
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CROT-KIAA1109 (FusionGDB2 ID:19539) |
Fusion Gene Summary for CROT-KIAA1109 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CROT-KIAA1109 | Fusion gene ID: 19539 | Hgene | Tgene | Gene symbol | CROT | KIAA1109 | Gene ID | 54677 | 84162 |
| Gene name | carnitine O-octanoyltransferase | KIAA1109 | |
| Synonyms | COT | ALKKUCS|FSA|Tweek | |
| Cytomap | 7q21.12 | 4q27 | |
| Type of gene | protein-coding | protein-coding | |
| Description | peroxisomal carnitine O-octanoyltransferaseperoxisomal carnitine acyltransferase | transmembrane protein KIAA1109fragile site-associated protein | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q9UKG9 | Q2LD37 | |
| Ensembl transtripts involved in fusion gene | ENST00000331536, ENST00000419147, ENST00000442291, ENST00000412227, | ENST00000388738, ENST00000455637, ENST00000495260, ENST00000264501, | |
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 4 X 5 X 3=60 |
| # samples | 3 | 5 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CROT [Title/Abstract] AND KIAA1109 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CROT(87022383)-KIAA1109(123283186), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CROT | GO:0006091 | generation of precursor metabolites and energy | 10486279 |
| Hgene | CROT | GO:0006635 | fatty acid beta-oxidation | 21619872 |
| Hgene | CROT | GO:0051791 | medium-chain fatty acid metabolic process | 10486279 |
 Fusion gene breakpoints across CROT (5'-gene) Fusion gene breakpoints across CROT (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
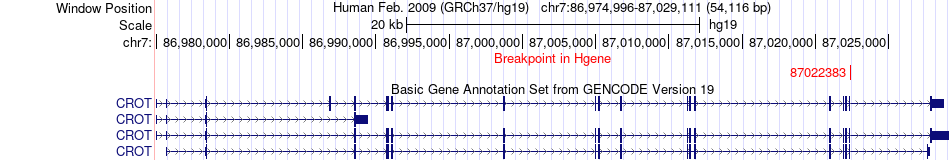 |
 Fusion gene breakpoints across KIAA1109 (3'-gene) Fusion gene breakpoints across KIAA1109 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
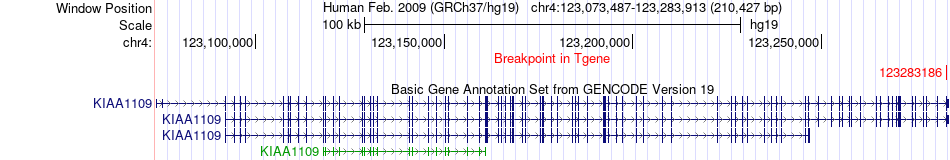 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | TCGA-IN-8462-11A | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
Top |
Fusion Gene ORF analysis for CROT-KIAA1109 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000331536 | ENST00000388738 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000331536 | ENST00000455637 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000331536 | ENST00000495260 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000419147 | ENST00000388738 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000419147 | ENST00000455637 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000419147 | ENST00000495260 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000442291 | ENST00000388738 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000442291 | ENST00000455637 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| 5CDS-intron | ENST00000442291 | ENST00000495260 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| In-frame | ENST00000331536 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| In-frame | ENST00000419147 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| In-frame | ENST00000442291 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| intron-3CDS | ENST00000412227 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| intron-intron | ENST00000412227 | ENST00000388738 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| intron-intron | ENST00000412227 | ENST00000455637 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
| intron-intron | ENST00000412227 | ENST00000495260 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000419147 | CROT | chr7 | 87022383 | + | ENST00000264501 | KIAA1109 | chr4 | 123283186 | + | 2723 | 2001 | 175 | 2217 | 680 |
| ENST00000331536 | CROT | chr7 | 87022383 | + | ENST00000264501 | KIAA1109 | chr4 | 123283186 | + | 2625 | 1903 | 161 | 2119 | 652 |
| ENST00000442291 | CROT | chr7 | 87022383 | + | ENST00000264501 | KIAA1109 | chr4 | 123283186 | + | 2557 | 1835 | 93 | 2051 | 652 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000419147 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + | 0.000253316 | 0.9997466 |
| ENST00000331536 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + | 0.000209208 | 0.99979085 |
| ENST00000442291 | ENST00000264501 | CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283186 | + | 0.000176802 | 0.9998233 |
Top |
Fusion Genomic Features for CROT-KIAA1109 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283185 | + | 0.05702558 | 0.9429744 |
| CROT | chr7 | 87022383 | + | KIAA1109 | chr4 | 123283185 | + | 0.05702558 | 0.9429744 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
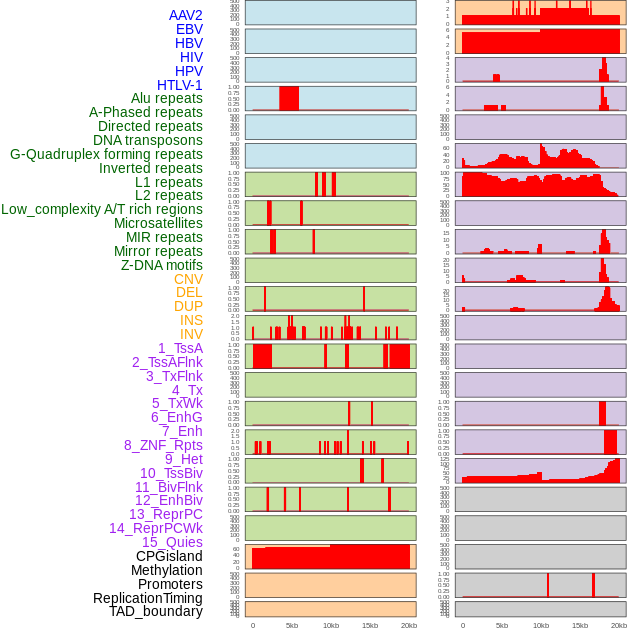 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
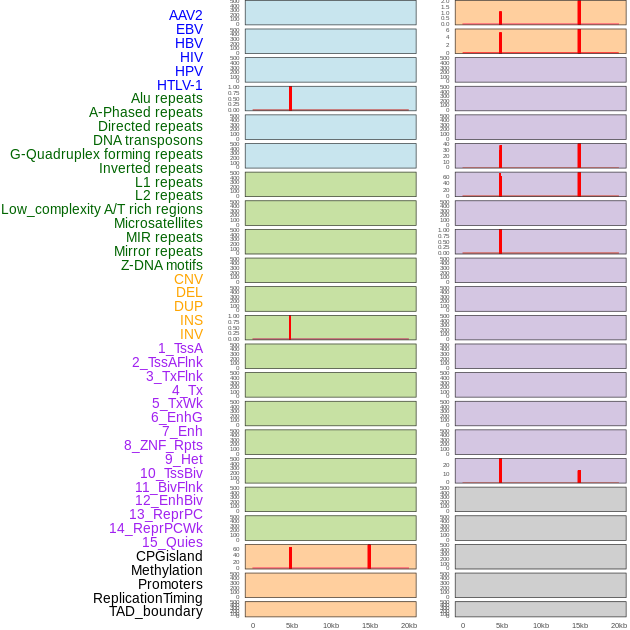 |
Top |
Fusion Protein Features for CROT-KIAA1109 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:87022383/chr4:123283186) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CROT | KIAA1109 |
| FUNCTION: Beta-oxidation of fatty acids. The highest activity concerns the C6 to C10 chain length substrate. Converts the end product of pristanic acid beta oxidation, 4,8-dimethylnonanoyl-CoA, to its corresponding carnitine ester. {ECO:0000269|PubMed:10486279}. | FUNCTION: Plays a role in endosomal trafficking and endosome recycling. Also involved in the actin cytoskeleton and cilia structural dynamics (PubMed:30906834). Acts as regulator of phagocytosis (PubMed:31540829). {ECO:0000269|PubMed:30906834, ECO:0000269|PubMed:31540829}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000331536 | + | 17 | 18 | 534_537 | 572 | 613.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000419147 | + | 18 | 19 | 534_537 | 600 | 641.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000331536 | + | 17 | 18 | 410_417 | 572 | 613.0 | Region | Coenzyme A binding |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000419147 | + | 18 | 19 | 410_417 | 600 | 641.0 | Region | Coenzyme A binding |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 1154_1157 | 0 | 3681.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 1226_1233 | 0 | 3681.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 1237_1240 | 0 | 3681.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 1417_1421 | 0 | 3681.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 1544_1547 | 0 | 3681.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 1991_1994 | 0 | 3681.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 3838_3841 | 0 | 3681.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 4110_4158 | 0 | 3681.0 | Compositional bias | Note=Ser-rich | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 772_779 | 0 | 3681.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000455637 | 0 | 64 | 26_46 | 0 | 3681.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000412227 | + | 1 | 4 | 534_537 | 0 | 88.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000331536 | + | 17 | 18 | 610_612 | 572 | 613.0 | Motif | Microbody targeting signal |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000412227 | + | 1 | 4 | 610_612 | 0 | 88.0 | Motif | Microbody targeting signal |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000419147 | + | 18 | 19 | 610_612 | 600 | 641.0 | Motif | Microbody targeting signal |
| Hgene | CROT | chr7:87022383 | chr4:123283186 | ENST00000412227 | + | 1 | 4 | 410_417 | 0 | 88.0 | Region | Coenzyme A binding |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 1154_1157 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 1226_1233 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 1237_1240 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 1417_1421 | 4933 | 5006.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 1544_1547 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 1991_1994 | 4933 | 5006.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 3838_3841 | 4933 | 5006.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 4110_4158 | 4933 | 5006.0 | Compositional bias | Note=Ser-rich | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 772_779 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 1154_1157 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 1226_1233 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 1237_1240 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 1417_1421 | 4933 | 5006.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 1544_1547 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 1991_1994 | 4933 | 5006.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 3838_3841 | 4933 | 5006.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 4110_4158 | 4933 | 5006.0 | Compositional bias | Note=Ser-rich | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 772_779 | 4933 | 5006.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000264501 | 84 | 86 | 26_46 | 4933 | 5006.0 | Transmembrane | Helical | |
| Tgene | KIAA1109 | chr7:87022383 | chr4:123283186 | ENST00000388738 | 82 | 84 | 26_46 | 4933 | 5006.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CROT-KIAA1109 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19539_19539_1_CROT-KIAA1109_CROT_chr7_87022383_ENST00000331536_KIAA1109_chr4_123283186_ENST00000264501_length(transcript)=2625nt_BP=1903nt GGGAGCGAGCCGGTGCTGCTGCAGGCTGAGGCTGCGGCAGAGGCGGCGAGGCGCGGGCGGTGAGGACGGACAGTCACCGACTTAGTCCAG TTCCCTGTGATCTCAAAACAATTGTTGCAGCAGGCTCCTGGCAGTCTCAAGCAGTTCATCTTCTTGGTGTACTGGTTTCCTATTGTGATT TTATCATGGAAAATCAATTGGCTAAATCAACTGAAGAACGAACATTTCAGTACCAGGATTCTCTTCCATCACTGCCTGTTCCTTCACTTG AAGAATCATTAAAAAAATACCTTGAATCAGTGAAACCATTTGCAAATCAAGAAGAATATAAGAAAACTGAAGAAATAGTTCAAAAATTTC AAAGTGGGATTGGAGAAAAATTGCACCAGAAATTGCTTGAAAGAGCAAAAGGAAAAAGAAATTGGCTGGAAGAGTGGTGGCTGAATGTTG CCTATCTGGATGTTCGTATACCATCACAATTGAATGTCAACTTTGCGGGTCCTGCAGCTCATTTTGAACACTACTGGCCTCCAAAGGAAG GGACTCAATTAGAAAGAGGAAGTATAACTCTTTGGCATAACTTGAACTACTGGCAGCTATTAAGAAAAGAAAAAGTGCCTGTTCATAAAG TTGGAAATACTCCTCTAGATATGAATCAATTCCGAATGCTATTTTCTACCTGCAAGGTTCCAGGAATTACTAGAGACTCCATTATGAATT ATTTTAGGACTGAGAGTGAAGGGCGTTCCCCAAACCACATTGTAGTGCTGTGTCGAGGCCGAGCTTTTGTCTTTGATGTAATACATGAAG GATGTTTGGTCACCCCGCCAGAGCTTCTCAGACAACTGACATATATCCACAAGAAGTGCCATAGTGAACCTGATGGACCTGGGATTGCAG CATTAACTAGTGAGGAGCGAACTCGATGGGCTAAGGCACGAGAATATCTGATTGGTCTTGATCCAGAGAACTTGGCTTTGTTAGAAAAAA TTCAGAGTAGTTTACTGGTATATTCCATGGAGGATAGCAGTCCACATGTAACACCAGAGGATTATTCTGAGATTATTGCAGCCATCCTTA TTGGAGATCCAACAGTACGCTGGGGTGACAAATCCTATAACTTGATTTCCTTTTCTAATGGAGTATTTGGCTGTAATTGTGATCATGCTC CTTTTGATGCAATGATTATGGTGAACATCAGTTATTATGTGGATGAGAAAATTTTTCAGAATGAAGGAAGATGGAAGGGTTCAGAGAAGG TACGAGATATACCACTTCCAGAAGAGCTCATTTTCATTGTGGATGAGAAAGTTTTAAATGACATCAACCAAGCTAAAGCCCAGTATCTCA GGGAGGCATCTGATCTACAGATTGCGGCTTATGCCTTTACATCTTTTGGCAAAAAGCTAACCAAGAACAAGATGCTTCACCCGGATACGT TTATTCAGCTTGCACTTCAGCTGGCCTATTACAGACTTCATGGACACCCTGGTTGTTGCTATGAAACAGCTATGACAAGACATTTTTATC ATGGCCGTACAGAGACTATGCGATCATGCACAGTTGAAGCAGTGAGGTGGTGCCAGTCCATGCAGGATCCTTCTGTCAATCTTCGTGAGC GGCAGCAAAAGATGTTACAAGCTTTTGCAAAGCATAATAAAATGATGAAAGATTGTTCAGCTGGAAAAGGATTTGATCGTCACCTTTTAG GTCTCTTACTCATAGCAAAAGAGGAAGGTCTTCCTGTTCCAGAACTCTTTACGGACCCACTTTTTTCCAAAAGCGGAGGAGGTGGAAATT TTGTTCTCTCAACAAGTCTGGTTGGCTATTTACGAGTCCAGGGAGTGGTAGTTCCCATGGTACACAATGGTTATGGATTTTTCTACCATA TCAGAGATGACAGATTAATTTCTTGGACTGGAAGAAAGATTGATCCAGTAGGTGTTGATTATATTCTTCAAAAATTGGGCTTTCATCATG CTAGGACTACTATTCCTAAATGGCTTCAAAGAGGAGTCATGGATCCACTGGACAAGGTTCTGTCAGTTCTTATCAAAAAGCTCGGTACTG CACTACAGGATGAAAAGGAAAAGAAAGGCAAAGACAAAGAAGAACACTAAAAAAGTAATTTGATCTGTGAACAAATTATGATTGTGTCTG TTTTATTACACTGGAGTGTTTTTTTAGTATAATAATTTGAAATATAACTTTAAAATAATTCTAAATTTGTGGCTATAATTAAAAGTTTGT AAGTTAACCTGTTCTAGTTCCATCATTCTGTGTACAGTGAAGTATTGCATGATAATGTAAATTTTGTGAAAAACTAGATTAAAATATATA ACTGCTTGTTATGGTTTATAATTATATAATGTGCAATACAATTCCTGCATCTTTAAAATGTCTGCAGAATAACTGTGAATTTTTTTGTTA TTGGATTGGCCGTAACTTTTAGAAAAAAATCTTGTTGATGATAATGTGATTTTGGGGAGGTCATTAATTGCTTTTTCTTTTTTAAATGTA GACTTATATAAATACCTGTTTGTATATAGCTTGAGTAATTGTGATATGATTGTATACCACTAAAATATTGTTAACTATTATAATAAAGTC >19539_19539_1_CROT-KIAA1109_CROT_chr7_87022383_ENST00000331536_KIAA1109_chr4_123283186_ENST00000264501_length(amino acids)=652AA_BP=580 MVSYCDFIMENQLAKSTEERTFQYQDSLPSLPVPSLEESLKKYLESVKPFANQEEYKKTEEIVQKFQSGIGEKLHQKLLERAKGKRNWLE EWWLNVAYLDVRIPSQLNVNFAGPAAHFEHYWPPKEGTQLERGSITLWHNLNYWQLLRKEKVPVHKVGNTPLDMNQFRMLFSTCKVPGIT RDSIMNYFRTESEGRSPNHIVVLCRGRAFVFDVIHEGCLVTPPELLRQLTYIHKKCHSEPDGPGIAALTSEERTRWAKAREYLIGLDPEN LALLEKIQSSLLVYSMEDSSPHVTPEDYSEIIAAILIGDPTVRWGDKSYNLISFSNGVFGCNCDHAPFDAMIMVNISYYVDEKIFQNEGR WKGSEKVRDIPLPEELIFIVDEKVLNDINQAKAQYLREASDLQIAAYAFTSFGKKLTKNKMLHPDTFIQLALQLAYYRLHGHPGCCYETA MTRHFYHGRTETMRSCTVEAVRWCQSMQDPSVNLRERQQKMLQAFAKHNKMMKDCSAGKGFDRHLLGLLLIAKEEGLPVPELFTDPLFSK SGGGGNFVLSTSLVGYLRVQGVVVPMVHNGYGFFYHIRDDRLISWTGRKIDPVGVDYILQKLGFHHARTTIPKWLQRGVMDPLDKVLSVL -------------------------------------------------------------- >19539_19539_2_CROT-KIAA1109_CROT_chr7_87022383_ENST00000419147_KIAA1109_chr4_123283186_ENST00000264501_length(transcript)=2723nt_BP=2001nt TTTGAGGCCCAAGGGGGAGCGAGCCGGTGCTGCTGCAGGCTGAGGCTGCGGCAGAGGCGGCGAGGCGCGGGCGGTGAGGACGGACAGTCA CCGACTTAGTCCAGTTCCCTGTGATCTCAAAACAATTGTTGCAGCAGGCTCCTGGCAGTCTCAAGCAGTTCATCTTCTTGGTGTACTGGT TTCCTATTGTGATTTTATCATGGAAAATCAATTGGCTAAATCAACTGAAGAACGAACATTTCAGTACCAGGATTCTCTTCCATCACTGCC TGTTCCTTCACTTGAAGAATCATTAAAAAAATACCTTGAATCAGTCACCAGAACATGCTACCAGATAAGGGGTCTTGATCCAGATGCTAA GAGAGGGTTCTTGGATCTCACGCGGGAAGGAATTCAAGTGAAACCATTTGCAAATCAAGAAGAATATAAGAAAACTGAAGAAATAGTTCA AAAATTTCAAAGTGGGATTGGAGAAAAATTGCACCAGAAATTGCTTGAAAGAGCAAAAGGAAAAAGAAATTGGCTGGAAGAGTGGTGGCT GAATGTTGCCTATCTGGATGTTCGTATACCATCACAATTGAATGTCAACTTTGCGGGTCCTGCAGCTCATTTTGAACACTACTGGCCTCC AAAGGAAGGGACTCAATTAGAAAGAGGAAGTATAACTCTTTGGCATAACTTGAACTACTGGCAGCTATTAAGAAAAGAAAAAGTGCCTGT TCATAAAGTTGGAAATACTCCTCTAGATATGAATCAATTCCGAATGCTATTTTCTACCTGCAAGGTTCCAGGAATTACTAGAGACTCCAT TATGAATTATTTTAGGACTGAGAGTGAAGGGCGTTCCCCAAACCACATTGTAGTGCTGTGTCGAGGCCGAGCTTTTGTCTTTGATGTAAT ACATGAAGGATGTTTGGTCACCCCGCCAGAGCTTCTCAGACAACTGACATATATCCACAAGAAGTGCCATAGTGAACCTGATGGACCTGG GATTGCAGCATTAACTAGTGAGGAGCGAACTCGATGGGCTAAGGCACGAGAATATCTGATTGGTCTTGATCCAGAGAACTTGGCTTTGTT AGAAAAAATTCAGAGTAGTTTACTGGTATATTCCATGGAGGATAGCAGTCCACATGTAACACCAGAGGATTATTCTGAGATTATTGCAGC CATCCTTATTGGAGATCCAACAGTACGCTGGGGTGACAAATCCTATAACTTGATTTCCTTTTCTAATGGAGTATTTGGCTGTAATTGTGA TCATGCTCCTTTTGATGCAATGATTATGGTGAACATCAGTTATTATGTGGATGAGAAAATTTTTCAGAATGAAGGAAGATGGAAGGGTTC AGAGAAGGTACGAGATATACCACTTCCAGAAGAGCTCATTTTCATTGTGGATGAGAAAGTTTTAAATGACATCAACCAAGCTAAAGCCCA GTATCTCAGGGAGGCATCTGATCTACAGATTGCGGCTTATGCCTTTACATCTTTTGGCAAAAAGCTAACCAAGAACAAGATGCTTCACCC GGATACGTTTATTCAGCTTGCACTTCAGCTGGCCTATTACAGACTTCATGGACACCCTGGTTGTTGCTATGAAACAGCTATGACAAGACA TTTTTATCATGGCCGTACAGAGACTATGCGATCATGCACAGTTGAAGCAGTGAGGTGGTGCCAGTCCATGCAGGATCCTTCTGTCAATCT TCGTGAGCGGCAGCAAAAGATGTTACAAGCTTTTGCAAAGCATAATAAAATGATGAAAGATTGTTCAGCTGGAAAAGGATTTGATCGTCA CCTTTTAGGTCTCTTACTCATAGCAAAAGAGGAAGGTCTTCCTGTTCCAGAACTCTTTACGGACCCACTTTTTTCCAAAAGCGGAGGAGG TGGAAATTTTGTTCTCTCAACAAGTCTGGTTGGCTATTTACGAGTCCAGGGAGTGGTAGTTCCCATGGTACACAATGGTTATGGATTTTT CTACCATATCAGAGATGACAGATTAATTTCTTGGACTGGAAGAAAGATTGATCCAGTAGGTGTTGATTATATTCTTCAAAAATTGGGCTT TCATCATGCTAGGACTACTATTCCTAAATGGCTTCAAAGAGGAGTCATGGATCCACTGGACAAGGTTCTGTCAGTTCTTATCAAAAAGCT CGGTACTGCACTACAGGATGAAAAGGAAAAGAAAGGCAAAGACAAAGAAGAACACTAAAAAAGTAATTTGATCTGTGAACAAATTATGAT TGTGTCTGTTTTATTACACTGGAGTGTTTTTTTAGTATAATAATTTGAAATATAACTTTAAAATAATTCTAAATTTGTGGCTATAATTAA AAGTTTGTAAGTTAACCTGTTCTAGTTCCATCATTCTGTGTACAGTGAAGTATTGCATGATAATGTAAATTTTGTGAAAAACTAGATTAA AATATATAACTGCTTGTTATGGTTTATAATTATATAATGTGCAATACAATTCCTGCATCTTTAAAATGTCTGCAGAATAACTGTGAATTT TTTTGTTATTGGATTGGCCGTAACTTTTAGAAAAAAATCTTGTTGATGATAATGTGATTTTGGGGAGGTCATTAATTGCTTTTTCTTTTT TAAATGTAGACTTATATAAATACCTGTTTGTATATAGCTTGAGTAATTGTGATATGATTGTATACCACTAAAATATTGTTAACTATTATA >19539_19539_2_CROT-KIAA1109_CROT_chr7_87022383_ENST00000419147_KIAA1109_chr4_123283186_ENST00000264501_length(amino acids)=680AA_BP=608 MVSYCDFIMENQLAKSTEERTFQYQDSLPSLPVPSLEESLKKYLESVTRTCYQIRGLDPDAKRGFLDLTREGIQVKPFANQEEYKKTEEI VQKFQSGIGEKLHQKLLERAKGKRNWLEEWWLNVAYLDVRIPSQLNVNFAGPAAHFEHYWPPKEGTQLERGSITLWHNLNYWQLLRKEKV PVHKVGNTPLDMNQFRMLFSTCKVPGITRDSIMNYFRTESEGRSPNHIVVLCRGRAFVFDVIHEGCLVTPPELLRQLTYIHKKCHSEPDG PGIAALTSEERTRWAKAREYLIGLDPENLALLEKIQSSLLVYSMEDSSPHVTPEDYSEIIAAILIGDPTVRWGDKSYNLISFSNGVFGCN CDHAPFDAMIMVNISYYVDEKIFQNEGRWKGSEKVRDIPLPEELIFIVDEKVLNDINQAKAQYLREASDLQIAAYAFTSFGKKLTKNKML HPDTFIQLALQLAYYRLHGHPGCCYETAMTRHFYHGRTETMRSCTVEAVRWCQSMQDPSVNLRERQQKMLQAFAKHNKMMKDCSAGKGFD RHLLGLLLIAKEEGLPVPELFTDPLFSKSGGGGNFVLSTSLVGYLRVQGVVVPMVHNGYGFFYHIRDDRLISWTGRKIDPVGVDYILQKL -------------------------------------------------------------- >19539_19539_3_CROT-KIAA1109_CROT_chr7_87022383_ENST00000442291_KIAA1109_chr4_123283186_ENST00000264501_length(transcript)=2557nt_BP=1835nt CACAGTCACCGACTTAGTCCAGTTCCCTGTGATCTCAAAACAATTGTTGCAGCAGGCTCCTGGCAGTCTCAAGCAGTTCATCTTCTTGGT GTACTGGTTTCCTATTGTGATTTTATCATGGAAAATCAATTGGCTAAATCAACTGAAGAACGAACATTTCAGTACCAGGATTCTCTTCCA TCACTGCCTGTTCCTTCACTTGAAGAATCATTAAAAAAATACCTTGAATCAGTGAAACCATTTGCAAATCAAGAAGAATATAAGAAAACT GAAGAAATAGTTCAAAAATTTCAAAGTGGGATTGGAGAAAAATTGCACCAGAAATTGCTTGAAAGAGCAAAAGGAAAAAGAAATTGGCTG GAAGAGTGGTGGCTGAATGTTGCCTATCTGGATGTTCGTATACCATCACAATTGAATGTCAACTTTGCGGGTCCTGCAGCTCATTTTGAA CACTACTGGCCTCCAAAGGAAGGGACTCAATTAGAAAGAGGAAGTATAACTCTTTGGCATAACTTGAACTACTGGCAGCTATTAAGAAAA GAAAAAGTGCCTGTTCATAAAGTTGGAAATACTCCTCTAGATATGAATCAATTCCGAATGCTATTTTCTACCTGCAAGGTTCCAGGAATT ACTAGAGACTCCATTATGAATTATTTTAGGACTGAGAGTGAAGGGCGTTCCCCAAACCACATTGTAGTGCTGTGTCGAGGCCGAGCTTTT GTCTTTGATGTAATACATGAAGGATGTTTGGTCACCCCGCCAGAGCTTCTCAGACAACTGACATATATCCACAAGAAGTGCCATAGTGAA CCTGATGGACCTGGGATTGCAGCATTAACTAGTGAGGAGCGAACTCGATGGGCTAAGGCACGAGAATATCTGATTGGTCTTGATCCAGAG AACTTGGCTTTGTTAGAAAAAATTCAGAGTAGTTTACTGGTATATTCCATGGAGGATAGCAGTCCACATGTAACACCAGAGGATTATTCT GAGATTATTGCAGCCATCCTTATTGGAGATCCAACAGTACGCTGGGGTGACAAATCCTATAACTTGATTTCCTTTTCTAATGGAGTATTT GGCTGTAATTGTGATCATGCTCCTTTTGATGCAATGATTATGGTGAACATCAGTTATTATGTGGATGAGAAAATTTTTCAGAATGAAGGA AGATGGAAGGGTTCAGAGAAGGTACGAGATATACCACTTCCAGAAGAGCTCATTTTCATTGTGGATGAGAAAGTTTTAAATGACATCAAC CAAGCTAAAGCCCAGTATCTCAGGGAGGCATCTGATCTACAGATTGCGGCTTATGCCTTTACATCTTTTGGCAAAAAGCTAACCAAGAAC AAGATGCTTCACCCGGATACGTTTATTCAGCTTGCACTTCAGCTGGCCTATTACAGACTTCATGGACACCCTGGTTGTTGCTATGAAACA GCTATGACAAGACATTTTTATCATGGCCGTACAGAGACTATGCGATCATGCACAGTTGAAGCAGTGAGGTGGTGCCAGTCCATGCAGGAT CCTTCTGTCAATCTTCGTGAGCGGCAGCAAAAGATGTTACAAGCTTTTGCAAAGCATAATAAAATGATGAAAGATTGTTCAGCTGGAAAA GGATTTGATCGTCACCTTTTAGGTCTCTTACTCATAGCAAAAGAGGAAGGTCTTCCTGTTCCAGAACTCTTTACGGACCCACTTTTTTCC AAAAGCGGAGGAGGTGGAAATTTTGTTCTCTCAACAAGTCTGGTTGGCTATTTACGAGTCCAGGGAGTGGTAGTTCCCATGGTACACAAT GGTTATGGATTTTTCTACCATATCAGAGATGACAGATTAATTTCTTGGACTGGAAGAAAGATTGATCCAGTAGGTGTTGATTATATTCTT CAAAAATTGGGCTTTCATCATGCTAGGACTACTATTCCTAAATGGCTTCAAAGAGGAGTCATGGATCCACTGGACAAGGTTCTGTCAGTT CTTATCAAAAAGCTCGGTACTGCACTACAGGATGAAAAGGAAAAGAAAGGCAAAGACAAAGAAGAACACTAAAAAAGTAATTTGATCTGT GAACAAATTATGATTGTGTCTGTTTTATTACACTGGAGTGTTTTTTTAGTATAATAATTTGAAATATAACTTTAAAATAATTCTAAATTT GTGGCTATAATTAAAAGTTTGTAAGTTAACCTGTTCTAGTTCCATCATTCTGTGTACAGTGAAGTATTGCATGATAATGTAAATTTTGTG AAAAACTAGATTAAAATATATAACTGCTTGTTATGGTTTATAATTATATAATGTGCAATACAATTCCTGCATCTTTAAAATGTCTGCAGA ATAACTGTGAATTTTTTTGTTATTGGATTGGCCGTAACTTTTAGAAAAAAATCTTGTTGATGATAATGTGATTTTGGGGAGGTCATTAAT TGCTTTTTCTTTTTTAAATGTAGACTTATATAAATACCTGTTTGTATATAGCTTGAGTAATTGTGATATGATTGTATACCACTAAAATAT >19539_19539_3_CROT-KIAA1109_CROT_chr7_87022383_ENST00000442291_KIAA1109_chr4_123283186_ENST00000264501_length(amino acids)=652AA_BP=580 MVSYCDFIMENQLAKSTEERTFQYQDSLPSLPVPSLEESLKKYLESVKPFANQEEYKKTEEIVQKFQSGIGEKLHQKLLERAKGKRNWLE EWWLNVAYLDVRIPSQLNVNFAGPAAHFEHYWPPKEGTQLERGSITLWHNLNYWQLLRKEKVPVHKVGNTPLDMNQFRMLFSTCKVPGIT RDSIMNYFRTESEGRSPNHIVVLCRGRAFVFDVIHEGCLVTPPELLRQLTYIHKKCHSEPDGPGIAALTSEERTRWAKAREYLIGLDPEN LALLEKIQSSLLVYSMEDSSPHVTPEDYSEIIAAILIGDPTVRWGDKSYNLISFSNGVFGCNCDHAPFDAMIMVNISYYVDEKIFQNEGR WKGSEKVRDIPLPEELIFIVDEKVLNDINQAKAQYLREASDLQIAAYAFTSFGKKLTKNKMLHPDTFIQLALQLAYYRLHGHPGCCYETA MTRHFYHGRTETMRSCTVEAVRWCQSMQDPSVNLRERQQKMLQAFAKHNKMMKDCSAGKGFDRHLLGLLLIAKEEGLPVPELFTDPLFSK SGGGGNFVLSTSLVGYLRVQGVVVPMVHNGYGFFYHIRDDRLISWTGRKIDPVGVDYILQKLGFHHARTTIPKWLQRGVMDPLDKVLSVL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CROT-KIAA1109 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CROT-KIAA1109 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CROT-KIAA1109 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
