
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CRELD1-FANCD2 (FusionGDB2 ID:19413) |
Fusion Gene Summary for CRELD1-FANCD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CRELD1-FANCD2 | Fusion gene ID: 19413 | Hgene | Tgene | Gene symbol | CRELD1 | FANCD2 | Gene ID | 78987 | 2177 |
| Gene name | cysteine rich with EGF like domains 1 | FA complementation group D2 | |
| Synonyms | AVSD2|CIRRIN | FA-D2|FA4|FACD|FAD|FAD2|FANCD | |
| Cytomap | 3p25.3 | 3p25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein disulfide isomerase CRELD1 | Fanconi anemia group D2 proteinFanconi anemia complementation group D2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96HD1 | Q96PS1 | |
| Ensembl transtripts involved in fusion gene | ENST00000489674, ENST00000326434, ENST00000383811, ENST00000397170, ENST00000452070, | ENST00000287647, ENST00000383806, ENST00000383807, ENST00000419585, ENST00000431693, ENST00000438741, | |
| Fusion gene scores | * DoF score | 3 X 4 X 3=36 | 9 X 13 X 7=819 |
| # samples | 4 | 11 | |
| ** MAII score | log2(4/36*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(11/819*10)=-2.89635992811635 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CRELD1 [Title/Abstract] AND FANCD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CRELD1(9985199)-FANCD2(10133865), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CRELD1-FANCD2 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | FANCD2 | GO:0010332 | response to gamma radiation | 12874027 |
 Fusion gene breakpoints across CRELD1 (5'-gene) Fusion gene breakpoints across CRELD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
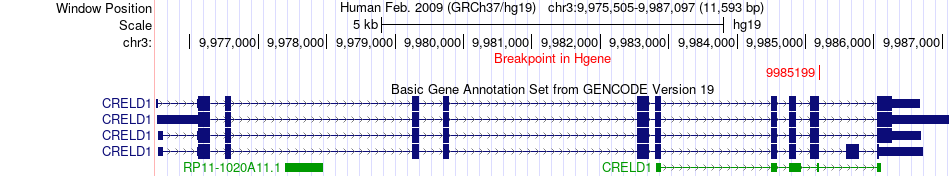 |
 Fusion gene breakpoints across FANCD2 (3'-gene) Fusion gene breakpoints across FANCD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
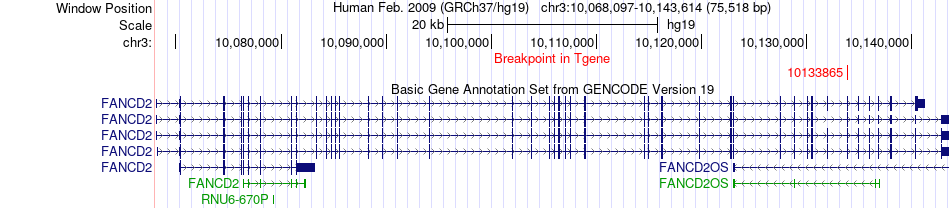 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-S9-A6WM-01A | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
Top |
Fusion Gene ORF analysis for CRELD1-FANCD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000489674 | ENST00000287647 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 3UTR-3CDS | ENST00000489674 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 3UTR-3CDS | ENST00000489674 | ENST00000383807 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 3UTR-3CDS | ENST00000489674 | ENST00000419585 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 3UTR-intron | ENST00000489674 | ENST00000431693 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 3UTR-intron | ENST00000489674 | ENST00000438741 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000326434 | ENST00000431693 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000326434 | ENST00000438741 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000383811 | ENST00000431693 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000383811 | ENST00000438741 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000397170 | ENST00000431693 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000397170 | ENST00000438741 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000452070 | ENST00000431693 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| 5CDS-intron | ENST00000452070 | ENST00000438741 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000326434 | ENST00000287647 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000326434 | ENST00000383807 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000326434 | ENST00000419585 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000383811 | ENST00000287647 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000383811 | ENST00000383807 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000383811 | ENST00000419585 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000397170 | ENST00000287647 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000397170 | ENST00000383807 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000397170 | ENST00000419585 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000452070 | ENST00000287647 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000452070 | ENST00000383807 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| Frame-shift | ENST00000452070 | ENST00000419585 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| In-frame | ENST00000326434 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| In-frame | ENST00000383811 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| In-frame | ENST00000397170 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
| In-frame | ENST00000452070 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000397170 | CRELD1 | chr3 | 9985199 | + | ENST00000383806 | FANCD2 | chr3 | 10133865 | + | 2345 | 1098 | 50 | 1105 | 351 |
| ENST00000383811 | CRELD1 | chr3 | 9985199 | + | ENST00000383806 | FANCD2 | chr3 | 10133865 | + | 2894 | 1647 | 599 | 1654 | 351 |
| ENST00000452070 | CRELD1 | chr3 | 9985199 | + | ENST00000383806 | FANCD2 | chr3 | 10133865 | + | 2390 | 1143 | 95 | 1150 | 351 |
| ENST00000326434 | CRELD1 | chr3 | 9985199 | + | ENST00000383806 | FANCD2 | chr3 | 10133865 | + | 2379 | 1132 | 84 | 1139 | 351 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000397170 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + | 0.004957873 | 0.9950421 |
| ENST00000383811 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + | 0.005008217 | 0.99499184 |
| ENST00000452070 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + | 0.004354091 | 0.99564594 |
| ENST00000326434 | ENST00000383806 | CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133865 | + | 0.004310237 | 0.9956898 |
Top |
Fusion Genomic Features for CRELD1-FANCD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133864 | + | 0.000106318 | 0.99989367 |
| CRELD1 | chr3 | 9985199 | + | FANCD2 | chr3 | 10133864 | + | 0.000106318 | 0.99989367 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for CRELD1-FANCD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:9985199/chr3:10133865) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CRELD1 | FANCD2 |
| FUNCTION: Protein disulfide isomerase (By similarity). Promotes the localization of acetylcholine receptors (AChRs) to the plasma membrane (By similarity). {ECO:0000250|UniProtKB:Q91XD7, ECO:0000250|UniProtKB:Q9CYA0}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 3_45 | 349 | 423.0 | Compositional bias | Note=Pro-rich |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 3_45 | 349 | 421.0 | Compositional bias | Note=Pro-rich |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 3_45 | 349 | 421.0 | Compositional bias | Note=Pro-rich |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 3_45 | 349 | 421.0 | Compositional bias | Note=Pro-rich |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 153_193 | 349 | 423.0 | Domain | EGF-like 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 305_344 | 349 | 423.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 153_193 | 349 | 421.0 | Domain | EGF-like 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 305_344 | 349 | 421.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 153_193 | 349 | 421.0 | Domain | EGF-like 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 305_344 | 349 | 421.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 153_193 | 349 | 421.0 | Domain | EGF-like 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 305_344 | 349 | 421.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 208_256 | 349 | 423.0 | Repeat | Note=FU 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 268_315 | 349 | 423.0 | Repeat | Note=FU 2 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 208_256 | 349 | 421.0 | Repeat | Note=FU 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 268_315 | 349 | 421.0 | Repeat | Note=FU 2 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 208_256 | 349 | 421.0 | Repeat | Note=FU 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 268_315 | 349 | 421.0 | Repeat | Note=FU 2 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 208_256 | 349 | 421.0 | Repeat | Note=FU 1 |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 268_315 | 349 | 421.0 | Repeat | Note=FU 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 30_362 | 349 | 423.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 384_384 | 349 | 423.0 | Topological domain | Cytoplasmic |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 406_420 | 349 | 423.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 30_362 | 349 | 421.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 384_384 | 349 | 421.0 | Topological domain | Cytoplasmic |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 406_420 | 349 | 421.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 30_362 | 349 | 421.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 384_384 | 349 | 421.0 | Topological domain | Cytoplasmic |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 406_420 | 349 | 421.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 30_362 | 349 | 421.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 384_384 | 349 | 421.0 | Topological domain | Cytoplasmic |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 406_420 | 349 | 421.0 | Topological domain | Extracellular |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 363_383 | 349 | 423.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000326434 | + | 10 | 12 | 385_405 | 349 | 423.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 363_383 | 349 | 421.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000383811 | + | 9 | 10 | 385_405 | 349 | 421.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 363_383 | 349 | 421.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000397170 | + | 10 | 11 | 385_405 | 349 | 421.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 363_383 | 349 | 421.0 | Transmembrane | Helical |
| Hgene | CRELD1 | chr3:9985199 | chr3:10133865 | ENST00000452070 | + | 10 | 11 | 385_405 | 349 | 421.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CRELD1-FANCD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19413_19413_1_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000326434_FANCD2_chr3_10133865_ENST00000383806_length(transcript)=2379nt_BP=1132nt AGGAGACGGCCCACGGCGCCCGCGGGCTGGGGCGGTCGCTTCTTCCTTCTCCGTGGCCTACGAGGGTCCCCAGCCTGGGTAAAGATGGCC CCATGGCCCCCGAAGGGCCTAGTCCCAGCTATGCTCTGGGGCCTCAGCCTCTTCCTCAACCTCCCAGGACCTATCTGGCTCCAGCCCTCT CCACCTCCCCAGTCTTCTCCCCCGCCTCAGCCCCATCCGTGTCATACCTGCCGGGGACTGGTTGACAGCTTTAACAAGGGCCTGGAGAGA ACCATCCGGGACAACTTTGGAGGTGGAAACACTGCCTGGGAGGAAGAGAATTTGTCCAAATACAAAGACAGTGAGACCCGCCTGGTAGAG GTGCTGGAGGGTGTGTGCAGCAAGTCAGACTTCGAGTGCCACCGCCTGCTGGAGCTGAGTGAGGAGCTGGTGGAGAGCTGGTGGTTTCAC AAGCAGCAGGAGGCCCCGGACCTCTTCCAGTGGCTGTGCTCAGATTCCCTGAAGCTCTGCTGCCCCGCAGGCACCTTCGGGCCCTCCTGC CTTCCCTGTCCTGGGGGAACAGAGAGGCCCTGCGGTGGCTACGGGCAGTGTGAAGGAGAAGGGACACGAGGGGGCAGCGGGCACTGTGAC TGCCAAGCCGGCTACGGGGGTGAGGCCTGTGGCCAGTGTGGCCTTGGCTACTTTGAGGCAGAACGCAACGCCAGCCATCTGGTATGTTCG GCTTGTTTTGGCCCCTGTGCCCGATGCTCAGGACCTGAGGAATCAAACTGTTTGCAATGCAAGAAGGGCTGGGCCCTGCATCACCTCAAG TGTGTAGACATTGATGAGTGTGGCACAGAGGGAGCCAACTGTGGAGCTGACCAATTCTGCGTGAACACTGAGGGCTCCTATGAGTGCCGA GACTGTGCCAAGGCCTGCCTAGGCTGCATGGGGGCAGGGCCAGGTCGCTGTAAGAAGTGTAGCCCTGGCTATCAGCAGGTGGGCTCCAAG TGTCTCGATGTGGATGAGTGTGAGACAGAGGTGTGTCCGGGAGAGAACAAGCAGTGTGAAAACACCGAGGGCGGTTATCGCTGCATCTGT GCCGAGGGCTACAAGCAGATGGAAGGCATCTGTGTGAAGGAGCAGATCCCAGATTCATGAAGAGAAACTCCTCTACTGGAACATGGCTGT TCGAGACTTCAGTATCCTCATCAACTTGATAAAGGTATTTGATAGTCATCCTGTTCTGCATGTATGTTTGAAGTATGGGCGTCTCTTTGT GGAAGCATTTCTGAAGCAATGTATGCCGCTCCTAGACTTCAGTTTTAGAAAACACCGGGAAGATGTTCTGAGCTTACTGGAAACCTTCCA GTTGGACACAAGGCTGCTTCATCACCTGTGTGGGCATTCCAAGATTCACCAGGACACGAGACTCACCCAACATGTGCCTCTGCTCAAAAA GACCCTGGAACTTTTAGTTTGCAGAGTCAAAGCTATGCTCACTCTCAACAATTGTAGAGAGGCTTTCTGGCTGGGCAATCTAAAAAACCG GGACTTGCAGGGTGAAGAGATTAAGTCCCAAAATTCCCAGGAGAGCACAGCAGATGAGAGTGAGGATGACATGTCATCCCAGGCCTCCAA GAGCAAAGCCACTGAGGATGGTGAAGAAGACGAAGTAAGTGCTGGAGAAAAGGAGCAAGATAGTGATGAGAGTTATGATGACTCTGATTA GACCCCAGATAAATTGTTGCCTGCTTCTGTGTCTCTGCCAGCCTGTGATCATTTTGTGTTAGAGTTTGAAATCCGCTGTTTGCCTTTCTT ACTGGTAGGATCCTTTTTTGTTCCTCTTTTTTTTTTTTTTTTTTTTTTTTTAAAGACGGGGACTCGCTGTGTTTCCCAGGCTGGAGTGCA GTGCTGCAATCTTGGCTCACTGCAACCTCCATCTCCTAGGTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACGACAGGCA CATGCCACCATGCCCAGCTAATTTTTGTATTTTTAGTAGATACGGGGTTTTACCATGTCGGCCAGATGGTCTCAATCTCCTGAACTCATG ATCCACCTGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCATGAGCCACCGCTCCCAGCCATATTTTGTTCTTAAAGTGGGGTCTTTAT TAACTTGTGGACATCATGGATTGTCTAACACCATCACAGTCCCTGGCTCAGGATTCTAATGTAGCATTATTTATTGGTTTGGATAAACCC AGCTGTGCTACACTGCAGAGTAAAATCTCTGAGTCATGATTCTGGACTTTGGGAGCTAGTTTTGAAACTCTGATTTATTGTAGAACTTAG >19413_19413_1_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000326434_FANCD2_chr3_10133865_ENST00000383806_length(amino acids)=351AA_BP= MAPWPPKGLVPAMLWGLSLFLNLPGPIWLQPSPPPQSSPPPQPHPCHTCRGLVDSFNKGLERTIRDNFGGGNTAWEEENLSKYKDSETRL VEVLEGVCSKSDFECHRLLELSEELVESWWFHKQQEAPDLFQWLCSDSLKLCCPAGTFGPSCLPCPGGTERPCGGYGQCEGEGTRGGSGH CDCQAGYGGEACGQCGLGYFEAERNASHLVCSACFGPCARCSGPEESNCLQCKKGWALHHLKCVDIDECGTEGANCGADQFCVNTEGSYE -------------------------------------------------------------- >19413_19413_2_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000383811_FANCD2_chr3_10133865_ENST00000383806_length(transcript)=2894nt_BP=1647nt TGGCAGCGACGCGGTGAGGAGACGGCCCACGGCGCCCGCGGGCTGGGGCGGTCGCTTCTTCCTTCTCCGTGGCCTACGAGGGTCTGGATC CTTCTCTGCCGGCTCGTGGGCCGTGCCTTTGCCCTTCTGCGAGGCCCTGAATCTGATCCCTTCCCTTCATATCCGGATCCGGGCTCCTCC CTCCAAGCCCGGGGTTCCGGACACCTCCCCCAAGACAACCCCTCTGGCCTCCTCTCCTTCAGTACTTGGAATCTGATCTCTTCTCCCTAA TTCTGCGGATCCGGCCCCTAATATTCTTTATCAGACCCTCAGACAAGAGGCTGACTTCTGCCCCCTTGTCAAGGAGCGAGGCCACTTTCC TCTCCACCCCATGCTAGCGAGGATAACTTATTTCTCTTCTGGAATTGCATCTTATGCGCCTTTCCCCACCCATCCCCACAGCCCCTGCAA TACCCAGTTTGGCCTCTTTTGCTTGTAATAACGCAGATCCCAGCGCCACGGCACCTTAGAACAGACCTTTTTCTTTCTCGCGTGGGGCCT GACTCCTTCAGTGAAGCCTCTCCACGCCCTCTATCTGCAGGTCCCCAGCCTGGGTAAAGATGGCCCCATGGCCCCCGAAGGGCCTAGTCC CAGCTATGCTCTGGGGCCTCAGCCTCTTCCTCAACCTCCCAGGACCTATCTGGCTCCAGCCCTCTCCACCTCCCCAGTCTTCTCCCCCGC CTCAGCCCCATCCGTGTCATACCTGCCGGGGACTGGTTGACAGCTTTAACAAGGGCCTGGAGAGAACCATCCGGGACAACTTTGGAGGTG GAAACACTGCCTGGGAGGAAGAGAATTTGTCCAAATACAAAGACAGTGAGACCCGCCTGGTAGAGGTGCTGGAGGGTGTGTGCAGCAAGT CAGACTTCGAGTGCCACCGCCTGCTGGAGCTGAGTGAGGAGCTGGTGGAGAGCTGGTGGTTTCACAAGCAGCAGGAGGCCCCGGACCTCT TCCAGTGGCTGTGCTCAGATTCCCTGAAGCTCTGCTGCCCCGCAGGCACCTTCGGGCCCTCCTGCCTTCCCTGTCCTGGGGGAACAGAGA GGCCCTGCGGTGGCTACGGGCAGTGTGAAGGAGAAGGGACACGAGGGGGCAGCGGGCACTGTGACTGCCAAGCCGGCTACGGGGGTGAGG CCTGTGGCCAGTGTGGCCTTGGCTACTTTGAGGCAGAACGCAACGCCAGCCATCTGGTATGTTCGGCTTGTTTTGGCCCCTGTGCCCGAT GCTCAGGACCTGAGGAATCAAACTGTTTGCAATGCAAGAAGGGCTGGGCCCTGCATCACCTCAAGTGTGTAGACATTGATGAGTGTGGCA CAGAGGGAGCCAACTGTGGAGCTGACCAATTCTGCGTGAACACTGAGGGCTCCTATGAGTGCCGAGACTGTGCCAAGGCCTGCCTAGGCT GCATGGGGGCAGGGCCAGGTCGCTGTAAGAAGTGTAGCCCTGGCTATCAGCAGGTGGGCTCCAAGTGTCTCGATGTGGATGAGTGTGAGA CAGAGGTGTGTCCGGGAGAGAACAAGCAGTGTGAAAACACCGAGGGCGGTTATCGCTGCATCTGTGCCGAGGGCTACAAGCAGATGGAAG GCATCTGTGTGAAGGAGCAGATCCCAGATTCATGAAGAGAAACTCCTCTACTGGAACATGGCTGTTCGAGACTTCAGTATCCTCATCAAC TTGATAAAGGTATTTGATAGTCATCCTGTTCTGCATGTATGTTTGAAGTATGGGCGTCTCTTTGTGGAAGCATTTCTGAAGCAATGTATG CCGCTCCTAGACTTCAGTTTTAGAAAACACCGGGAAGATGTTCTGAGCTTACTGGAAACCTTCCAGTTGGACACAAGGCTGCTTCATCAC CTGTGTGGGCATTCCAAGATTCACCAGGACACGAGACTCACCCAACATGTGCCTCTGCTCAAAAAGACCCTGGAACTTTTAGTTTGCAGA GTCAAAGCTATGCTCACTCTCAACAATTGTAGAGAGGCTTTCTGGCTGGGCAATCTAAAAAACCGGGACTTGCAGGGTGAAGAGATTAAG TCCCAAAATTCCCAGGAGAGCACAGCAGATGAGAGTGAGGATGACATGTCATCCCAGGCCTCCAAGAGCAAAGCCACTGAGGATGGTGAA GAAGACGAAGTAAGTGCTGGAGAAAAGGAGCAAGATAGTGATGAGAGTTATGATGACTCTGATTAGACCCCAGATAAATTGTTGCCTGCT TCTGTGTCTCTGCCAGCCTGTGATCATTTTGTGTTAGAGTTTGAAATCCGCTGTTTGCCTTTCTTACTGGTAGGATCCTTTTTTGTTCCT CTTTTTTTTTTTTTTTTTTTTTTTTTAAAGACGGGGACTCGCTGTGTTTCCCAGGCTGGAGTGCAGTGCTGCAATCTTGGCTCACTGCAA CCTCCATCTCCTAGGTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACGACAGGCACATGCCACCATGCCCAGCTAATTTT TGTATTTTTAGTAGATACGGGGTTTTACCATGTCGGCCAGATGGTCTCAATCTCCTGAACTCATGATCCACCTGCCTCAGCCTCCCAAAG TGCTGGGATTACAGGCATGAGCCACCGCTCCCAGCCATATTTTGTTCTTAAAGTGGGGTCTTTATTAACTTGTGGACATCATGGATTGTC TAACACCATCACAGTCCCTGGCTCAGGATTCTAATGTAGCATTATTTATTGGTTTGGATAAACCCAGCTGTGCTACACTGCAGAGTAAAA TCTCTGAGTCATGATTCTGGACTTTGGGAGCTAGTTTTGAAACTCTGATTTATTGTAGAACTTAGGCTTGTACCAATTTTACAAATAAAT >19413_19413_2_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000383811_FANCD2_chr3_10133865_ENST00000383806_length(amino acids)=351AA_BP= MAPWPPKGLVPAMLWGLSLFLNLPGPIWLQPSPPPQSSPPPQPHPCHTCRGLVDSFNKGLERTIRDNFGGGNTAWEEENLSKYKDSETRL VEVLEGVCSKSDFECHRLLELSEELVESWWFHKQQEAPDLFQWLCSDSLKLCCPAGTFGPSCLPCPGGTERPCGGYGQCEGEGTRGGSGH CDCQAGYGGEACGQCGLGYFEAERNASHLVCSACFGPCARCSGPEESNCLQCKKGWALHHLKCVDIDECGTEGANCGADQFCVNTEGSYE -------------------------------------------------------------- >19413_19413_3_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000397170_FANCD2_chr3_10133865_ENST00000383806_length(transcript)=2345nt_BP=1098nt TGCGTTTTACGCAGGCTGTGGCAGCGACGCGGTCCCCAGCCTGGGTAAAGATGGCCCCATGGCCCCCGAAGGGCCTAGTCCCAGCTATGC TCTGGGGCCTCAGCCTCTTCCTCAACCTCCCAGGACCTATCTGGCTCCAGCCCTCTCCACCTCCCCAGTCTTCTCCCCCGCCTCAGCCCC ATCCGTGTCATACCTGCCGGGGACTGGTTGACAGCTTTAACAAGGGCCTGGAGAGAACCATCCGGGACAACTTTGGAGGTGGAAACACTG CCTGGGAGGAAGAGAATTTGTCCAAATACAAAGACAGTGAGACCCGCCTGGTAGAGGTGCTGGAGGGTGTGTGCAGCAAGTCAGACTTCG AGTGCCACCGCCTGCTGGAGCTGAGTGAGGAGCTGGTGGAGAGCTGGTGGTTTCACAAGCAGCAGGAGGCCCCGGACCTCTTCCAGTGGC TGTGCTCAGATTCCCTGAAGCTCTGCTGCCCCGCAGGCACCTTCGGGCCCTCCTGCCTTCCCTGTCCTGGGGGAACAGAGAGGCCCTGCG GTGGCTACGGGCAGTGTGAAGGAGAAGGGACACGAGGGGGCAGCGGGCACTGTGACTGCCAAGCCGGCTACGGGGGTGAGGCCTGTGGCC AGTGTGGCCTTGGCTACTTTGAGGCAGAACGCAACGCCAGCCATCTGGTATGTTCGGCTTGTTTTGGCCCCTGTGCCCGATGCTCAGGAC CTGAGGAATCAAACTGTTTGCAATGCAAGAAGGGCTGGGCCCTGCATCACCTCAAGTGTGTAGACATTGATGAGTGTGGCACAGAGGGAG CCAACTGTGGAGCTGACCAATTCTGCGTGAACACTGAGGGCTCCTATGAGTGCCGAGACTGTGCCAAGGCCTGCCTAGGCTGCATGGGGG CAGGGCCAGGTCGCTGTAAGAAGTGTAGCCCTGGCTATCAGCAGGTGGGCTCCAAGTGTCTCGATGTGGATGAGTGTGAGACAGAGGTGT GTCCGGGAGAGAACAAGCAGTGTGAAAACACCGAGGGCGGTTATCGCTGCATCTGTGCCGAGGGCTACAAGCAGATGGAAGGCATCTGTG TGAAGGAGCAGATCCCAGATTCATGAAGAGAAACTCCTCTACTGGAACATGGCTGTTCGAGACTTCAGTATCCTCATCAACTTGATAAAG GTATTTGATAGTCATCCTGTTCTGCATGTATGTTTGAAGTATGGGCGTCTCTTTGTGGAAGCATTTCTGAAGCAATGTATGCCGCTCCTA GACTTCAGTTTTAGAAAACACCGGGAAGATGTTCTGAGCTTACTGGAAACCTTCCAGTTGGACACAAGGCTGCTTCATCACCTGTGTGGG CATTCCAAGATTCACCAGGACACGAGACTCACCCAACATGTGCCTCTGCTCAAAAAGACCCTGGAACTTTTAGTTTGCAGAGTCAAAGCT ATGCTCACTCTCAACAATTGTAGAGAGGCTTTCTGGCTGGGCAATCTAAAAAACCGGGACTTGCAGGGTGAAGAGATTAAGTCCCAAAAT TCCCAGGAGAGCACAGCAGATGAGAGTGAGGATGACATGTCATCCCAGGCCTCCAAGAGCAAAGCCACTGAGGATGGTGAAGAAGACGAA GTAAGTGCTGGAGAAAAGGAGCAAGATAGTGATGAGAGTTATGATGACTCTGATTAGACCCCAGATAAATTGTTGCCTGCTTCTGTGTCT CTGCCAGCCTGTGATCATTTTGTGTTAGAGTTTGAAATCCGCTGTTTGCCTTTCTTACTGGTAGGATCCTTTTTTGTTCCTCTTTTTTTT TTTTTTTTTTTTTTTTTAAAGACGGGGACTCGCTGTGTTTCCCAGGCTGGAGTGCAGTGCTGCAATCTTGGCTCACTGCAACCTCCATCT CCTAGGTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACGACAGGCACATGCCACCATGCCCAGCTAATTTTTGTATTTTT AGTAGATACGGGGTTTTACCATGTCGGCCAGATGGTCTCAATCTCCTGAACTCATGATCCACCTGCCTCAGCCTCCCAAAGTGCTGGGAT TACAGGCATGAGCCACCGCTCCCAGCCATATTTTGTTCTTAAAGTGGGGTCTTTATTAACTTGTGGACATCATGGATTGTCTAACACCAT CACAGTCCCTGGCTCAGGATTCTAATGTAGCATTATTTATTGGTTTGGATAAACCCAGCTGTGCTACACTGCAGAGTAAAATCTCTGAGT CATGATTCTGGACTTTGGGAGCTAGTTTTGAAACTCTGATTTATTGTAGAACTTAGGCTTGTACCAATTTTACAAATAAATTCTGTTCTA >19413_19413_3_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000397170_FANCD2_chr3_10133865_ENST00000383806_length(amino acids)=351AA_BP= MAPWPPKGLVPAMLWGLSLFLNLPGPIWLQPSPPPQSSPPPQPHPCHTCRGLVDSFNKGLERTIRDNFGGGNTAWEEENLSKYKDSETRL VEVLEGVCSKSDFECHRLLELSEELVESWWFHKQQEAPDLFQWLCSDSLKLCCPAGTFGPSCLPCPGGTERPCGGYGQCEGEGTRGGSGH CDCQAGYGGEACGQCGLGYFEAERNASHLVCSACFGPCARCSGPEESNCLQCKKGWALHHLKCVDIDECGTEGANCGADQFCVNTEGSYE -------------------------------------------------------------- >19413_19413_4_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000452070_FANCD2_chr3_10133865_ENST00000383806_length(transcript)=2390nt_BP=1143nt GCGACGCGGTGAGGAGACGGCCCACGGCGCCCGCGGGCTGGGGCGGTCGCTTCTTCCTTCTCCGTGGCCTACGAGGGTCCCCAGCCTGGG TAAAGATGGCCCCATGGCCCCCGAAGGGCCTAGTCCCAGCTATGCTCTGGGGCCTCAGCCTCTTCCTCAACCTCCCAGGACCTATCTGGC TCCAGCCCTCTCCACCTCCCCAGTCTTCTCCCCCGCCTCAGCCCCATCCGTGTCATACCTGCCGGGGACTGGTTGACAGCTTTAACAAGG GCCTGGAGAGAACCATCCGGGACAACTTTGGAGGTGGAAACACTGCCTGGGAGGAAGAGAATTTGTCCAAATACAAAGACAGTGAGACCC GCCTGGTAGAGGTGCTGGAGGGTGTGTGCAGCAAGTCAGACTTCGAGTGCCACCGCCTGCTGGAGCTGAGTGAGGAGCTGGTGGAGAGCT GGTGGTTTCACAAGCAGCAGGAGGCCCCGGACCTCTTCCAGTGGCTGTGCTCAGATTCCCTGAAGCTCTGCTGCCCCGCAGGCACCTTCG GGCCCTCCTGCCTTCCCTGTCCTGGGGGAACAGAGAGGCCCTGCGGTGGCTACGGGCAGTGTGAAGGAGAAGGGACACGAGGGGGCAGCG GGCACTGTGACTGCCAAGCCGGCTACGGGGGTGAGGCCTGTGGCCAGTGTGGCCTTGGCTACTTTGAGGCAGAACGCAACGCCAGCCATC TGGTATGTTCGGCTTGTTTTGGCCCCTGTGCCCGATGCTCAGGACCTGAGGAATCAAACTGTTTGCAATGCAAGAAGGGCTGGGCCCTGC ATCACCTCAAGTGTGTAGACATTGATGAGTGTGGCACAGAGGGAGCCAACTGTGGAGCTGACCAATTCTGCGTGAACACTGAGGGCTCCT ATGAGTGCCGAGACTGTGCCAAGGCCTGCCTAGGCTGCATGGGGGCAGGGCCAGGTCGCTGTAAGAAGTGTAGCCCTGGCTATCAGCAGG TGGGCTCCAAGTGTCTCGATGTGGATGAGTGTGAGACAGAGGTGTGTCCGGGAGAGAACAAGCAGTGTGAAAACACCGAGGGCGGTTATC GCTGCATCTGTGCCGAGGGCTACAAGCAGATGGAAGGCATCTGTGTGAAGGAGCAGATCCCAGATTCATGAAGAGAAACTCCTCTACTGG AACATGGCTGTTCGAGACTTCAGTATCCTCATCAACTTGATAAAGGTATTTGATAGTCATCCTGTTCTGCATGTATGTTTGAAGTATGGG CGTCTCTTTGTGGAAGCATTTCTGAAGCAATGTATGCCGCTCCTAGACTTCAGTTTTAGAAAACACCGGGAAGATGTTCTGAGCTTACTG GAAACCTTCCAGTTGGACACAAGGCTGCTTCATCACCTGTGTGGGCATTCCAAGATTCACCAGGACACGAGACTCACCCAACATGTGCCT CTGCTCAAAAAGACCCTGGAACTTTTAGTTTGCAGAGTCAAAGCTATGCTCACTCTCAACAATTGTAGAGAGGCTTTCTGGCTGGGCAAT CTAAAAAACCGGGACTTGCAGGGTGAAGAGATTAAGTCCCAAAATTCCCAGGAGAGCACAGCAGATGAGAGTGAGGATGACATGTCATCC CAGGCCTCCAAGAGCAAAGCCACTGAGGATGGTGAAGAAGACGAAGTAAGTGCTGGAGAAAAGGAGCAAGATAGTGATGAGAGTTATGAT GACTCTGATTAGACCCCAGATAAATTGTTGCCTGCTTCTGTGTCTCTGCCAGCCTGTGATCATTTTGTGTTAGAGTTTGAAATCCGCTGT TTGCCTTTCTTACTGGTAGGATCCTTTTTTGTTCCTCTTTTTTTTTTTTTTTTTTTTTTTTTAAAGACGGGGACTCGCTGTGTTTCCCAG GCTGGAGTGCAGTGCTGCAATCTTGGCTCACTGCAACCTCCATCTCCTAGGTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGG GACGACAGGCACATGCCACCATGCCCAGCTAATTTTTGTATTTTTAGTAGATACGGGGTTTTACCATGTCGGCCAGATGGTCTCAATCTC CTGAACTCATGATCCACCTGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCATGAGCCACCGCTCCCAGCCATATTTTGTTCTTAAAGT GGGGTCTTTATTAACTTGTGGACATCATGGATTGTCTAACACCATCACAGTCCCTGGCTCAGGATTCTAATGTAGCATTATTTATTGGTT TGGATAAACCCAGCTGTGCTACACTGCAGAGTAAAATCTCTGAGTCATGATTCTGGACTTTGGGAGCTAGTTTTGAAACTCTGATTTATT >19413_19413_4_CRELD1-FANCD2_CRELD1_chr3_9985199_ENST00000452070_FANCD2_chr3_10133865_ENST00000383806_length(amino acids)=351AA_BP= MAPWPPKGLVPAMLWGLSLFLNLPGPIWLQPSPPPQSSPPPQPHPCHTCRGLVDSFNKGLERTIRDNFGGGNTAWEEENLSKYKDSETRL VEVLEGVCSKSDFECHRLLELSEELVESWWFHKQQEAPDLFQWLCSDSLKLCCPAGTFGPSCLPCPGGTERPCGGYGQCEGEGTRGGSGH CDCQAGYGGEACGQCGLGYFEAERNASHLVCSACFGPCARCSGPEESNCLQCKKGWALHHLKCVDIDECGTEGANCGADQFCVNTEGSYE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CRELD1-FANCD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000431693 | 0 | 8 | 248_359 | 0 | 242.0 | BRCA2 | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000431693 | 0 | 8 | 1_291 | 0 | 242.0 | FANCE |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000287647 | 36 | 43 | 248_359 | 1259.0 | 1472.0 | BRCA2 | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000383806 | 35 | 43 | 248_359 | 1227.6666666666667 | 1469.6666666666667 | BRCA2 | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000383807 | 36 | 44 | 248_359 | 1259.0 | 1452.0 | BRCA2 | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000419585 | 36 | 44 | 248_359 | 1259.0 | 1452.0 | BRCA2 | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000287647 | 36 | 43 | 1_291 | 1259.0 | 1472.0 | FANCE | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000383806 | 35 | 43 | 1_291 | 1227.6666666666667 | 1469.6666666666667 | FANCE | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000383807 | 36 | 44 | 1_291 | 1259.0 | 1452.0 | FANCE | |
| Tgene | FANCD2 | chr3:9985199 | chr3:10133865 | ENST00000419585 | 36 | 44 | 1_291 | 1259.0 | 1452.0 | FANCE |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CRELD1-FANCD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CRELD1-FANCD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
