 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 892_912 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4016967 | ENST00000294016 | 9 | 11 | 921_941 | 956 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 118_138 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 142_162 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 172_192 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 216_235 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 242_259 | 628 | 1354.0 | Transmembrane | Helical | |
| Tgene | ADCY9 | chr16:3929833 | chr16:4043511 | ENST00000294016 | 2 | 11 | 281_301 | 628 | 1354.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CREBBP-ADCY9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19349_19349_1_CREBBP-ADCY9_CREBBP_chr16_3929833_ENST00000262367_ADCY9_chr16_4043511_ENST00000294016_length(transcript)=6197nt_BP=895nt TTCTTTTTTTTTTAATTGAGGAATCAACAGCCGCCATCTTGTCGCGGACCCGACCGGGGCTTCGAGCGCGATCTACTCGGCCCCGCCGGT CCCGGGCCCCACAACCGCCCGCGCACCCCGCTCCGCCCGGCCGGCCCGCTCCGCCCGGCCCTCGGCGCCCGCCCCGGCGGCCCCGCTCGC CTCTCGGCTCGGCCTCCCGGAGCCCGGCGGCGGCGGCGGCGGCAGCGGCGGCGGCGGCGGCGGAACGGGGGGTGGGGGGGCCGCGGCGGC GGCGGCGACCCCGCTCGGCGCATTGTTTTTCCTCACGGCGGCGGCGGCGGCGGGCCGCGGGCCGGGAGCGGAGCCCGGAGCCCCCTCGTC GTCGGGCCGCGAGCGAATTCATTAAGTGGGGCGCGGGGGGGGAGCGAGGCGGCGGCGGCGGCGGCACCATGTTCTCGGGGACTGCCTGAG CCGCCCGGCCGGGCGCCGTCGCTGCCAGCCGGGCCCGGGGGGGCGGCCGGGCCGCCGGGGCGCCCCCACCGCGGAGTGTCGCGCTCGGGA GGCGGGCAGGGGATGAGGGGGCCGCGGCCGGCGGCGGCGGCGGCGGCCGGGGGCGGGCGGTGAGCGCTGCGGGGCGCTGTTGCTGTGGCT GAGATTTGGCCGCCGCCTCCCCCACCCGGCCTGCGCCCTCCCTCTCCCTCGGCGCCCGCCCGCCCGCTCGCGGCCCGCGCTCGCTCCTCT CCCTCGCAGCCGGCAGGGCCCCCGACCCCCGTCCGGGCCCTCGCCGGCCCGGCCGCCCGTGCCCGGGGCTGTTTTCGCGAGCAGGTGAAA ATGGCTGAGAACTTGCTGGACGGACCGCCCAACCCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGACCTG CCCTTCGTGCGGAATCACATTTGCTCCCAAATCTGAAGCCGGCGCCGAGGGAGGAGCACCTCAAAACGGCTGCCAAGACGAGCATAAAAA CAGCACCAAGGCTTCTGGAGGACCTAATCCCAAAACTCAGAACGGGCTCCTCAGCCCTCCCCAAGAGGAGAAGCTCACCAACAGTCAGAC TTCTCTGTGTGAGATCTTGCAGGAGAAGGGAAGGTGGGCAGGGGTGAGCCTGGACCAGTCGGCTCTCCTTCCGCTGAGGTTCAAGAACAT CCGGGAGAAAACGGACGCCCACTTTGTGGACGTTATCAAAGAAGACAGCCTGATGAAAGATTACTTTTTTAAGCCGCCCATTAATCAGTT CAGCCTGAACTTCCTGGATCAGGAGCTGGAGCGATCCTACAGGACCAGCTATCAGGAAGAGGTCATAAAGAACTCCCCCGTGAAGACGTT TGCTAGTCCCACCTTCAGCTCCCTCCTGGATGTGTTTCTGTCGACCACAGTGTTTCTGACGCTGTCCACCACCTGCTTCCTGAAGTACGA GGCGGCCACCGTGCCTCCCCCGCCCGCCGCCCTGGCGGTCTTCAGTGCAGCCCTGCTGCTGGAGGTGCTGTCCCTCGCGGTGTCCATCAG GATGGTGTTCTTCCTGGAGGACGTCATGGCCTGCACCAAGCGCCTGCTGGAGTGGATCGCCGGCTGGCTACCACGTCACTGCATCGGGGC CATCCTGGTGTCGCTTCCCGCACTGGCCGTCTACTCCCATGTCACCTCCGAATATGAGACCAACATACACTTCCCAGTGTTCACAGGCTC GGCCGCGCTGATTGCCGTCGTGCACTACTGTAACTTCTGCCAGCTCAGCTCCTGGATGAGGTCCTCCCTCGCCACCGTCGTGGGGGCCGG GCCGCTGCTCCTGCTCTACGTCTCCCTGTGCCCAGACAGTTCTGTATTAACTTCGCCCCTTGACGCAGTACAGAATTTCAGTTCCGAGAG GAACCCGTGCAATAGTTCGGTGCCGCGTGACCTCCGGCGGCCCGCCAGCCTCATCGGCCAGGAGGTGGTTCTCGTCTTCTTTCTCCTGCT CTTGTTGGTCTGGTTCCTGAATCGCGAATTTGAAGTCAGCTACCGCCTCCACTACCACGGAGACGTGGAAGCGGATCTTCACCGCACCAA GATCCAGAGCATGCGGGACCAGGCAGACTGGCTGCTGAGGAACATCATCCCCTACCACGTGGCTGAGCAGCTGAAGGTGTCCCAGACCTA CTCCAAGAACCATGACAGCGGAGGGGTGATCTTCGCCAGCATCGTCAACTTCAGCGAGTTCTACGAGGAGAACTACGAGGGCGGCAAGGA GTGCTACCGGGTCCTCAACGAGCTCATCGGGGACTTTGACGAGCTCCTAAGCAAGCCGGACTACAGCAGCATCGAGAAGATCAAGACCAT CGGAGCCACGTACATGGCGGCGTCAGGGCTGAACACCGCGCAGGCCCAGGACGGCAGCCACCCGCAGGAGCACCTGCAGATCCTGTTCGA GTTCGCCAAGGAGATGATGCGCGTGGTGGACGACTTCAACAACAACATGCTGTGGTTCAACTTCAAGCTCCGCGTCGGCTTCAACCATGG GCCCCTCACGGCCGGGGTCATCGGCACCACCAAGCTGCTGTACGACATCTGGGGAGACACCGTCAACATCGCCAGCAGGATGGACACCAC CGGCGTGGAGTGCCGCATCCAGGTGAGCGAAGAGAGCTACCGCGTCTTGAGCAAGATGGGCTATGACTTCGACTACAGAGGGACCGTGAA TGTCAAGGGGAAAGGCCAGATGAAGACCTACCTGTACCCAAAGTGCACGGATCACAGGGTCATCCCACAGCACCAGCTGTCCATCTCCCC AGACATCCGCGTCCAGGTGGATGGCAGCATCGGACGGTCTCCCACAGACGAGATTGCCAACCTGGTGCCTTCTGTCCAGTATGTGGACAA GACATCTCTGGGTTCTGACAGCAGCACGCAGGCCAAGGATGCCCACCTGTCCCCCAAGAGACCGTGGAAGGAGCCCGTCAAAGCCGAAGA AAGGGGTCGATTTGGCAAAGCCATAGAGAAAGACGACTGTGACGAAACAGGAATAGAAGAAGCCAACGAACTCACCAAGCTCAACGTTTC AAAGAGTGTGTGAGGCGGCGCCCACCCGCTGCCCGAGGTGCTCTGTTTGTCGAAACACAGTAATATTTGTATTTGGCTGTTGTGCTTTCC AAGCGCCACAGTTGCCCTCCCCGGACGTGGTGTTATGTGGTCATTTCAGCCCTAACTTCTGTGTGGATCACAGTTATTCAGGGTTCATTT TCATCCATTCTTCCCTTTCGCTCCCTTCCCTGGAAACCCCGCTGCCTCTGGGTCATCCGTTCAGCACGTGGTGGAGAACAAGTGCCTTCA GGGCTGGCCTCGGCCTCGAGTCTCGGGACAGAGGCCGCCAGTGGAGATCATGGCTTTGGGTATTATTTGACTTTTAGAACAAAAGCTGTG GTTAAGATCTCATTTTTATTGCTTTTTCCCACGTCCCACGAGACACTATTTTCGGTTCTCTGGCTAATACCCTGTTTTTGAGTTTATTTT GTTTCTGTCTATGTCACAGTGTTCCTCTACGACCCGACCTCTCTATGTAAGCACACATGCGCACACACACTTGCATTCATGAATCTGATA TAAAGTGCCAGTAATCCGCCAAGAGGGGGTGCGAAGGGGGCATGTCACGACAGCTCCGCCACCCCCCATTGCCCACCCGCACTTTCCCGA GCACCGCGCCCCGTGGGCTGTGGGTGAGCCGCGCTCCCTGCACTGAGCGGGTTTAGGGGCTCGCCCACATGCATGCAGGCCAAGACAGCA AATGCCAGCCGGGCACGACGCCCGTGTGCCCAGGCCTCGGGGGTCTCAGAGCCGCCTCTCACCCCCGACCCTCCACCCAGGGGTCTCCCC ATCGGGAGTGGAGGTGTTGGTCCTGGAAGCTGACTCATCGGAGAGGGAAATACCAAATAAACATCCGAGGTTGCATTTATTTATTTATTT ATTTACCCGGAAGCATGTGCGCTCCCGGGAGGGGTGCTGGGTCCTCAGGGTGGGGGGTCATAGCCACAGGGGAGGGCAGTTCCCAGGAAG GTAACCAGATGTCTCTTTTGTGTTTTGGCCACCTGGGGCCGCCCCCAACCTCCCCACCCCAAGAGGGGTCTGAGTGGGACCTCAAAGCAT GGCCAGTGCAATTCTGAGGCTAGGGGAATAGGAGAAGCCCACCAGCTGCCCCTGCCACCCACGGGGGCTCCTCCCTGCCGGGGTCCAGCT CAGAACCACGGGGGTCTGTGCCGGCAGCACCGGTGCCATCTTGGCTTCCACAAATTCCAGAAGTTTCTTGGCTGCCTCCCGCTCTCAGAT GGCACAGGGCCTATCAGTGGCAGGATTTAATCTGGGGAGCAAATTCCAGCTCTGAAATGGAGTTCTCACACCGTATCTAGGGACAGAGGC ATCTGGATGCAAACGCGGGGTTTGTTAGGGGCGGAATCCCGAGGGTGGTGTGGGGGAGCTCCGTCCGGGCAGTTGGCTGCTGGTCCTGTC TGATTATGTTTTGCCATATTGAGTCTGAAGTTTGTGTGGAAGTGAGCTCGCCCGAGTTGTCCGGTCGGAGTTGTAGCCAGTCTTTCAGGG GACCCCGGGACGCTGATGAGGCCCAGGGTGCACTCCTTGTCAGTCTTGCCCGTTGTCCCTGTGCATTTTGCTTTCCGCTGTCAAATCTCA CTGTCTCGTTTTCACACCGCATCAGTCCCCATCATCACTACTCCTCCCCAGAGACATACAGATTGCCCTCCCCGGCATGAAGAAGTCACA ATTCTGACCCGCCACCTCTCCAGGGGCGGTGCCTGGGATGGCCACCCATCAGGACAGCCAGAGCCAGGGTCCCGGCGTCCTGAGAGTCCA GGGCACCTCCCCCCATTAGCTGTAAGGTTTCCTGTCAGGTGTGACTGTCAGTTTGCTGGTCTTCCAATACCGAAGAAAGGGTGGTTGTGA CAATACCTCTTGCTTCTAAAGAATGTATTATAAAACACCGCAGATTTTTTTTTTTCCTTAAAAAACACTACCTGATGCTTTCCTTGTTCG TGGGGATTGTGGTCACATGAAGCTCTTTCTGCATCAGTATTAAGGTGTATATTTGAATGTCCTCCCCTCCCCTTTCCCCTCCAGGCTGTG TAGCTTTGAGGGGCTGGGCGTTTGCTCACGACCTTGCTGTCTCGCTCAGAACATGCTCCGCAAAGTTCTCCGCACACACTTCTTCCCCAT CAAGCCCATTTCCTTCCCCAACCACAAAGGTGTTTGTGATTCCTCACCCCGGGAAACCAAGGAGCTGCAAAGTGGAGTCTGGTTCAGCCC CGTGCAGACTCACCCAGAGCTTAACGTTGTCTTTCAAACACCCTGAGCCTTCCTAAACAGCCAGTGCAGACGTTCTCTCTGGGCCACGAA GCCCCTCGGGTCCTCCCCGTCCCCTGCTCCGATGCATACCTCAGTGCAGAACCACAGAATCTCTGCAGCGGAAACGCCGTGCATCTCTTG TCTGTTGGCAGCGAGCACATCGTGCTGGAGACACGAGTTTCTAAGCAGCTGGCACGAGGGCTGCTGACGGCATGGGTCGTGCTTCAGGGT GGCAATACCTCTTAGGAACTTAGGGCAGGAAGCAATACTTCAGCATTGAATGTGTGTAAATAGTTGCTTTGAGTTGCAATTGCTATTTTC TTCTCAGTCCCAGCTCAGATCGAATTATATATCCATATATATATATATATATATATATATATATATATATATATATATATGGTAAACAAG CACACACAATTTTATCCAATGCAAACAAATGTAGAGCATCAGTTACAAAACCCTCGAATAGCTTGAGAGCCCCACAGGCTCTGCCACACC CGTGACTTCATCCACACTGACGTCACCCGCGGGGGCTCCCCCTGCACATTTGCACACGATCCGGAGAGCCGAAGGCCGCGTGCTTCCTGT CACATGGGCTGTAATCATTTGTAGTTTCCAAAGACACGTCTGCATTTGAATTTCTAGATTTTCGAGGTAAGGAGTTTTTTTTAATTGGTT GTTTGGAAAATCACATCATGCCTAGAATCTGAAATTGAATTAGCAAGAACCGACTGTTTGCATTTTCCATATATCCTTTTATCTGCTCTT >19349_19349_1_CREBBP-ADCY9_CREBBP_chr16_3929833_ENST00000262367_ADCY9_chr16_4043511_ENST00000294016_length(amino acids)=752AA_BP=27 MRTCWTDRPTPKEPNSARPVSRRMTAQTCPSCGITFAPKSEAGAEGGAPQNGCQDEHKNSTKASGGPNPKTQNGLLSPPQEEKLTNSQTS LCEILQEKGRWAGVSLDQSALLPLRFKNIREKTDAHFVDVIKEDSLMKDYFFKPPINQFSLNFLDQELERSYRTSYQEEVIKNSPVKTFA SPTFSSLLDVFLSTTVFLTLSTTCFLKYEAATVPPPPAALAVFSAALLLEVLSLAVSIRMVFFLEDVMACTKRLLEWIAGWLPRHCIGAI LVSLPALAVYSHVTSEYETNIHFPVFTGSAALIAVVHYCNFCQLSSWMRSSLATVVGAGPLLLLYVSLCPDSSVLTSPLDAVQNFSSERN PCNSSVPRDLRRPASLIGQEVVLVFFLLLLLVWFLNREFEVSYRLHYHGDVEADLHRTKIQSMRDQADWLLRNIIPYHVAEQLKVSQTYS KNHDSGGVIFASIVNFSEFYEENYEGGKECYRVLNELIGDFDELLSKPDYSSIEKIKTIGATYMAASGLNTAQAQDGSHPQEHLQILFEF AKEMMRVVDDFNNNMLWFNFKLRVGFNHGPLTAGVIGTTKLLYDIWGDTVNIASRMDTTGVECRIQVSEESYRVLSKMGYDFDYRGTVNV KGKGQMKTYLYPKCTDHRVIPQHQLSISPDIRVQVDGSIGRSPTDEIANLVPSVQYVDKTSLGSDSSTQAKDAHLSPKRPWKEPVKAEER -------------------------------------------------------------- >19349_19349_2_CREBBP-ADCY9_CREBBP_chr16_3929833_ENST00000382070_ADCY9_chr16_4043511_ENST00000294016_length(transcript)=5591nt_BP=289nt CTGCGGGGCGCTGTTGCTGTGGCTGAGATTTGGCCGCCGCCTCCCCCACCCGGCCTGCGCCCTCCCTCTCCCTCGGCGCCCGCCCGCCCG CTCGCGGCCCGCGCTCGCTCCTCTCCCTCGCAGCCGGCAGGGCCCCCGACCCCCGTCCGGGCCCTCGCCGGCCCGGCCGCCCGTGCCCGG GGCTGTTTTCGCGAGCAGGTGAAAATGGCTGAGAACTTGCTGGACGGACCGCCCAACCCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTC TCGGCGAATGACAGCACAGACCTGCCCTTCGTGCGGAATCACATTTGCTCCCAAATCTGAAGCCGGCGCCGAGGGAGGAGCACCTCAAAA CGGCTGCCAAGACGAGCATAAAAACAGCACCAAGGCTTCTGGAGGACCTAATCCCAAAACTCAGAACGGGCTCCTCAGCCCTCCCCAAGA GGAGAAGCTCACCAACAGTCAGACTTCTCTGTGTGAGATCTTGCAGGAGAAGGGAAGGTGGGCAGGGGTGAGCCTGGACCAGTCGGCTCT CCTTCCGCTGAGGTTCAAGAACATCCGGGAGAAAACGGACGCCCACTTTGTGGACGTTATCAAAGAAGACAGCCTGATGAAAGATTACTT TTTTAAGCCGCCCATTAATCAGTTCAGCCTGAACTTCCTGGATCAGGAGCTGGAGCGATCCTACAGGACCAGCTATCAGGAAGAGGTCAT AAAGAACTCCCCCGTGAAGACGTTTGCTAGTCCCACCTTCAGCTCCCTCCTGGATGTGTTTCTGTCGACCACAGTGTTTCTGACGCTGTC CACCACCTGCTTCCTGAAGTACGAGGCGGCCACCGTGCCTCCCCCGCCCGCCGCCCTGGCGGTCTTCAGTGCAGCCCTGCTGCTGGAGGT GCTGTCCCTCGCGGTGTCCATCAGGATGGTGTTCTTCCTGGAGGACGTCATGGCCTGCACCAAGCGCCTGCTGGAGTGGATCGCCGGCTG GCTACCACGTCACTGCATCGGGGCCATCCTGGTGTCGCTTCCCGCACTGGCCGTCTACTCCCATGTCACCTCCGAATATGAGACCAACAT ACACTTCCCAGTGTTCACAGGCTCGGCCGCGCTGATTGCCGTCGTGCACTACTGTAACTTCTGCCAGCTCAGCTCCTGGATGAGGTCCTC CCTCGCCACCGTCGTGGGGGCCGGGCCGCTGCTCCTGCTCTACGTCTCCCTGTGCCCAGACAGTTCTGTATTAACTTCGCCCCTTGACGC AGTACAGAATTTCAGTTCCGAGAGGAACCCGTGCAATAGTTCGGTGCCGCGTGACCTCCGGCGGCCCGCCAGCCTCATCGGCCAGGAGGT GGTTCTCGTCTTCTTTCTCCTGCTCTTGTTGGTCTGGTTCCTGAATCGCGAATTTGAAGTCAGCTACCGCCTCCACTACCACGGAGACGT GGAAGCGGATCTTCACCGCACCAAGATCCAGAGCATGCGGGACCAGGCAGACTGGCTGCTGAGGAACATCATCCCCTACCACGTGGCTGA GCAGCTGAAGGTGTCCCAGACCTACTCCAAGAACCATGACAGCGGAGGGGTGATCTTCGCCAGCATCGTCAACTTCAGCGAGTTCTACGA GGAGAACTACGAGGGCGGCAAGGAGTGCTACCGGGTCCTCAACGAGCTCATCGGGGACTTTGACGAGCTCCTAAGCAAGCCGGACTACAG CAGCATCGAGAAGATCAAGACCATCGGAGCCACGTACATGGCGGCGTCAGGGCTGAACACCGCGCAGGCCCAGGACGGCAGCCACCCGCA GGAGCACCTGCAGATCCTGTTCGAGTTCGCCAAGGAGATGATGCGCGTGGTGGACGACTTCAACAACAACATGCTGTGGTTCAACTTCAA GCTCCGCGTCGGCTTCAACCATGGGCCCCTCACGGCCGGGGTCATCGGCACCACCAAGCTGCTGTACGACATCTGGGGAGACACCGTCAA CATCGCCAGCAGGATGGACACCACCGGCGTGGAGTGCCGCATCCAGGTGAGCGAAGAGAGCTACCGCGTCTTGAGCAAGATGGGCTATGA CTTCGACTACAGAGGGACCGTGAATGTCAAGGGGAAAGGCCAGATGAAGACCTACCTGTACCCAAAGTGCACGGATCACAGGGTCATCCC ACAGCACCAGCTGTCCATCTCCCCAGACATCCGCGTCCAGGTGGATGGCAGCATCGGACGGTCTCCCACAGACGAGATTGCCAACCTGGT GCCTTCTGTCCAGTATGTGGACAAGACATCTCTGGGTTCTGACAGCAGCACGCAGGCCAAGGATGCCCACCTGTCCCCCAAGAGACCGTG GAAGGAGCCCGTCAAAGCCGAAGAAAGGGGTCGATTTGGCAAAGCCATAGAGAAAGACGACTGTGACGAAACAGGAATAGAAGAAGCCAA CGAACTCACCAAGCTCAACGTTTCAAAGAGTGTGTGAGGCGGCGCCCACCCGCTGCCCGAGGTGCTCTGTTTGTCGAAACACAGTAATAT TTGTATTTGGCTGTTGTGCTTTCCAAGCGCCACAGTTGCCCTCCCCGGACGTGGTGTTATGTGGTCATTTCAGCCCTAACTTCTGTGTGG ATCACAGTTATTCAGGGTTCATTTTCATCCATTCTTCCCTTTCGCTCCCTTCCCTGGAAACCCCGCTGCCTCTGGGTCATCCGTTCAGCA CGTGGTGGAGAACAAGTGCCTTCAGGGCTGGCCTCGGCCTCGAGTCTCGGGACAGAGGCCGCCAGTGGAGATCATGGCTTTGGGTATTAT TTGACTTTTAGAACAAAAGCTGTGGTTAAGATCTCATTTTTATTGCTTTTTCCCACGTCCCACGAGACACTATTTTCGGTTCTCTGGCTA ATACCCTGTTTTTGAGTTTATTTTGTTTCTGTCTATGTCACAGTGTTCCTCTACGACCCGACCTCTCTATGTAAGCACACATGCGCACAC ACACTTGCATTCATGAATCTGATATAAAGTGCCAGTAATCCGCCAAGAGGGGGTGCGAAGGGGGCATGTCACGACAGCTCCGCCACCCCC CATTGCCCACCCGCACTTTCCCGAGCACCGCGCCCCGTGGGCTGTGGGTGAGCCGCGCTCCCTGCACTGAGCGGGTTTAGGGGCTCGCCC ACATGCATGCAGGCCAAGACAGCAAATGCCAGCCGGGCACGACGCCCGTGTGCCCAGGCCTCGGGGGTCTCAGAGCCGCCTCTCACCCCC GACCCTCCACCCAGGGGTCTCCCCATCGGGAGTGGAGGTGTTGGTCCTGGAAGCTGACTCATCGGAGAGGGAAATACCAAATAAACATCC GAGGTTGCATTTATTTATTTATTTATTTACCCGGAAGCATGTGCGCTCCCGGGAGGGGTGCTGGGTCCTCAGGGTGGGGGGTCATAGCCA CAGGGGAGGGCAGTTCCCAGGAAGGTAACCAGATGTCTCTTTTGTGTTTTGGCCACCTGGGGCCGCCCCCAACCTCCCCACCCCAAGAGG GGTCTGAGTGGGACCTCAAAGCATGGCCAGTGCAATTCTGAGGCTAGGGGAATAGGAGAAGCCCACCAGCTGCCCCTGCCACCCACGGGG GCTCCTCCCTGCCGGGGTCCAGCTCAGAACCACGGGGGTCTGTGCCGGCAGCACCGGTGCCATCTTGGCTTCCACAAATTCCAGAAGTTT CTTGGCTGCCTCCCGCTCTCAGATGGCACAGGGCCTATCAGTGGCAGGATTTAATCTGGGGAGCAAATTCCAGCTCTGAAATGGAGTTCT CACACCGTATCTAGGGACAGAGGCATCTGGATGCAAACGCGGGGTTTGTTAGGGGCGGAATCCCGAGGGTGGTGTGGGGGAGCTCCGTCC GGGCAGTTGGCTGCTGGTCCTGTCTGATTATGTTTTGCCATATTGAGTCTGAAGTTTGTGTGGAAGTGAGCTCGCCCGAGTTGTCCGGTC GGAGTTGTAGCCAGTCTTTCAGGGGACCCCGGGACGCTGATGAGGCCCAGGGTGCACTCCTTGTCAGTCTTGCCCGTTGTCCCTGTGCAT TTTGCTTTCCGCTGTCAAATCTCACTGTCTCGTTTTCACACCGCATCAGTCCCCATCATCACTACTCCTCCCCAGAGACATACAGATTGC CCTCCCCGGCATGAAGAAGTCACAATTCTGACCCGCCACCTCTCCAGGGGCGGTGCCTGGGATGGCCACCCATCAGGACAGCCAGAGCCA GGGTCCCGGCGTCCTGAGAGTCCAGGGCACCTCCCCCCATTAGCTGTAAGGTTTCCTGTCAGGTGTGACTGTCAGTTTGCTGGTCTTCCA ATACCGAAGAAAGGGTGGTTGTGACAATACCTCTTGCTTCTAAAGAATGTATTATAAAACACCGCAGATTTTTTTTTTTCCTTAAAAAAC ACTACCTGATGCTTTCCTTGTTCGTGGGGATTGTGGTCACATGAAGCTCTTTCTGCATCAGTATTAAGGTGTATATTTGAATGTCCTCCC CTCCCCTTTCCCCTCCAGGCTGTGTAGCTTTGAGGGGCTGGGCGTTTGCTCACGACCTTGCTGTCTCGCTCAGAACATGCTCCGCAAAGT TCTCCGCACACACTTCTTCCCCATCAAGCCCATTTCCTTCCCCAACCACAAAGGTGTTTGTGATTCCTCACCCCGGGAAACCAAGGAGCT GCAAAGTGGAGTCTGGTTCAGCCCCGTGCAGACTCACCCAGAGCTTAACGTTGTCTTTCAAACACCCTGAGCCTTCCTAAACAGCCAGTG CAGACGTTCTCTCTGGGCCACGAAGCCCCTCGGGTCCTCCCCGTCCCCTGCTCCGATGCATACCTCAGTGCAGAACCACAGAATCTCTGC AGCGGAAACGCCGTGCATCTCTTGTCTGTTGGCAGCGAGCACATCGTGCTGGAGACACGAGTTTCTAAGCAGCTGGCACGAGGGCTGCTG ACGGCATGGGTCGTGCTTCAGGGTGGCAATACCTCTTAGGAACTTAGGGCAGGAAGCAATACTTCAGCATTGAATGTGTGTAAATAGTTG CTTTGAGTTGCAATTGCTATTTTCTTCTCAGTCCCAGCTCAGATCGAATTATATATCCATATATATATATATATATATATATATATATAT ATATATATATATATGGTAAACAAGCACACACAATTTTATCCAATGCAAACAAATGTAGAGCATCAGTTACAAAACCCTCGAATAGCTTGA GAGCCCCACAGGCTCTGCCACACCCGTGACTTCATCCACACTGACGTCACCCGCGGGGGCTCCCCCTGCACATTTGCACACGATCCGGAG AGCCGAAGGCCGCGTGCTTCCTGTCACATGGGCTGTAATCATTTGTAGTTTCCAAAGACACGTCTGCATTTGAATTTCTAGATTTTCGAG GTAAGGAGTTTTTTTTAATTGGTTGTTTGGAAAATCACATCATGCCTAGAATCTGAAATTGAATTAGCAAGAACCGACTGTTTGCATTTT CCATATATCCTTTTATCTGCTCTTTTTAAATTGTTAATTCTAATAATTTCAAAATGCATTCACTGAAGAAATGGACATTAAAATATTCTA >19349_19349_2_CREBBP-ADCY9_CREBBP_chr16_3929833_ENST00000382070_ADCY9_chr16_4043511_ENST00000294016_length(amino acids)=752AA_BP=27 MRTCWTDRPTPKEPNSARPVSRRMTAQTCPSCGITFAPKSEAGAEGGAPQNGCQDEHKNSTKASGGPNPKTQNGLLSPPQEEKLTNSQTS LCEILQEKGRWAGVSLDQSALLPLRFKNIREKTDAHFVDVIKEDSLMKDYFFKPPINQFSLNFLDQELERSYRTSYQEEVIKNSPVKTFA SPTFSSLLDVFLSTTVFLTLSTTCFLKYEAATVPPPPAALAVFSAALLLEVLSLAVSIRMVFFLEDVMACTKRLLEWIAGWLPRHCIGAI LVSLPALAVYSHVTSEYETNIHFPVFTGSAALIAVVHYCNFCQLSSWMRSSLATVVGAGPLLLLYVSLCPDSSVLTSPLDAVQNFSSERN PCNSSVPRDLRRPASLIGQEVVLVFFLLLLLVWFLNREFEVSYRLHYHGDVEADLHRTKIQSMRDQADWLLRNIIPYHVAEQLKVSQTYS KNHDSGGVIFASIVNFSEFYEENYEGGKECYRVLNELIGDFDELLSKPDYSSIEKIKTIGATYMAASGLNTAQAQDGSHPQEHLQILFEF AKEMMRVVDDFNNNMLWFNFKLRVGFNHGPLTAGVIGTTKLLYDIWGDTVNIASRMDTTGVECRIQVSEESYRVLSKMGYDFDYRGTVNV KGKGQMKTYLYPKCTDHRVIPQHQLSISPDIRVQVDGSIGRSPTDEIANLVPSVQYVDKTSLGSDSSTQAKDAHLSPKRPWKEPVKAEER -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CREBBP-ADCY9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1162_1180 | 28.333333333333332 | 2443.0 | ASF1A |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1162_1180 | 28.333333333333332 | 2405.0 | ASF1A |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1162_1180 | 28.333333333333332 | 2443.0 | ASF1A |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1162_1180 | 28.333333333333332 | 2405.0 | ASF1A |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1433_1435 | 28.333333333333332 | 2443.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1433_1435 | 28.333333333333332 | 2405.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1433_1435 | 28.333333333333332 | 2443.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1433_1435 | 28.333333333333332 | 2405.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1124_1170 | 28.333333333333332 | 2443.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1124_1170 | 28.333333333333332 | 2405.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1124_1170 | 28.333333333333332 | 2443.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1124_1170 | 28.333333333333332 | 2405.0 | histone |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 227_410 | 28.333333333333332 | 2443.0 | SRCAP |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 227_410 | 28.333333333333332 | 2405.0 | SRCAP |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 227_410 | 28.333333333333332 | 2443.0 | SRCAP |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 227_410 | 28.333333333333332 | 2405.0 | SRCAP |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000262367 | - | 1 | 31 | 1460_1891 | 28.333333333333332 | 2443.0 | TRERF1 |
| Hgene | CREBBP | chr16:3929833 | chr16:4016967 | ENST00000382070 | - | 1 | 30 | 1460_1891 | 28.333333333333332 | 2405.0 | TRERF1 |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000262367 | - | 1 | 31 | 1460_1891 | 28.333333333333332 | 2443.0 | TRERF1 |
| Hgene | CREBBP | chr16:3929833 | chr16:4043511 | ENST00000382070 | - | 1 | 30 | 1460_1891 | 28.333333333333332 | 2405.0 | TRERF1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CREBBP-ADCY9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CREBBP-ADCY9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
