
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CRY1-HPSE2 (FusionGDB2 ID:19578) |
Fusion Gene Summary for CRY1-HPSE2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CRY1-HPSE2 | Fusion gene ID: 19578 | Hgene | Tgene | Gene symbol | CRY1 | HPSE2 | Gene ID | 1407 | 60495 |
| Gene name | cryptochrome circadian regulator 1 | heparanase 2 (inactive) | |
| Synonyms | DSPD|PHLL1 | HPA2|HPR2|UFS|UFS1 | |
| Cytomap | 12q23.3 | 10q24.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cryptochrome-1cryptochrome 1 (photolyase-like)cryptochrome circadian clock 1 | inactive heparanase-2heparanase 3heparanase-like protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q16526 | Q8WWQ2 | |
| Ensembl transtripts involved in fusion gene | ENST00000550633, ENST00000008527, | ENST00000370546, ENST00000370549, ENST00000370552, ENST00000404542, | |
| Fusion gene scores | * DoF score | 3 X 2 X 3=18 | 9 X 8 X 3=216 |
| # samples | 3 | 9 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(9/216*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CRY1 [Title/Abstract] AND HPSE2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CRY1(107486582)-HPSE2(100249953), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CRY1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 12397359|14672706|15147242 |
| Hgene | CRY1 | GO:0045892 | negative regulation of transcription, DNA-templated | 12397359|23133559 |
 Fusion gene breakpoints across CRY1 (5'-gene) Fusion gene breakpoints across CRY1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
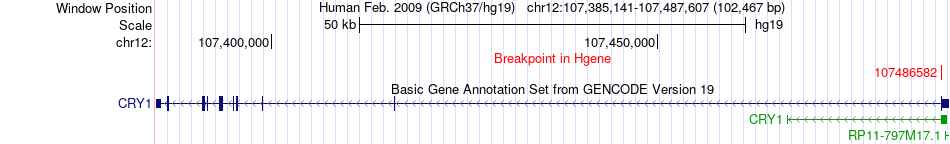 |
 Fusion gene breakpoints across HPSE2 (3'-gene) Fusion gene breakpoints across HPSE2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
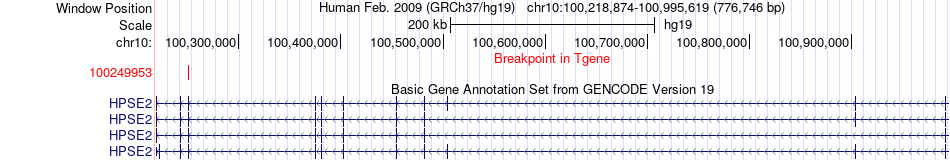 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-5527-01A | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
Top |
Fusion Gene ORF analysis for CRY1-HPSE2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000550633 | ENST00000370546 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| 5UTR-3CDS | ENST00000550633 | ENST00000370549 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| 5UTR-3CDS | ENST00000550633 | ENST00000370552 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| 5UTR-3CDS | ENST00000550633 | ENST00000404542 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| In-frame | ENST00000008527 | ENST00000370546 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| In-frame | ENST00000008527 | ENST00000370549 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| In-frame | ENST00000008527 | ENST00000370552 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
| In-frame | ENST00000008527 | ENST00000404542 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000008527 | CRY1 | chr12 | 107486582 | - | ENST00000370549 | HPSE2 | chr10 | 100249953 | - | 1941 | 1026 | 888 | 1484 | 198 |
| ENST00000008527 | CRY1 | chr12 | 107486582 | - | ENST00000370552 | HPSE2 | chr10 | 100249953 | - | 1941 | 1026 | 888 | 1484 | 198 |
| ENST00000008527 | CRY1 | chr12 | 107486582 | - | ENST00000370546 | HPSE2 | chr10 | 100249953 | - | 1555 | 1026 | 888 | 1352 | 154 |
| ENST00000008527 | CRY1 | chr12 | 107486582 | - | ENST00000404542 | HPSE2 | chr10 | 100249953 | - | 1485 | 1026 | 888 | 1484 | 198 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000008527 | ENST00000370549 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - | 0.19841191 | 0.80158806 |
| ENST00000008527 | ENST00000370552 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - | 0.19841191 | 0.80158806 |
| ENST00000008527 | ENST00000370546 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - | 0.1887707 | 0.81122935 |
| ENST00000008527 | ENST00000404542 | CRY1 | chr12 | 107486582 | - | HPSE2 | chr10 | 100249953 | - | 0.5248987 | 0.47510126 |
Top |
Fusion Genomic Features for CRY1-HPSE2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
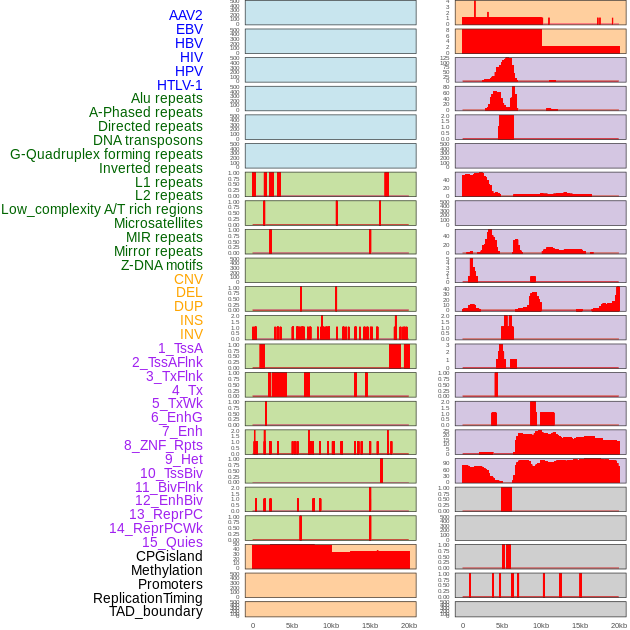 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CRY1-HPSE2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:107486582/chr10:100249953) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CRY1 | HPSE2 |
| FUNCTION: Transcriptional repressor which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. CRY1 and CRY2 have redundant functions but also differential and selective contributions at least in defining the pace of the SCN circadian clock and its circadian transcriptional outputs. More potent transcriptional repressor in cerebellum and liver than CRY2, though more effective in lengthening the period of the SCN oscillator. On its side, CRY2 seems to play a critical role in tuning SCN circadian period by opposing the action of CRY1. With CRY2, is dispensable for circadian rhythm generation but necessary for the development of intercellular networks for rhythm synchrony. Capable of translocating circadian clock core proteins such as PER proteins to the nucleus. Interacts with CLOCK-ARNTL/BMAL1 independently of PER proteins and is found at CLOCK-ARNTL/BMAL1-bound sites, suggesting that CRY may act as a molecular gatekeeper to maintain CLOCK-ARNTL/BMAL1 in a poised and repressed state until the proper time for transcriptional activation. Represses the CLOCK-ARNTL/BMAL1 induced transcription of BHLHE40/DEC1. Represses the CLOCK-ARNTL/BMAL1 induced transcription of ATF4, MTA1, KLF10 and NAMPT (By similarity). May repress circadian target genes expression in collaboration with HDAC1 and HDAC2 through histone deacetylation. Mediates the clock-control activation of ATR and modulates ATR-mediated DNA damage checkpoint. In liver, mediates circadian regulation of cAMP signaling and gluconeogenesis by binding to membrane-coupled G proteins and blocking glucagon-mediated increases in intracellular cAMP concentrations and CREB1 phosphorylation. Inhibits hepatic gluconeogenesis by decreasing nuclear FOXO1 levels that downregulates gluconeogenic gene expression (By similarity). Besides its role in the maintenance of the circadian clock, is also involved in the regulation of other processes. Represses glucocorticoid receptor NR3C1/GR-induced transcriptional activity by binding to glucocorticoid response elements (GREs). Plays a key role in glucose and lipid metabolism modulation, in part, through the transcriptional regulation of genes involved in these pathways, such as LEP or ACSL4 (By similarity). Represses PPARD and its target genes in the skeletal muscle and limits exercise capacity (By similarity). Plays an essential role in the generation of circadian rhythms in the retina (By similarity). Represses the transcriptional activity of NR1I2 (By similarity). {ECO:0000250|UniProtKB:P97784, ECO:0000269|PubMed:10531061, ECO:0000269|PubMed:14672706, ECO:0000269|PubMed:22170608, ECO:0000269|PubMed:23133559, ECO:0000269|PubMed:28388406}. | FUNCTION: Binds heparin and heparan sulfate with high affinity, but lacks heparanase activity. Inhibits HPSE, possibly by competing for its substrates (in vitro). {ECO:0000269|PubMed:20576607}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 50_54 | 52 | 456.0 | Motif | LIR 1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 3_132 | 52 | 456.0 | Domain | Note=Photolyase/cryptochrome alpha/beta |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 151_156 | 52 | 456.0 | Motif | LIR 3 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 255_260 | 52 | 456.0 | Motif | LIR 4 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 271_276 | 52 | 456.0 | Motif | LIR 5 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 285_290 | 52 | 456.0 | Motif | LIR 6 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 335_339 | 52 | 456.0 | Motif | LIR 7 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 379_384 | 52 | 456.0 | Motif | LIR 8 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 395_400 | 52 | 456.0 | Motif | LIR 9 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 411_416 | 52 | 456.0 | Motif | LIR 10 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 430_435 | 52 | 456.0 | Motif | LIR 11 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 486_491 | 52 | 456.0 | Motif | LIR 12 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 492_497 | 52 | 456.0 | Motif | LIR 13 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 82_87 | 52 | 456.0 | Motif | LIR 2 |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 387_389 | 52 | 456.0 | Nucleotide binding | FAD |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 371_470 | 52 | 456.0 | Region | Required for inhibition of CLOCK-ARNTL/BMAL1-mediated transcription |
Top |
Fusion Gene Sequence for CRY1-HPSE2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19578_19578_1_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000370546_length(transcript)=1555nt_BP=1026nt GGGGCCTGTGGTCAACGCGATTTGCTTCCAAGGGACGGCCACCAGTCGGCACAGGAAAGGGGCAGAGGCAGTGAGTTCAGCGTGTGGACG AGGGTCAACAAGTTTGGGATCAAGCGGCTGCCGCTCCTCCAAAAGCGACCGAAGCGCGAGCAGATTACCCCTCCGAGCCAGTGTAGTAAA CACACTTCAGAAACGTGAGGTGCCGGTGGTCACGAGGGGAGCGCGCCCTCCAATGAGGAGCCGGGGGCGGGGCCGAGGCCGCTGACGCGG CGGCGGCGGCAGAGTCACCCGGGCAGCCTCGGGACCGGTCACCGGCCGGCAACCGTCCAGCGGCCTCGACCACCGCCTCTAGCCTCCGTT CCCGGTCCTTTCTCCCGGGCCGAGAGACAGCGTCGCCGACAGGGGCTCATTCCCCTCCGGTTCTCCTCGGTGACTCACCTCGGGCGGGCC GTTTTGTCTTTAGGGGCCGCCTTGGTGGGGCGAGGTTTCCGTGACGAATCTCCTGGGGCCGTCCGTGCCGGCTCGGGCCGTCGTGGCGGC TCGAGCTCCTGGAACTTGCTCAGGCTCCGGAGGTCCGAGGCCCTCGAAGTTATGCGTCGCCTCCAGGCGGTTGCGGCGGGCGCGGGCTCC TAAAGGGCGTCACACCCGGACTCCGCCGACTAGGCAACCTCCATTCATCTTTCCACTGCGCCTCCGGCGCCCCCGCCTTCTCCGGTCCCC TCCTCGGAGTCATTTTTTCCTGTTCCCCCTCTGCCGCCCTTTCCTCACGCCCCGGGTGAGGCAATTCTCTTGGAAGCGAAGGTGTCGGCT ATGAGCCGGAGCCTCCTTCCTTGAATTTCTCCGTGGAGGACCCGCCGCGCCCCCCGGCATGGGGGTGAACGCCGTGCACTGGTTCCGAAA GGGGCTCCGGCTCCACGACAACCCCGCCCTGAAGGAGTGCATTCAGGGCGCCGACACCATCCGCTGCGTCTACATCCTGGACCCCTGGTT CGCCGGCTCCTCCAATGTGGGCATCAACAGGTGGCGGACTACTGGCTCTCTCTCCTCTACAAGCGCCTGATCGGCCCCAAAGTCTTGGCT GTGCATGTGGCTGGGCTCCAGCGGAAGCCACGGCCTGGCCGAGTGATCCGGGACAAACTAAGGATTTATGCTCACTGCACAAACCACCAC AACCACAACTACGTTCGTGGGTCCATTACACTTTTTATCATCAACTTGCATCGATCAAGAAAGAAAATCAAGCTGGCTGGGACTCTCAGA GACAAGCTGGTTCACCAGTACCTGCTGCAGCCCTATGGGCAGGAGGGCCTAAAGTCCAAAACCCAAAGATGTCAATACTGTGGGATCATC TGAATCTAAAAGGGCATTACCTAATTCTTCAGAAGAAAGGTCAGTGCAACTGAATGGCCAGCCCTTAGTGATGGTGGACGACGGGACCCT CCCAGAATTGAAGCCCCGCCCCCTTCGGGCCGGCCGGACATTGGTCATCCCTCCAGTCACCATGGGCTTTTATGTGGTCAAGAATGTCAA >19578_19578_1_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000370546_length(amino acids)=154AA_BP=46 MVPKGAPAPRQPRPEGVHSGRRHHPLRLHPGPLVRRLLQCGHQQVADYWLSLLYKRLIGPKVLAVHVAGLQRKPRPGRVIRDKLRIYAHC -------------------------------------------------------------- >19578_19578_2_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000370549_length(transcript)=1941nt_BP=1026nt GGGGCCTGTGGTCAACGCGATTTGCTTCCAAGGGACGGCCACCAGTCGGCACAGGAAAGGGGCAGAGGCAGTGAGTTCAGCGTGTGGACG AGGGTCAACAAGTTTGGGATCAAGCGGCTGCCGCTCCTCCAAAAGCGACCGAAGCGCGAGCAGATTACCCCTCCGAGCCAGTGTAGTAAA CACACTTCAGAAACGTGAGGTGCCGGTGGTCACGAGGGGAGCGCGCCCTCCAATGAGGAGCCGGGGGCGGGGCCGAGGCCGCTGACGCGG CGGCGGCGGCAGAGTCACCCGGGCAGCCTCGGGACCGGTCACCGGCCGGCAACCGTCCAGCGGCCTCGACCACCGCCTCTAGCCTCCGTT CCCGGTCCTTTCTCCCGGGCCGAGAGACAGCGTCGCCGACAGGGGCTCATTCCCCTCCGGTTCTCCTCGGTGACTCACCTCGGGCGGGCC GTTTTGTCTTTAGGGGCCGCCTTGGTGGGGCGAGGTTTCCGTGACGAATCTCCTGGGGCCGTCCGTGCCGGCTCGGGCCGTCGTGGCGGC TCGAGCTCCTGGAACTTGCTCAGGCTCCGGAGGTCCGAGGCCCTCGAAGTTATGCGTCGCCTCCAGGCGGTTGCGGCGGGCGCGGGCTCC TAAAGGGCGTCACACCCGGACTCCGCCGACTAGGCAACCTCCATTCATCTTTCCACTGCGCCTCCGGCGCCCCCGCCTTCTCCGGTCCCC TCCTCGGAGTCATTTTTTCCTGTTCCCCCTCTGCCGCCCTTTCCTCACGCCCCGGGTGAGGCAATTCTCTTGGAAGCGAAGGTGTCGGCT ATGAGCCGGAGCCTCCTTCCTTGAATTTCTCCGTGGAGGACCCGCCGCGCCCCCCGGCATGGGGGTGAACGCCGTGCACTGGTTCCGAAA GGGGCTCCGGCTCCACGACAACCCCGCCCTGAAGGAGTGCATTCAGGGCGCCGACACCATCCGCTGCGTCTACATCCTGGACCCCTGGTT CGCCGGCTCCTCCAATGTGGGCATCAACAGGTGGCGGACTACTGGCTCTCTCTCCTCTACAAGCGCCTGATCGGCCCCAAAGTCTTGGCT GTGCATGTGGCTGGGCTCCAGCGGAAGCCACGGCCTGGCCGAGTGATCCGGGACAAACTAAGGATTTATGCTCACTGCACAAACCACCAC AACCACAACTACGTTCGTGGGTCCATTACACTTTTTATCATCAACTTGCATCGATCAAGAAAGAAAATCAAGCTGGCTGGGACTCTCAGA GACAAGCTGGTTCACCAGTACCTGCTGCAGCCCTATGGGCAGGAGGGCCTAAAGTCCAAGTCAGTGCAACTGAATGGCCAGCCCTTAGTG ATGGTGGACGACGGGACCCTCCCAGAATTGAAGCCCCGCCCCCTTCGGGCCGGCCGGACATTGGTCATCCCTCCAGTCACCATGGGCTTT TATGTGGTCAAGAATGTCAATGCTTTGGCCTGCCGCTACCGATAAGCTATCCTCACACTCACGGCTACCAGTGGGCCTGCTGGGCTGCTT CCACTCCTCCACTCCAGTAGTATCCTCTGTTTTCAGACATCCTAGCAACCAGCCCCTGCTGCCCCATCCTGCTGGAATCAACACAGACTT GCTCTCCAAAGAGACTAAATGTCATAGCGTGATCTTAGCCTAGGTAGGCCACATCCATCCCAAAGGAAAATGTAGACATCACCTGTACCT ATATAAGGATAAAGGCATGTGTATAGAGCAGAATGTTTCCCTTCATGTGCACTATGAAAACGAGCTGACAGCACACTCCCAGGAGAAATG TTTCCAGACAACTCCCCATGATCCTGTCACACAGCATTATAACCACAAATCCAAACCTTAGCCTGCTGCTGCTGCTGCCCTCAGAGGAAG >19578_19578_2_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000370549_length(amino acids)=198AA_BP=46 MVPKGAPAPRQPRPEGVHSGRRHHPLRLHPGPLVRRLLQCGHQQVADYWLSLLYKRLIGPKVLAVHVAGLQRKPRPGRVIRDKLRIYAHC TNHHNHNYVRGSITLFIINLHRSRKKIKLAGTLRDKLVHQYLLQPYGQEGLKSKSVQLNGQPLVMVDDGTLPELKPRPLRAGRTLVIPPV -------------------------------------------------------------- >19578_19578_3_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000370552_length(transcript)=1941nt_BP=1026nt GGGGCCTGTGGTCAACGCGATTTGCTTCCAAGGGACGGCCACCAGTCGGCACAGGAAAGGGGCAGAGGCAGTGAGTTCAGCGTGTGGACG AGGGTCAACAAGTTTGGGATCAAGCGGCTGCCGCTCCTCCAAAAGCGACCGAAGCGCGAGCAGATTACCCCTCCGAGCCAGTGTAGTAAA CACACTTCAGAAACGTGAGGTGCCGGTGGTCACGAGGGGAGCGCGCCCTCCAATGAGGAGCCGGGGGCGGGGCCGAGGCCGCTGACGCGG CGGCGGCGGCAGAGTCACCCGGGCAGCCTCGGGACCGGTCACCGGCCGGCAACCGTCCAGCGGCCTCGACCACCGCCTCTAGCCTCCGTT CCCGGTCCTTTCTCCCGGGCCGAGAGACAGCGTCGCCGACAGGGGCTCATTCCCCTCCGGTTCTCCTCGGTGACTCACCTCGGGCGGGCC GTTTTGTCTTTAGGGGCCGCCTTGGTGGGGCGAGGTTTCCGTGACGAATCTCCTGGGGCCGTCCGTGCCGGCTCGGGCCGTCGTGGCGGC TCGAGCTCCTGGAACTTGCTCAGGCTCCGGAGGTCCGAGGCCCTCGAAGTTATGCGTCGCCTCCAGGCGGTTGCGGCGGGCGCGGGCTCC TAAAGGGCGTCACACCCGGACTCCGCCGACTAGGCAACCTCCATTCATCTTTCCACTGCGCCTCCGGCGCCCCCGCCTTCTCCGGTCCCC TCCTCGGAGTCATTTTTTCCTGTTCCCCCTCTGCCGCCCTTTCCTCACGCCCCGGGTGAGGCAATTCTCTTGGAAGCGAAGGTGTCGGCT ATGAGCCGGAGCCTCCTTCCTTGAATTTCTCCGTGGAGGACCCGCCGCGCCCCCCGGCATGGGGGTGAACGCCGTGCACTGGTTCCGAAA GGGGCTCCGGCTCCACGACAACCCCGCCCTGAAGGAGTGCATTCAGGGCGCCGACACCATCCGCTGCGTCTACATCCTGGACCCCTGGTT CGCCGGCTCCTCCAATGTGGGCATCAACAGGTGGCGGACTACTGGCTCTCTCTCCTCTACAAGCGCCTGATCGGCCCCAAAGTCTTGGCT GTGCATGTGGCTGGGCTCCAGCGGAAGCCACGGCCTGGCCGAGTGATCCGGGACAAACTAAGGATTTATGCTCACTGCACAAACCACCAC AACCACAACTACGTTCGTGGGTCCATTACACTTTTTATCATCAACTTGCATCGATCAAGAAAGAAAATCAAGCTGGCTGGGACTCTCAGA GACAAGCTGGTTCACCAGTACCTGCTGCAGCCCTATGGGCAGGAGGGCCTAAAGTCCAAGTCAGTGCAACTGAATGGCCAGCCCTTAGTG ATGGTGGACGACGGGACCCTCCCAGAATTGAAGCCCCGCCCCCTTCGGGCCGGCCGGACATTGGTCATCCCTCCAGTCACCATGGGCTTT TATGTGGTCAAGAATGTCAATGCTTTGGCCTGCCGCTACCGATAAGCTATCCTCACACTCACGGCTACCAGTGGGCCTGCTGGGCTGCTT CCACTCCTCCACTCCAGTAGTATCCTCTGTTTTCAGACATCCTAGCAACCAGCCCCTGCTGCCCCATCCTGCTGGAATCAACACAGACTT GCTCTCCAAAGAGACTAAATGTCATAGCGTGATCTTAGCCTAGGTAGGCCACATCCATCCCAAAGGAAAATGTAGACATCACCTGTACCT ATATAAGGATAAAGGCATGTGTATAGAGCAGAATGTTTCCCTTCATGTGCACTATGAAAACGAGCTGACAGCACACTCCCAGGAGAAATG TTTCCAGACAACTCCCCATGATCCTGTCACACAGCATTATAACCACAAATCCAAACCTTAGCCTGCTGCTGCTGCTGCCCTCAGAGGAAG >19578_19578_3_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000370552_length(amino acids)=198AA_BP=46 MVPKGAPAPRQPRPEGVHSGRRHHPLRLHPGPLVRRLLQCGHQQVADYWLSLLYKRLIGPKVLAVHVAGLQRKPRPGRVIRDKLRIYAHC TNHHNHNYVRGSITLFIINLHRSRKKIKLAGTLRDKLVHQYLLQPYGQEGLKSKSVQLNGQPLVMVDDGTLPELKPRPLRAGRTLVIPPV -------------------------------------------------------------- >19578_19578_4_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000404542_length(transcript)=1485nt_BP=1026nt GGGGCCTGTGGTCAACGCGATTTGCTTCCAAGGGACGGCCACCAGTCGGCACAGGAAAGGGGCAGAGGCAGTGAGTTCAGCGTGTGGACG AGGGTCAACAAGTTTGGGATCAAGCGGCTGCCGCTCCTCCAAAAGCGACCGAAGCGCGAGCAGATTACCCCTCCGAGCCAGTGTAGTAAA CACACTTCAGAAACGTGAGGTGCCGGTGGTCACGAGGGGAGCGCGCCCTCCAATGAGGAGCCGGGGGCGGGGCCGAGGCCGCTGACGCGG CGGCGGCGGCAGAGTCACCCGGGCAGCCTCGGGACCGGTCACCGGCCGGCAACCGTCCAGCGGCCTCGACCACCGCCTCTAGCCTCCGTT CCCGGTCCTTTCTCCCGGGCCGAGAGACAGCGTCGCCGACAGGGGCTCATTCCCCTCCGGTTCTCCTCGGTGACTCACCTCGGGCGGGCC GTTTTGTCTTTAGGGGCCGCCTTGGTGGGGCGAGGTTTCCGTGACGAATCTCCTGGGGCCGTCCGTGCCGGCTCGGGCCGTCGTGGCGGC TCGAGCTCCTGGAACTTGCTCAGGCTCCGGAGGTCCGAGGCCCTCGAAGTTATGCGTCGCCTCCAGGCGGTTGCGGCGGGCGCGGGCTCC TAAAGGGCGTCACACCCGGACTCCGCCGACTAGGCAACCTCCATTCATCTTTCCACTGCGCCTCCGGCGCCCCCGCCTTCTCCGGTCCCC TCCTCGGAGTCATTTTTTCCTGTTCCCCCTCTGCCGCCCTTTCCTCACGCCCCGGGTGAGGCAATTCTCTTGGAAGCGAAGGTGTCGGCT ATGAGCCGGAGCCTCCTTCCTTGAATTTCTCCGTGGAGGACCCGCCGCGCCCCCCGGCATGGGGGTGAACGCCGTGCACTGGTTCCGAAA GGGGCTCCGGCTCCACGACAACCCCGCCCTGAAGGAGTGCATTCAGGGCGCCGACACCATCCGCTGCGTCTACATCCTGGACCCCTGGTT CGCCGGCTCCTCCAATGTGGGCATCAACAGGTGGCGGACTACTGGCTCTCTCTCCTCTACAAGCGCCTGATCGGCCCCAAAGTCTTGGCT GTGCATGTGGCTGGGCTCCAGCGGAAGCCACGGCCTGGCCGAGTGATCCGGGACAAACTAAGGATTTATGCTCACTGCACAAACCACCAC AACCACAACTACGTTCGTGGGTCCATTACACTTTTTATCATCAACTTGCATCGATCAAGAAAGAAAATCAAGCTGGCTGGGACTCTCAGA GACAAGCTGGTTCACCAGTACCTGCTGCAGCCCTATGGGCAGGAGGGCCTAAAGTCCAAGTCAGTGCAACTGAATGGCCAGCCCTTAGTG ATGGTGGACGACGGGACCCTCCCAGAATTGAAGCCCCGCCCCCTTCGGGCCGGCCGGACATTGGTCATCCCTCCAGTCACCATGGGCTTT >19578_19578_4_CRY1-HPSE2_CRY1_chr12_107486582_ENST00000008527_HPSE2_chr10_100249953_ENST00000404542_length(amino acids)=198AA_BP=46 MVPKGAPAPRQPRPEGVHSGRRHHPLRLHPGPLVRRLLQCGHQQVADYWLSLLYKRLIGPKVLAVHVAGLQRKPRPGRVIRDKLRIYAHC TNHHNHNYVRGSITLFIINLHRSRKKIKLAGTLRDKLVHQYLLQPYGQEGLKSKSVQLNGQPLVMVDDGTLPELKPRPLRAGRTLVIPPV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CRY1-HPSE2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CRY1 | chr12:107486582 | chr10:100249953 | ENST00000008527 | - | 1 | 13 | 471_493 | 52.666666666666664 | 456.0 | TIMELESS |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CRY1-HPSE2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CRY1-HPSE2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
