
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CRTC1-HNRNPM (FusionGDB2 ID:19556) |
Fusion Gene Summary for CRTC1-HNRNPM |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CRTC1-HNRNPM | Fusion gene ID: 19556 | Hgene | Tgene | Gene symbol | CRTC1 | HNRNPM | Gene ID | 23373 | 4670 |
| Gene name | CREB regulated transcription coactivator 1 | heterogeneous nuclear ribonucleoprotein M | |
| Synonyms | MAML2|MECT1|Mam-2|TORC-1|TORC1|WAMTP1 | CEAR|HNRNPM4|HNRPM|HNRPM4|HTGR1|NAGR1|hnRNP M | |
| Cytomap | 19p13.11 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CREB-regulated transcription coactivator 1Mastermind-like protein 2mucoepidermoid carcinoma translocated protein 1transducer of regulated cAMP response element-binding protein (CREB) 1 | heterogeneous nuclear ribonucleoprotein MCEA receptorN-acetylglucosamine receptor 1heterogenous nuclear ribonucleoprotein M4hnRNA-binding protein M4 | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q6UUV9 | P52272 | |
| Ensembl transtripts involved in fusion gene | ENST00000321949, ENST00000338797, ENST00000594658, ENST00000601916, | ENST00000602219, ENST00000325495, ENST00000348943, | |
| Fusion gene scores | * DoF score | 10 X 7 X 7=490 | 16 X 16 X 5=1280 |
| # samples | 10 | 18 | |
| ** MAII score | log2(10/490*10)=-2.29278174922785 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/1280*10)=-2.83007499855769 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CRTC1 [Title/Abstract] AND HNRNPM [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CRTC1(18860685)-HNRNPM(8548042), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CRTC1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 14536081 |
 Fusion gene breakpoints across CRTC1 (5'-gene) Fusion gene breakpoints across CRTC1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
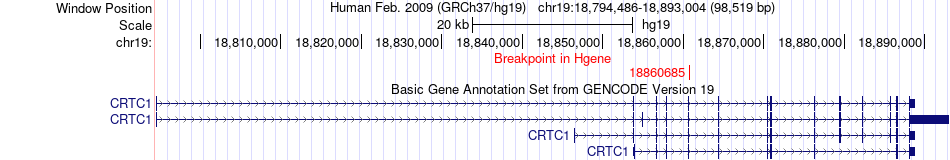 |
 Fusion gene breakpoints across HNRNPM (3'-gene) Fusion gene breakpoints across HNRNPM (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
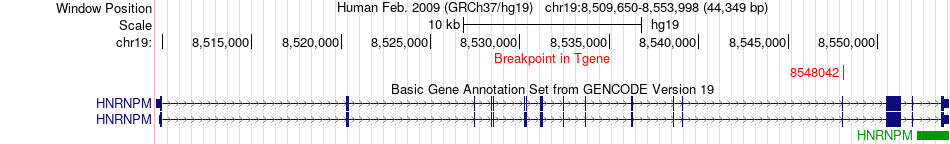 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-19-A6J5-01A | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
Top |
Fusion Gene ORF analysis for CRTC1-HNRNPM |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000321949 | ENST00000602219 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| 5CDS-intron | ENST00000338797 | ENST00000602219 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| 5CDS-intron | ENST00000594658 | ENST00000602219 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| 5CDS-intron | ENST00000601916 | ENST00000602219 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000321949 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000321949 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000338797 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000338797 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000594658 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000594658 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000601916 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
| In-frame | ENST00000601916 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000321949 | CRTC1 | chr19 | 18860685 | + | ENST00000348943 | HNRNPM | chr19 | 8548042 | + | 1897 | 564 | 26 | 1636 | 536 |
| ENST00000321949 | CRTC1 | chr19 | 18860685 | + | ENST00000325495 | HNRNPM | chr19 | 8548042 | + | 1897 | 564 | 26 | 1636 | 536 |
| ENST00000338797 | CRTC1 | chr19 | 18860685 | + | ENST00000348943 | HNRNPM | chr19 | 8548042 | + | 1944 | 611 | 25 | 1683 | 552 |
| ENST00000338797 | CRTC1 | chr19 | 18860685 | + | ENST00000325495 | HNRNPM | chr19 | 8548042 | + | 1944 | 611 | 25 | 1683 | 552 |
| ENST00000594658 | CRTC1 | chr19 | 18860685 | + | ENST00000348943 | HNRNPM | chr19 | 8548042 | + | 1780 | 447 | 17 | 1519 | 500 |
| ENST00000594658 | CRTC1 | chr19 | 18860685 | + | ENST00000325495 | HNRNPM | chr19 | 8548042 | + | 1780 | 447 | 17 | 1519 | 500 |
| ENST00000601916 | CRTC1 | chr19 | 18860685 | + | ENST00000348943 | HNRNPM | chr19 | 8548042 | + | 1745 | 412 | 24 | 1484 | 486 |
| ENST00000601916 | CRTC1 | chr19 | 18860685 | + | ENST00000325495 | HNRNPM | chr19 | 8548042 | + | 1745 | 412 | 24 | 1484 | 486 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000321949 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.020536793 | 0.9794632 |
| ENST00000321949 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.020536793 | 0.9794632 |
| ENST00000338797 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.019548103 | 0.9804519 |
| ENST00000338797 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.019548103 | 0.9804519 |
| ENST00000594658 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.018640637 | 0.98135936 |
| ENST00000594658 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.018640637 | 0.98135936 |
| ENST00000601916 | ENST00000348943 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.020500282 | 0.9794997 |
| ENST00000601916 | ENST00000325495 | CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548042 | + | 0.020500282 | 0.9794997 |
Top |
Fusion Genomic Features for CRTC1-HNRNPM |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548041 | + | 1.39E-06 | 0.99999857 |
| CRTC1 | chr19 | 18860685 | + | HNRNPM | chr19 | 8548041 | + | 1.39E-06 | 0.99999857 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
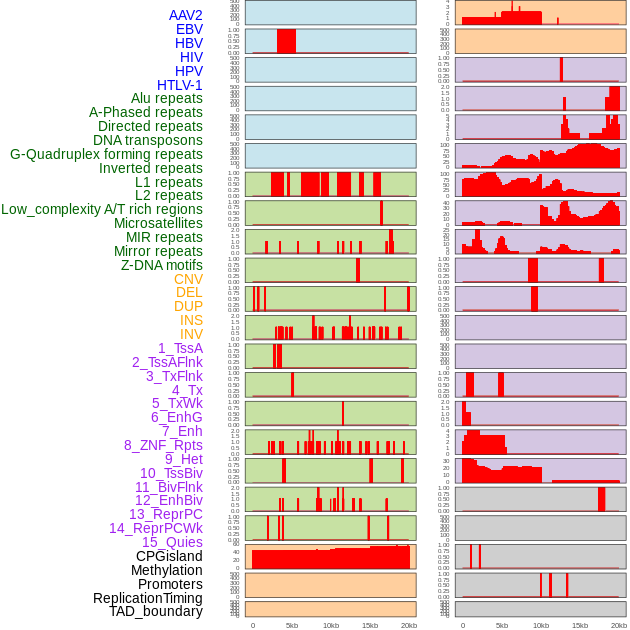 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
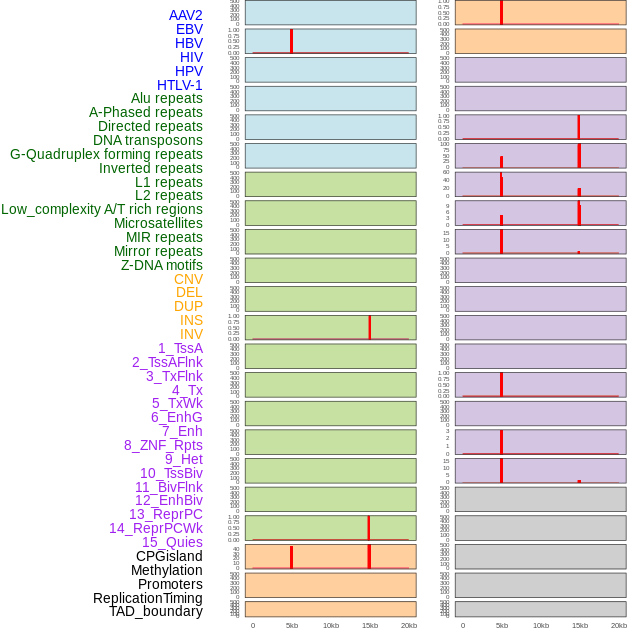 |
Top |
Fusion Protein Features for CRTC1-HNRNPM |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:18860685/chr19:8548042) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CRTC1 | HNRNPM |
| FUNCTION: Transcriptional coactivator for CREB1 which activates transcription through both consensus and variant cAMP response element (CRE) sites. Acts as a coactivator, in the SIK/TORC signaling pathway, being active when dephosphorylated and acts independently of CREB1 'Ser-133' phosphorylation. Enhances the interaction of CREB1 with TAF4. Regulates the expression of specific CREB-activated genes such as the steroidogenic gene, StAR. Potent coactivator of PGC1alpha and inducer of mitochondrial biogenesis in muscle cells. In the hippocampus, involved in late-phase long-term potentiation (L-LTP) maintenance at the Schaffer collateral-CA1 synapses. May be required for dendritic growth of developing cortical neurons (By similarity). In concert with SIK1, regulates the light-induced entrainment of the circadian clock. In response to light stimulus, coactivates the CREB-mediated transcription of PER1 which plays an important role in the photic entrainment of the circadian clock. {ECO:0000250|UniProtKB:Q157S1, ECO:0000250|UniProtKB:Q68ED7, ECO:0000269|PubMed:23699513}.; FUNCTION: (Microbial infection) Plays a role of coactivator for TAX activation of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeats (LTR). {ECO:0000269|PubMed:16809310}. | FUNCTION: Pre-mRNA binding protein in vivo, binds avidly to poly(G) and poly(U) RNA homopolymers in vitro. Involved in splicing. Acts as a receptor for carcinoembryonic antigen in Kupffer cells, may initiate a series of signaling events leading to tyrosine phosphorylation of proteins and induction of IL-1 alpha, IL-6, IL-10 and tumor necrosis factor alpha cytokines. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 390_396 | 373 | 731.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 612_616 | 373 | 731.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 390_396 | 334 | 692.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 612_616 | 334 | 692.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 653_729 | 373 | 731.0 | Domain | RRM 3 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 653_729 | 334 | 692.0 | Domain | RRM 3 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 400_608 | 373 | 731.0 | Region | Note=27 X 6 AA repeats of [GEVSTPAN]-[ILMV]-[DE]-[RH]-[MLVI]-[GAV] | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 400_608 | 334 | 692.0 | Region | Note=27 X 6 AA repeats of [GEVSTPAN]-[ILMV]-[DE]-[RH]-[MLVI]-[GAV] | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 400_405 | 373 | 731.0 | Repeat | Note=1 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 407_412 | 373 | 731.0 | Repeat | Note=2 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 415_420 | 373 | 731.0 | Repeat | Note=3 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 426_431 | 373 | 731.0 | Repeat | Note=4 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 433_438 | 373 | 731.0 | Repeat | Note=5 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 440_445 | 373 | 731.0 | Repeat | Note=6 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 446_451 | 373 | 731.0 | Repeat | Note=7 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 453_458 | 373 | 731.0 | Repeat | Note=8 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 461_466 | 373 | 731.0 | Repeat | Note=9 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 468_473 | 373 | 731.0 | Repeat | Note=10 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 475_480 | 373 | 731.0 | Repeat | Note=11 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 482_487 | 373 | 731.0 | Repeat | Note=12 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 493_498 | 373 | 731.0 | Repeat | Note=13 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 500_505 | 373 | 731.0 | Repeat | Note=14 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 507_512 | 373 | 731.0 | Repeat | Note=15 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 514_519 | 373 | 731.0 | Repeat | Note=16 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 521_526 | 373 | 731.0 | Repeat | Note=17 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 528_533 | 373 | 731.0 | Repeat | Note=18 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 540_545 | 373 | 731.0 | Repeat | Note=19 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 547_552 | 373 | 731.0 | Repeat | Note=20 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 554_559 | 373 | 731.0 | Repeat | Note=21 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 562_566 | 373 | 731.0 | Repeat | Note=22 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 567_572 | 373 | 731.0 | Repeat | Note=23 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 575_579 | 373 | 731.0 | Repeat | Note=24 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 580_585 | 373 | 731.0 | Repeat | Note=25 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 588_593 | 373 | 731.0 | Repeat | Note=26 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 603_608 | 373 | 731.0 | Repeat | Note=27 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 400_405 | 334 | 692.0 | Repeat | Note=1 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 407_412 | 334 | 692.0 | Repeat | Note=2 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 415_420 | 334 | 692.0 | Repeat | Note=3 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 426_431 | 334 | 692.0 | Repeat | Note=4 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 433_438 | 334 | 692.0 | Repeat | Note=5 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 440_445 | 334 | 692.0 | Repeat | Note=6 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 446_451 | 334 | 692.0 | Repeat | Note=7 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 453_458 | 334 | 692.0 | Repeat | Note=8 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 461_466 | 334 | 692.0 | Repeat | Note=9 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 468_473 | 334 | 692.0 | Repeat | Note=10 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 475_480 | 334 | 692.0 | Repeat | Note=11 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 482_487 | 334 | 692.0 | Repeat | Note=12 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 493_498 | 334 | 692.0 | Repeat | Note=13 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 500_505 | 334 | 692.0 | Repeat | Note=14 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 507_512 | 334 | 692.0 | Repeat | Note=15 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 514_519 | 334 | 692.0 | Repeat | Note=16 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 521_526 | 334 | 692.0 | Repeat | Note=17 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 528_533 | 334 | 692.0 | Repeat | Note=18 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 540_545 | 334 | 692.0 | Repeat | Note=19 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 547_552 | 334 | 692.0 | Repeat | Note=20 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 554_559 | 334 | 692.0 | Repeat | Note=21 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 562_566 | 334 | 692.0 | Repeat | Note=22 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 567_572 | 334 | 692.0 | Repeat | Note=23 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 575_579 | 334 | 692.0 | Repeat | Note=24 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 580_585 | 334 | 692.0 | Repeat | Note=25 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 588_593 | 334 | 692.0 | Repeat | Note=26 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 603_608 | 334 | 692.0 | Repeat | Note=27 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRTC1 | chr19:18860685 | chr19:8548042 | ENST00000321949 | + | 5 | 14 | 299_366 | 179 | 635.0 | Compositional bias | Note=Ser-rich |
| Hgene | CRTC1 | chr19:18860685 | chr19:8548042 | ENST00000338797 | + | 6 | 15 | 299_366 | 195 | 651.0 | Compositional bias | Note=Ser-rich |
| Hgene | CRTC1 | chr19:18860685 | chr19:8548042 | ENST00000321949 | + | 5 | 14 | 242_258 | 179 | 635.0 | Motif | Nuclear export signal |
| Hgene | CRTC1 | chr19:18860685 | chr19:8548042 | ENST00000338797 | + | 6 | 15 | 242_258 | 195 | 651.0 | Motif | Nuclear export signal |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 204_281 | 373 | 731.0 | Domain | RRM 2 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000325495 | 11 | 16 | 71_149 | 373 | 731.0 | Domain | RRM 1 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 204_281 | 334 | 692.0 | Domain | RRM 2 | |
| Tgene | HNRNPM | chr19:18860685 | chr19:8548042 | ENST00000348943 | 12 | 17 | 71_149 | 334 | 692.0 | Domain | RRM 1 |
Top |
Fusion Gene Sequence for CRTC1-HNRNPM |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19556_19556_1_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000321949_HNRNPM_chr19_8548042_ENST00000325495_length(transcript)=1897nt_BP=564nt GAGGAGGAGGAGGTGGCGGCGAGAAGATGGCGACTTCGAACAATCCGCGGAAATTCAGCGAGAAGATCGCGCTGCACAATCAGAAGCAGG CGGAGGAGACGGCGGCCTTCGAGGAGGTCATGAAGGACCTGAGCCTGACGCGGGCCGCGCGGCTCCAGCTCCAGAAATCCCAGTACCTGC AACTGGGCCCCAGCCGAGGCCAGTACTATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGGAGTGGCACCATGGACCTGCCCTTCCAGA CCCCCTTCCAATCCTCGGGCCTGGACACCAGCCGGACCACCCGGCACCATGGGCTGGTGGACAGGGTGTACCGGGAGCGTGGCCGGCTCG GCTCCCCACACCGCCGGCCCCTGTCAGTGGACAAACACGGACGGCAGGCCGACAGCTGCCCCTATGGCACCATGTACCTCTCACCACCCG CGGACACCAGCTGGAGAAGGACCAATTCTGACTCCGCCCTGCACCAGAGCACAATGACGCCCACGCAGCCAGAATCCTTTAGCAGTGGGT CCCAGGACGTGCACCAGAAAAGAGAAATCCTAAGTAATGCACTGAAGAGAGGAGAGATCATTGCAAAGCAGGGAGGAGGTGGAGGTGGAG GAAGCGTCCCTGGGATCGAGAGGATGGGTCCTGGCATTGACCGCCTCGGGGGTGCCGGCATGGAGCGCATGGGCGCGGGCCTGGGCCACG GCATGGATCGCGTGGGCTCCGAGATCGAGCGCATGGGCCTGGTCATGGACCGCATGGGCTCCGTGGAGCGCATGGGCTCCGGCATTGAGC GCATGGGCCCGCTGGGCCTCGACCACATGGCCTCCAGCATTGAGCGCATGGGCCAGACCATGGAGCGCATTGGCTCTGGCGTGGAGCGCA TGGGTGCCGGCATGGGCTTCGGCCTTGAGCGCATGGCCGCTCCCATCGACCGTGTGGGCCAGACCATTGAGCGCATGGGCTCTGGCGTGG AGCGCATGGGCCCTGCCATCGAGCGCATGGGCCTGAGCATGGAGCGCATGGTGCCCGCAGGTATGGGAGCTGGCCTGGAGCGCATGGGCC CCGTGATGGATCGCATGGCCACCGGCCTGGAGCGCATGGGCGCCAACAATCTGGAGCGGATGGGCCTGGAGCGCATGGGCGCCAACAGCC TCGAGCGCATGGGCCTGGAGCGCATGGGTGCCAACAGCCTCGAGCGCATGGGCCCTGCCATGGGCCCGGCCCTGGGCGCTGGCATTGAGC GCATGGGCCTGGCCATGGGTGGCGGTGGCGGTGCCAGCTTTGACCGTGCCATCGAGATGGAGCGTGGCAACTTCGGAGGAAGCTTCGCAG GTTCCTTTGGTGGAGCTGGAGGCCATGCTCCTGGGGTGGCCAGGAAGGCCTGCCAGATATTTGTGAGAAATCTGCCATTCGATTTCACAT GGAAGATGCTAAAGGACAAATTCAACGAGTGCGGCCACGTGCTGTACGCCGACATCAAGATGGAGAATGGGAAGTCCAAGGGGTGTGGCG TGGTTAAGTTCGAGTCGCCAGAGGTGGCCGAGAGAGCCTGCCGGATGATGAATGGCATGAAGCTGAGTGGCCGAGAGATTGACGTTCGAA TTGATAGAAACGCTTAAGCAGTTGCCTTTTTTAAACATCGATACGAGACCTCTGAATTTGTATTTTTTCTTGTTAACCATTTTAATTTGT TGGCTGGATGTATAAAGATGTTTAAAAAATTCAGTTGCTTTTTGGGGTAATTTGAATTACTTTTTTAATGACTGGGGTTCCATTTGACTG TTTGCATTGAGATTGCAATGTGCGCAATTTTTTTTGTAGTTGTGGCATCTTGTTGACATCGAATATGACTTTGATAATAAATACCGGTTC >19556_19556_1_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000321949_HNRNPM_chr19_8548042_ENST00000325495_length(amino acids)=536AA_BP=179 MATSNNPRKFSEKIALHNQKQAEETAAFEEVMKDLSLTRAARLQLQKSQYLQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQTPFQSSGLD TSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGRQADSCPYGTMYLSPPADTSWRRTNSDSALHQSTMTPTQPESFSSGSQDVHQKRE ILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDRLGGAGMERMGAGLGHGMDRVGSEIERMGLVMDRMGSVERMGSGIERMGPLGLDH MASSIERMGQTMERIGSGVERMGAGMGFGLERMAAPIDRVGQTIERMGSGVERMGPAIERMGLSMERMVPAGMGAGLERMGPVMDRMATG LERMGANNLERMGLERMGANSLERMGLERMGANSLERMGPAMGPALGAGIERMGLAMGGGGGASFDRAIEMERGNFGGSFAGSFGGAGGH -------------------------------------------------------------- >19556_19556_2_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000321949_HNRNPM_chr19_8548042_ENST00000348943_length(transcript)=1897nt_BP=564nt GAGGAGGAGGAGGTGGCGGCGAGAAGATGGCGACTTCGAACAATCCGCGGAAATTCAGCGAGAAGATCGCGCTGCACAATCAGAAGCAGG CGGAGGAGACGGCGGCCTTCGAGGAGGTCATGAAGGACCTGAGCCTGACGCGGGCCGCGCGGCTCCAGCTCCAGAAATCCCAGTACCTGC AACTGGGCCCCAGCCGAGGCCAGTACTATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGGAGTGGCACCATGGACCTGCCCTTCCAGA CCCCCTTCCAATCCTCGGGCCTGGACACCAGCCGGACCACCCGGCACCATGGGCTGGTGGACAGGGTGTACCGGGAGCGTGGCCGGCTCG GCTCCCCACACCGCCGGCCCCTGTCAGTGGACAAACACGGACGGCAGGCCGACAGCTGCCCCTATGGCACCATGTACCTCTCACCACCCG CGGACACCAGCTGGAGAAGGACCAATTCTGACTCCGCCCTGCACCAGAGCACAATGACGCCCACGCAGCCAGAATCCTTTAGCAGTGGGT CCCAGGACGTGCACCAGAAAAGAGAAATCCTAAGTAATGCACTGAAGAGAGGAGAGATCATTGCAAAGCAGGGAGGAGGTGGAGGTGGAG GAAGCGTCCCTGGGATCGAGAGGATGGGTCCTGGCATTGACCGCCTCGGGGGTGCCGGCATGGAGCGCATGGGCGCGGGCCTGGGCCACG GCATGGATCGCGTGGGCTCCGAGATCGAGCGCATGGGCCTGGTCATGGACCGCATGGGCTCCGTGGAGCGCATGGGCTCCGGCATTGAGC GCATGGGCCCGCTGGGCCTCGACCACATGGCCTCCAGCATTGAGCGCATGGGCCAGACCATGGAGCGCATTGGCTCTGGCGTGGAGCGCA TGGGTGCCGGCATGGGCTTCGGCCTTGAGCGCATGGCCGCTCCCATCGACCGTGTGGGCCAGACCATTGAGCGCATGGGCTCTGGCGTGG AGCGCATGGGCCCTGCCATCGAGCGCATGGGCCTGAGCATGGAGCGCATGGTGCCCGCAGGTATGGGAGCTGGCCTGGAGCGCATGGGCC CCGTGATGGATCGCATGGCCACCGGCCTGGAGCGCATGGGCGCCAACAATCTGGAGCGGATGGGCCTGGAGCGCATGGGCGCCAACAGCC TCGAGCGCATGGGCCTGGAGCGCATGGGTGCCAACAGCCTCGAGCGCATGGGCCCTGCCATGGGCCCGGCCCTGGGCGCTGGCATTGAGC GCATGGGCCTGGCCATGGGTGGCGGTGGCGGTGCCAGCTTTGACCGTGCCATCGAGATGGAGCGTGGCAACTTCGGAGGAAGCTTCGCAG GTTCCTTTGGTGGAGCTGGAGGCCATGCTCCTGGGGTGGCCAGGAAGGCCTGCCAGATATTTGTGAGAAATCTGCCATTCGATTTCACAT GGAAGATGCTAAAGGACAAATTCAACGAGTGCGGCCACGTGCTGTACGCCGACATCAAGATGGAGAATGGGAAGTCCAAGGGGTGTGGCG TGGTTAAGTTCGAGTCGCCAGAGGTGGCCGAGAGAGCCTGCCGGATGATGAATGGCATGAAGCTGAGTGGCCGAGAGATTGACGTTCGAA TTGATAGAAACGCTTAAGCAGTTGCCTTTTTTAAACATCGATACGAGACCTCTGAATTTGTATTTTTTCTTGTTAACCATTTTAATTTGT TGGCTGGATGTATAAAGATGTTTAAAAAATTCAGTTGCTTTTTGGGGTAATTTGAATTACTTTTTTAATGACTGGGGTTCCATTTGACTG TTTGCATTGAGATTGCAATGTGCGCAATTTTTTTTGTAGTTGTGGCATCTTGTTGACATCGAATATGACTTTGATAATAAATACCGGTTC >19556_19556_2_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000321949_HNRNPM_chr19_8548042_ENST00000348943_length(amino acids)=536AA_BP=179 MATSNNPRKFSEKIALHNQKQAEETAAFEEVMKDLSLTRAARLQLQKSQYLQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQTPFQSSGLD TSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGRQADSCPYGTMYLSPPADTSWRRTNSDSALHQSTMTPTQPESFSSGSQDVHQKRE ILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDRLGGAGMERMGAGLGHGMDRVGSEIERMGLVMDRMGSVERMGSGIERMGPLGLDH MASSIERMGQTMERIGSGVERMGAGMGFGLERMAAPIDRVGQTIERMGSGVERMGPAIERMGLSMERMVPAGMGAGLERMGPVMDRMATG LERMGANNLERMGLERMGANSLERMGLERMGANSLERMGPAMGPALGAGIERMGLAMGGGGGASFDRAIEMERGNFGGSFAGSFGGAGGH -------------------------------------------------------------- >19556_19556_3_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000338797_HNRNPM_chr19_8548042_ENST00000325495_length(transcript)=1944nt_BP=611nt AGGAGGAGGAGGTGGCGGCGAGAAGATGGCGACTTCGAACAATCCGCGGAAATTCAGCGAGAAGATCGCGCTGCACAATCAGAAGCAGGC GGAGGAGACGGCGGCCTTCGAGGAGGTCATGAAGGACCTGAGCCTGACGCGGGCCGCGCGGCTCCAGCTCCAGAAATCCCAGTACCTGCA ACTGGGCCCCAGCCGAGGCCAGTACTATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGGAGTGGCACCATGGACCTGCCCTTCCAGCC CAGCGGATTTCTGGGGGAGGCCCTGGCAGCGGCTCCTGTCTCTCTGACCCCCTTCCAATCCTCGGGCCTGGACACCAGCCGGACCACCCG GCACCATGGGCTGGTGGACAGGGTGTACCGGGAGCGTGGCCGGCTCGGCTCCCCACACCGCCGGCCCCTGTCAGTGGACAAACACGGACG GCAGGCCGACAGCTGCCCCTATGGCACCATGTACCTCTCACCACCCGCGGACACCAGCTGGAGAAGGACCAATTCTGACTCCGCCCTGCA CCAGAGCACAATGACGCCCACGCAGCCAGAATCCTTTAGCAGTGGGTCCCAGGACGTGCACCAGAAAAGAGAAATCCTAAGTAATGCACT GAAGAGAGGAGAGATCATTGCAAAGCAGGGAGGAGGTGGAGGTGGAGGAAGCGTCCCTGGGATCGAGAGGATGGGTCCTGGCATTGACCG CCTCGGGGGTGCCGGCATGGAGCGCATGGGCGCGGGCCTGGGCCACGGCATGGATCGCGTGGGCTCCGAGATCGAGCGCATGGGCCTGGT CATGGACCGCATGGGCTCCGTGGAGCGCATGGGCTCCGGCATTGAGCGCATGGGCCCGCTGGGCCTCGACCACATGGCCTCCAGCATTGA GCGCATGGGCCAGACCATGGAGCGCATTGGCTCTGGCGTGGAGCGCATGGGTGCCGGCATGGGCTTCGGCCTTGAGCGCATGGCCGCTCC CATCGACCGTGTGGGCCAGACCATTGAGCGCATGGGCTCTGGCGTGGAGCGCATGGGCCCTGCCATCGAGCGCATGGGCCTGAGCATGGA GCGCATGGTGCCCGCAGGTATGGGAGCTGGCCTGGAGCGCATGGGCCCCGTGATGGATCGCATGGCCACCGGCCTGGAGCGCATGGGCGC CAACAATCTGGAGCGGATGGGCCTGGAGCGCATGGGCGCCAACAGCCTCGAGCGCATGGGCCTGGAGCGCATGGGTGCCAACAGCCTCGA GCGCATGGGCCCTGCCATGGGCCCGGCCCTGGGCGCTGGCATTGAGCGCATGGGCCTGGCCATGGGTGGCGGTGGCGGTGCCAGCTTTGA CCGTGCCATCGAGATGGAGCGTGGCAACTTCGGAGGAAGCTTCGCAGGTTCCTTTGGTGGAGCTGGAGGCCATGCTCCTGGGGTGGCCAG GAAGGCCTGCCAGATATTTGTGAGAAATCTGCCATTCGATTTCACATGGAAGATGCTAAAGGACAAATTCAACGAGTGCGGCCACGTGCT GTACGCCGACATCAAGATGGAGAATGGGAAGTCCAAGGGGTGTGGCGTGGTTAAGTTCGAGTCGCCAGAGGTGGCCGAGAGAGCCTGCCG GATGATGAATGGCATGAAGCTGAGTGGCCGAGAGATTGACGTTCGAATTGATAGAAACGCTTAAGCAGTTGCCTTTTTTAAACATCGATA CGAGACCTCTGAATTTGTATTTTTTCTTGTTAACCATTTTAATTTGTTGGCTGGATGTATAAAGATGTTTAAAAAATTCAGTTGCTTTTT GGGGTAATTTGAATTACTTTTTTAATGACTGGGGTTCCATTTGACTGTTTGCATTGAGATTGCAATGTGCGCAATTTTTTTTGTAGTTGT >19556_19556_3_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000338797_HNRNPM_chr19_8548042_ENST00000325495_length(amino acids)=552AA_BP=195 MATSNNPRKFSEKIALHNQKQAEETAAFEEVMKDLSLTRAARLQLQKSQYLQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQPSGFLGEAL AAAPVSLTPFQSSGLDTSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGRQADSCPYGTMYLSPPADTSWRRTNSDSALHQSTMTPTQ PESFSSGSQDVHQKREILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDRLGGAGMERMGAGLGHGMDRVGSEIERMGLVMDRMGSVE RMGSGIERMGPLGLDHMASSIERMGQTMERIGSGVERMGAGMGFGLERMAAPIDRVGQTIERMGSGVERMGPAIERMGLSMERMVPAGMG AGLERMGPVMDRMATGLERMGANNLERMGLERMGANSLERMGLERMGANSLERMGPAMGPALGAGIERMGLAMGGGGGASFDRAIEMERG NFGGSFAGSFGGAGGHAPGVARKACQIFVRNLPFDFTWKMLKDKFNECGHVLYADIKMENGKSKGCGVVKFESPEVAERACRMMNGMKLS -------------------------------------------------------------- >19556_19556_4_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000338797_HNRNPM_chr19_8548042_ENST00000348943_length(transcript)=1944nt_BP=611nt AGGAGGAGGAGGTGGCGGCGAGAAGATGGCGACTTCGAACAATCCGCGGAAATTCAGCGAGAAGATCGCGCTGCACAATCAGAAGCAGGC GGAGGAGACGGCGGCCTTCGAGGAGGTCATGAAGGACCTGAGCCTGACGCGGGCCGCGCGGCTCCAGCTCCAGAAATCCCAGTACCTGCA ACTGGGCCCCAGCCGAGGCCAGTACTATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGGAGTGGCACCATGGACCTGCCCTTCCAGCC CAGCGGATTTCTGGGGGAGGCCCTGGCAGCGGCTCCTGTCTCTCTGACCCCCTTCCAATCCTCGGGCCTGGACACCAGCCGGACCACCCG GCACCATGGGCTGGTGGACAGGGTGTACCGGGAGCGTGGCCGGCTCGGCTCCCCACACCGCCGGCCCCTGTCAGTGGACAAACACGGACG GCAGGCCGACAGCTGCCCCTATGGCACCATGTACCTCTCACCACCCGCGGACACCAGCTGGAGAAGGACCAATTCTGACTCCGCCCTGCA CCAGAGCACAATGACGCCCACGCAGCCAGAATCCTTTAGCAGTGGGTCCCAGGACGTGCACCAGAAAAGAGAAATCCTAAGTAATGCACT GAAGAGAGGAGAGATCATTGCAAAGCAGGGAGGAGGTGGAGGTGGAGGAAGCGTCCCTGGGATCGAGAGGATGGGTCCTGGCATTGACCG CCTCGGGGGTGCCGGCATGGAGCGCATGGGCGCGGGCCTGGGCCACGGCATGGATCGCGTGGGCTCCGAGATCGAGCGCATGGGCCTGGT CATGGACCGCATGGGCTCCGTGGAGCGCATGGGCTCCGGCATTGAGCGCATGGGCCCGCTGGGCCTCGACCACATGGCCTCCAGCATTGA GCGCATGGGCCAGACCATGGAGCGCATTGGCTCTGGCGTGGAGCGCATGGGTGCCGGCATGGGCTTCGGCCTTGAGCGCATGGCCGCTCC CATCGACCGTGTGGGCCAGACCATTGAGCGCATGGGCTCTGGCGTGGAGCGCATGGGCCCTGCCATCGAGCGCATGGGCCTGAGCATGGA GCGCATGGTGCCCGCAGGTATGGGAGCTGGCCTGGAGCGCATGGGCCCCGTGATGGATCGCATGGCCACCGGCCTGGAGCGCATGGGCGC CAACAATCTGGAGCGGATGGGCCTGGAGCGCATGGGCGCCAACAGCCTCGAGCGCATGGGCCTGGAGCGCATGGGTGCCAACAGCCTCGA GCGCATGGGCCCTGCCATGGGCCCGGCCCTGGGCGCTGGCATTGAGCGCATGGGCCTGGCCATGGGTGGCGGTGGCGGTGCCAGCTTTGA CCGTGCCATCGAGATGGAGCGTGGCAACTTCGGAGGAAGCTTCGCAGGTTCCTTTGGTGGAGCTGGAGGCCATGCTCCTGGGGTGGCCAG GAAGGCCTGCCAGATATTTGTGAGAAATCTGCCATTCGATTTCACATGGAAGATGCTAAAGGACAAATTCAACGAGTGCGGCCACGTGCT GTACGCCGACATCAAGATGGAGAATGGGAAGTCCAAGGGGTGTGGCGTGGTTAAGTTCGAGTCGCCAGAGGTGGCCGAGAGAGCCTGCCG GATGATGAATGGCATGAAGCTGAGTGGCCGAGAGATTGACGTTCGAATTGATAGAAACGCTTAAGCAGTTGCCTTTTTTAAACATCGATA CGAGACCTCTGAATTTGTATTTTTTCTTGTTAACCATTTTAATTTGTTGGCTGGATGTATAAAGATGTTTAAAAAATTCAGTTGCTTTTT GGGGTAATTTGAATTACTTTTTTAATGACTGGGGTTCCATTTGACTGTTTGCATTGAGATTGCAATGTGCGCAATTTTTTTTGTAGTTGT >19556_19556_4_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000338797_HNRNPM_chr19_8548042_ENST00000348943_length(amino acids)=552AA_BP=195 MATSNNPRKFSEKIALHNQKQAEETAAFEEVMKDLSLTRAARLQLQKSQYLQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQPSGFLGEAL AAAPVSLTPFQSSGLDTSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGRQADSCPYGTMYLSPPADTSWRRTNSDSALHQSTMTPTQ PESFSSGSQDVHQKREILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDRLGGAGMERMGAGLGHGMDRVGSEIERMGLVMDRMGSVE RMGSGIERMGPLGLDHMASSIERMGQTMERIGSGVERMGAGMGFGLERMAAPIDRVGQTIERMGSGVERMGPAIERMGLSMERMVPAGMG AGLERMGPVMDRMATGLERMGANNLERMGLERMGANSLERMGLERMGANSLERMGPAMGPALGAGIERMGLAMGGGGGASFDRAIEMERG NFGGSFAGSFGGAGGHAPGVARKACQIFVRNLPFDFTWKMLKDKFNECGHVLYADIKMENGKSKGCGVVKFESPEVAERACRMMNGMKLS -------------------------------------------------------------- >19556_19556_5_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000594658_HNRNPM_chr19_8548042_ENST00000325495_length(transcript)=1780nt_BP=447nt CCCTTAGGGGAAGAGTCCTGCTCTGGCTGTTGATGCTCCAGCTCCAGAAATCCCAGTACCTGCAACTGGGCCCCAGCCGAGGCCAGTACT ATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGGAGTGGCACCATGGACCTGCCCTTCCAGACCCCCTTCCAATCCTCGGGCCTGGACA CCAGCCGGACCACCCGGCACCATGGGCTGGTGGACAGGGTGTACCGGGAGCGTGGCCGGCTCGGCTCCCCACACCGCCGGCCCCTGTCAG TGGACAAACACGGACGGCAGGCCGACAGCTGCCCCTATGGCACCATGTACCTCTCACCACCCGCGGACACCAGCTGGAGAAGGACCAATT CTGACTCCGCCCTGCACCAGAGCACAATGACGCCCACGCAGCCAGAATCCTTTAGCAGTGGGTCCCAGGACGTGCACCAGAAAAGAGAAA TCCTAAGTAATGCACTGAAGAGAGGAGAGATCATTGCAAAGCAGGGAGGAGGTGGAGGTGGAGGAAGCGTCCCTGGGATCGAGAGGATGG GTCCTGGCATTGACCGCCTCGGGGGTGCCGGCATGGAGCGCATGGGCGCGGGCCTGGGCCACGGCATGGATCGCGTGGGCTCCGAGATCG AGCGCATGGGCCTGGTCATGGACCGCATGGGCTCCGTGGAGCGCATGGGCTCCGGCATTGAGCGCATGGGCCCGCTGGGCCTCGACCACA TGGCCTCCAGCATTGAGCGCATGGGCCAGACCATGGAGCGCATTGGCTCTGGCGTGGAGCGCATGGGTGCCGGCATGGGCTTCGGCCTTG AGCGCATGGCCGCTCCCATCGACCGTGTGGGCCAGACCATTGAGCGCATGGGCTCTGGCGTGGAGCGCATGGGCCCTGCCATCGAGCGCA TGGGCCTGAGCATGGAGCGCATGGTGCCCGCAGGTATGGGAGCTGGCCTGGAGCGCATGGGCCCCGTGATGGATCGCATGGCCACCGGCC TGGAGCGCATGGGCGCCAACAATCTGGAGCGGATGGGCCTGGAGCGCATGGGCGCCAACAGCCTCGAGCGCATGGGCCTGGAGCGCATGG GTGCCAACAGCCTCGAGCGCATGGGCCCTGCCATGGGCCCGGCCCTGGGCGCTGGCATTGAGCGCATGGGCCTGGCCATGGGTGGCGGTG GCGGTGCCAGCTTTGACCGTGCCATCGAGATGGAGCGTGGCAACTTCGGAGGAAGCTTCGCAGGTTCCTTTGGTGGAGCTGGAGGCCATG CTCCTGGGGTGGCCAGGAAGGCCTGCCAGATATTTGTGAGAAATCTGCCATTCGATTTCACATGGAAGATGCTAAAGGACAAATTCAACG AGTGCGGCCACGTGCTGTACGCCGACATCAAGATGGAGAATGGGAAGTCCAAGGGGTGTGGCGTGGTTAAGTTCGAGTCGCCAGAGGTGG CCGAGAGAGCCTGCCGGATGATGAATGGCATGAAGCTGAGTGGCCGAGAGATTGACGTTCGAATTGATAGAAACGCTTAAGCAGTTGCCT TTTTTAAACATCGATACGAGACCTCTGAATTTGTATTTTTTCTTGTTAACCATTTTAATTTGTTGGCTGGATGTATAAAGATGTTTAAAA AATTCAGTTGCTTTTTGGGGTAATTTGAATTACTTTTTTAATGACTGGGGTTCCATTTGACTGTTTGCATTGAGATTGCAATGTGCGCAA >19556_19556_5_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000594658_HNRNPM_chr19_8548042_ENST00000325495_length(amino acids)=500AA_BP=143 MLWLLMLQLQKSQYLQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQTPFQSSGLDTSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGR QADSCPYGTMYLSPPADTSWRRTNSDSALHQSTMTPTQPESFSSGSQDVHQKREILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDR LGGAGMERMGAGLGHGMDRVGSEIERMGLVMDRMGSVERMGSGIERMGPLGLDHMASSIERMGQTMERIGSGVERMGAGMGFGLERMAAP IDRVGQTIERMGSGVERMGPAIERMGLSMERMVPAGMGAGLERMGPVMDRMATGLERMGANNLERMGLERMGANSLERMGLERMGANSLE RMGPAMGPALGAGIERMGLAMGGGGGASFDRAIEMERGNFGGSFAGSFGGAGGHAPGVARKACQIFVRNLPFDFTWKMLKDKFNECGHVL -------------------------------------------------------------- >19556_19556_6_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000594658_HNRNPM_chr19_8548042_ENST00000348943_length(transcript)=1780nt_BP=447nt CCCTTAGGGGAAGAGTCCTGCTCTGGCTGTTGATGCTCCAGCTCCAGAAATCCCAGTACCTGCAACTGGGCCCCAGCCGAGGCCAGTACT ATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGGAGTGGCACCATGGACCTGCCCTTCCAGACCCCCTTCCAATCCTCGGGCCTGGACA CCAGCCGGACCACCCGGCACCATGGGCTGGTGGACAGGGTGTACCGGGAGCGTGGCCGGCTCGGCTCCCCACACCGCCGGCCCCTGTCAG TGGACAAACACGGACGGCAGGCCGACAGCTGCCCCTATGGCACCATGTACCTCTCACCACCCGCGGACACCAGCTGGAGAAGGACCAATT CTGACTCCGCCCTGCACCAGAGCACAATGACGCCCACGCAGCCAGAATCCTTTAGCAGTGGGTCCCAGGACGTGCACCAGAAAAGAGAAA TCCTAAGTAATGCACTGAAGAGAGGAGAGATCATTGCAAAGCAGGGAGGAGGTGGAGGTGGAGGAAGCGTCCCTGGGATCGAGAGGATGG GTCCTGGCATTGACCGCCTCGGGGGTGCCGGCATGGAGCGCATGGGCGCGGGCCTGGGCCACGGCATGGATCGCGTGGGCTCCGAGATCG AGCGCATGGGCCTGGTCATGGACCGCATGGGCTCCGTGGAGCGCATGGGCTCCGGCATTGAGCGCATGGGCCCGCTGGGCCTCGACCACA TGGCCTCCAGCATTGAGCGCATGGGCCAGACCATGGAGCGCATTGGCTCTGGCGTGGAGCGCATGGGTGCCGGCATGGGCTTCGGCCTTG AGCGCATGGCCGCTCCCATCGACCGTGTGGGCCAGACCATTGAGCGCATGGGCTCTGGCGTGGAGCGCATGGGCCCTGCCATCGAGCGCA TGGGCCTGAGCATGGAGCGCATGGTGCCCGCAGGTATGGGAGCTGGCCTGGAGCGCATGGGCCCCGTGATGGATCGCATGGCCACCGGCC TGGAGCGCATGGGCGCCAACAATCTGGAGCGGATGGGCCTGGAGCGCATGGGCGCCAACAGCCTCGAGCGCATGGGCCTGGAGCGCATGG GTGCCAACAGCCTCGAGCGCATGGGCCCTGCCATGGGCCCGGCCCTGGGCGCTGGCATTGAGCGCATGGGCCTGGCCATGGGTGGCGGTG GCGGTGCCAGCTTTGACCGTGCCATCGAGATGGAGCGTGGCAACTTCGGAGGAAGCTTCGCAGGTTCCTTTGGTGGAGCTGGAGGCCATG CTCCTGGGGTGGCCAGGAAGGCCTGCCAGATATTTGTGAGAAATCTGCCATTCGATTTCACATGGAAGATGCTAAAGGACAAATTCAACG AGTGCGGCCACGTGCTGTACGCCGACATCAAGATGGAGAATGGGAAGTCCAAGGGGTGTGGCGTGGTTAAGTTCGAGTCGCCAGAGGTGG CCGAGAGAGCCTGCCGGATGATGAATGGCATGAAGCTGAGTGGCCGAGAGATTGACGTTCGAATTGATAGAAACGCTTAAGCAGTTGCCT TTTTTAAACATCGATACGAGACCTCTGAATTTGTATTTTTTCTTGTTAACCATTTTAATTTGTTGGCTGGATGTATAAAGATGTTTAAAA AATTCAGTTGCTTTTTGGGGTAATTTGAATTACTTTTTTAATGACTGGGGTTCCATTTGACTGTTTGCATTGAGATTGCAATGTGCGCAA >19556_19556_6_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000594658_HNRNPM_chr19_8548042_ENST00000348943_length(amino acids)=500AA_BP=143 MLWLLMLQLQKSQYLQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQTPFQSSGLDTSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGR QADSCPYGTMYLSPPADTSWRRTNSDSALHQSTMTPTQPESFSSGSQDVHQKREILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDR LGGAGMERMGAGLGHGMDRVGSEIERMGLVMDRMGSVERMGSGIERMGPLGLDHMASSIERMGQTMERIGSGVERMGAGMGFGLERMAAP IDRVGQTIERMGSGVERMGPAIERMGLSMERMVPAGMGAGLERMGPVMDRMATGLERMGANNLERMGLERMGANSLERMGLERMGANSLE RMGPAMGPALGAGIERMGLAMGGGGGASFDRAIEMERGNFGGSFAGSFGGAGGHAPGVARKACQIFVRNLPFDFTWKMLKDKFNECGHVL -------------------------------------------------------------- >19556_19556_7_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000601916_HNRNPM_chr19_8548042_ENST00000325495_length(transcript)=1745nt_BP=412nt CTCCAGCTCCAGAAATCCCAGTACCTGCAACTGGGCCCCAGCCGAGGCCAGTACTATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGG AGTGGCACCATGGACCTGCCCTTCCAGACCCCCTTCCAATCCTCGGGCCTGGACACCAGCCGGACCACCCGGCACCATGGGCTGGTGGAC AGGGTGTACCGGGAGCGTGGCCGGCTCGGCTCCCCACACCGCCGGCCCCTGTCAGTGGACAAACACGGACGGCAGGCCGACAGCTGCCCC TATGGCACCATGTACCTCTCACCACCCGCGGACACCAGCTGGAGAAGGACCAATTCTGACTCCGCCCTGCACCAGAGCACAATGACGCCC ACGCAGCCAGAATCCTTTAGCAGTGGGTCCCAGGACGTGCACCAGAAAAGAGAAATCCTAAGTAATGCACTGAAGAGAGGAGAGATCATT GCAAAGCAGGGAGGAGGTGGAGGTGGAGGAAGCGTCCCTGGGATCGAGAGGATGGGTCCTGGCATTGACCGCCTCGGGGGTGCCGGCATG GAGCGCATGGGCGCGGGCCTGGGCCACGGCATGGATCGCGTGGGCTCCGAGATCGAGCGCATGGGCCTGGTCATGGACCGCATGGGCTCC GTGGAGCGCATGGGCTCCGGCATTGAGCGCATGGGCCCGCTGGGCCTCGACCACATGGCCTCCAGCATTGAGCGCATGGGCCAGACCATG GAGCGCATTGGCTCTGGCGTGGAGCGCATGGGTGCCGGCATGGGCTTCGGCCTTGAGCGCATGGCCGCTCCCATCGACCGTGTGGGCCAG ACCATTGAGCGCATGGGCTCTGGCGTGGAGCGCATGGGCCCTGCCATCGAGCGCATGGGCCTGAGCATGGAGCGCATGGTGCCCGCAGGT ATGGGAGCTGGCCTGGAGCGCATGGGCCCCGTGATGGATCGCATGGCCACCGGCCTGGAGCGCATGGGCGCCAACAATCTGGAGCGGATG GGCCTGGAGCGCATGGGCGCCAACAGCCTCGAGCGCATGGGCCTGGAGCGCATGGGTGCCAACAGCCTCGAGCGCATGGGCCCTGCCATG GGCCCGGCCCTGGGCGCTGGCATTGAGCGCATGGGCCTGGCCATGGGTGGCGGTGGCGGTGCCAGCTTTGACCGTGCCATCGAGATGGAG CGTGGCAACTTCGGAGGAAGCTTCGCAGGTTCCTTTGGTGGAGCTGGAGGCCATGCTCCTGGGGTGGCCAGGAAGGCCTGCCAGATATTT GTGAGAAATCTGCCATTCGATTTCACATGGAAGATGCTAAAGGACAAATTCAACGAGTGCGGCCACGTGCTGTACGCCGACATCAAGATG GAGAATGGGAAGTCCAAGGGGTGTGGCGTGGTTAAGTTCGAGTCGCCAGAGGTGGCCGAGAGAGCCTGCCGGATGATGAATGGCATGAAG CTGAGTGGCCGAGAGATTGACGTTCGAATTGATAGAAACGCTTAAGCAGTTGCCTTTTTTAAACATCGATACGAGACCTCTGAATTTGTA TTTTTTCTTGTTAACCATTTTAATTTGTTGGCTGGATGTATAAAGATGTTTAAAAAATTCAGTTGCTTTTTGGGGTAATTTGAATTACTT TTTTAATGACTGGGGTTCCATTTGACTGTTTGCATTGAGATTGCAATGTGCGCAATTTTTTTTGTAGTTGTGGCATCTTGTTGACATCGA >19556_19556_7_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000601916_HNRNPM_chr19_8548042_ENST00000325495_length(amino acids)=486AA_BP=129 MQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQTPFQSSGLDTSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGRQADSCPYGTMYLSP PADTSWRRTNSDSALHQSTMTPTQPESFSSGSQDVHQKREILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDRLGGAGMERMGAGLG HGMDRVGSEIERMGLVMDRMGSVERMGSGIERMGPLGLDHMASSIERMGQTMERIGSGVERMGAGMGFGLERMAAPIDRVGQTIERMGSG VERMGPAIERMGLSMERMVPAGMGAGLERMGPVMDRMATGLERMGANNLERMGLERMGANSLERMGLERMGANSLERMGPAMGPALGAGI ERMGLAMGGGGGASFDRAIEMERGNFGGSFAGSFGGAGGHAPGVARKACQIFVRNLPFDFTWKMLKDKFNECGHVLYADIKMENGKSKGC -------------------------------------------------------------- >19556_19556_8_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000601916_HNRNPM_chr19_8548042_ENST00000348943_length(transcript)=1745nt_BP=412nt CTCCAGCTCCAGAAATCCCAGTACCTGCAACTGGGCCCCAGCCGAGGCCAGTACTATGGCGGGTCCCTGCCCAACGTGAACCAGATCGGG AGTGGCACCATGGACCTGCCCTTCCAGACCCCCTTCCAATCCTCGGGCCTGGACACCAGCCGGACCACCCGGCACCATGGGCTGGTGGAC AGGGTGTACCGGGAGCGTGGCCGGCTCGGCTCCCCACACCGCCGGCCCCTGTCAGTGGACAAACACGGACGGCAGGCCGACAGCTGCCCC TATGGCACCATGTACCTCTCACCACCCGCGGACACCAGCTGGAGAAGGACCAATTCTGACTCCGCCCTGCACCAGAGCACAATGACGCCC ACGCAGCCAGAATCCTTTAGCAGTGGGTCCCAGGACGTGCACCAGAAAAGAGAAATCCTAAGTAATGCACTGAAGAGAGGAGAGATCATT GCAAAGCAGGGAGGAGGTGGAGGTGGAGGAAGCGTCCCTGGGATCGAGAGGATGGGTCCTGGCATTGACCGCCTCGGGGGTGCCGGCATG GAGCGCATGGGCGCGGGCCTGGGCCACGGCATGGATCGCGTGGGCTCCGAGATCGAGCGCATGGGCCTGGTCATGGACCGCATGGGCTCC GTGGAGCGCATGGGCTCCGGCATTGAGCGCATGGGCCCGCTGGGCCTCGACCACATGGCCTCCAGCATTGAGCGCATGGGCCAGACCATG GAGCGCATTGGCTCTGGCGTGGAGCGCATGGGTGCCGGCATGGGCTTCGGCCTTGAGCGCATGGCCGCTCCCATCGACCGTGTGGGCCAG ACCATTGAGCGCATGGGCTCTGGCGTGGAGCGCATGGGCCCTGCCATCGAGCGCATGGGCCTGAGCATGGAGCGCATGGTGCCCGCAGGT ATGGGAGCTGGCCTGGAGCGCATGGGCCCCGTGATGGATCGCATGGCCACCGGCCTGGAGCGCATGGGCGCCAACAATCTGGAGCGGATG GGCCTGGAGCGCATGGGCGCCAACAGCCTCGAGCGCATGGGCCTGGAGCGCATGGGTGCCAACAGCCTCGAGCGCATGGGCCCTGCCATG GGCCCGGCCCTGGGCGCTGGCATTGAGCGCATGGGCCTGGCCATGGGTGGCGGTGGCGGTGCCAGCTTTGACCGTGCCATCGAGATGGAG CGTGGCAACTTCGGAGGAAGCTTCGCAGGTTCCTTTGGTGGAGCTGGAGGCCATGCTCCTGGGGTGGCCAGGAAGGCCTGCCAGATATTT GTGAGAAATCTGCCATTCGATTTCACATGGAAGATGCTAAAGGACAAATTCAACGAGTGCGGCCACGTGCTGTACGCCGACATCAAGATG GAGAATGGGAAGTCCAAGGGGTGTGGCGTGGTTAAGTTCGAGTCGCCAGAGGTGGCCGAGAGAGCCTGCCGGATGATGAATGGCATGAAG CTGAGTGGCCGAGAGATTGACGTTCGAATTGATAGAAACGCTTAAGCAGTTGCCTTTTTTAAACATCGATACGAGACCTCTGAATTTGTA TTTTTTCTTGTTAACCATTTTAATTTGTTGGCTGGATGTATAAAGATGTTTAAAAAATTCAGTTGCTTTTTGGGGTAATTTGAATTACTT TTTTAATGACTGGGGTTCCATTTGACTGTTTGCATTGAGATTGCAATGTGCGCAATTTTTTTTGTAGTTGTGGCATCTTGTTGACATCGA >19556_19556_8_CRTC1-HNRNPM_CRTC1_chr19_18860685_ENST00000601916_HNRNPM_chr19_8548042_ENST00000348943_length(amino acids)=486AA_BP=129 MQLGPSRGQYYGGSLPNVNQIGSGTMDLPFQTPFQSSGLDTSRTTRHHGLVDRVYRERGRLGSPHRRPLSVDKHGRQADSCPYGTMYLSP PADTSWRRTNSDSALHQSTMTPTQPESFSSGSQDVHQKREILSNALKRGEIIAKQGGGGGGGSVPGIERMGPGIDRLGGAGMERMGAGLG HGMDRVGSEIERMGLVMDRMGSVERMGSGIERMGPLGLDHMASSIERMGQTMERIGSGVERMGAGMGFGLERMAAPIDRVGQTIERMGSG VERMGPAIERMGLSMERMVPAGMGAGLERMGPVMDRMATGLERMGANNLERMGLERMGANSLERMGLERMGANSLERMGPAMGPALGAGI ERMGLAMGGGGGASFDRAIEMERGNFGGSFAGSFGGAGGHAPGVARKACQIFVRNLPFDFTWKMLKDKFNECGHVLYADIKMENGKSKGC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CRTC1-HNRNPM |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CRTC1-HNRNPM |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CRTC1-HNRNPM |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
