
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ADAMTS12-C1QTNF3 (FusionGDB2 ID:2041) |
Fusion Gene Summary for ADAMTS12-C1QTNF3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ADAMTS12-C1QTNF3 | Fusion gene ID: 2041 | Hgene | Tgene | Gene symbol | ADAMTS12 | C1QTNF3 | Gene ID | 81792 | 114899 |
| Gene name | ADAM metallopeptidase with thrombospondin type 1 motif 12 | C1q and TNF related 3 | |
| Synonyms | PRO4389 | C1ATNF3|CORCS|CORS|CORS-26|CORS26|CTRP3 | |
| Cytomap | 5p13.3-p13.2 | 5p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | A disintegrin and metalloproteinase with thrombospondin motifs 12ADAM-TS 12ADAM-TS12ADAMTS-12a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 12 | complement C1q tumor necrosis factor-related protein 3C1q and tumor necrosis factor related protein 3cartonectincollagenous repeat-containing sequence 26 kDa proteincollagenous repeat-containing sequence of 26-kDasecretory protein CORS26 | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | P58397 | Q9BXJ4 | |
| Ensembl transtripts involved in fusion gene | ENST00000352040, ENST00000504830, ENST00000515401, ENST00000504582, | ENST00000513065, ENST00000231338, ENST00000382065, | |
| Fusion gene scores | * DoF score | 6 X 6 X 4=144 | 7 X 3 X 6=126 |
| # samples | 6 | 8 | |
| ** MAII score | log2(6/144*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/126*10)=-0.655351828612554 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ADAMTS12 [Title/Abstract] AND C1QTNF3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ADAMTS12(33891835)-C1QTNF3(34035863), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ADAMTS12 | GO:0030167 | proteoglycan catabolic process | 17895370 |
| Hgene | ADAMTS12 | GO:0032331 | negative regulation of chondrocyte differentiation | 22247065 |
| Hgene | ADAMTS12 | GO:0050727 | regulation of inflammatory response | 23019333 |
| Hgene | ADAMTS12 | GO:0051603 | proteolysis involved in cellular protein catabolic process | 16611630 |
| Hgene | ADAMTS12 | GO:0071773 | cellular response to BMP stimulus | 22247065 |
| Hgene | ADAMTS12 | GO:1901509 | regulation of endothelial tube morphogenesis | 17895370 |
| Hgene | ADAMTS12 | GO:1902203 | negative regulation of hepatocyte growth factor receptor signaling pathway | 17895370 |
| Hgene | ADAMTS12 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 17895370 |
| Hgene | ADAMTS12 | GO:2001113 | negative regulation of cellular response to hepatocyte growth factor stimulus | 17895370 |
| Tgene | C1QTNF3 | GO:0035356 | cellular triglyceride homeostasis | 20739398 |
| Tgene | C1QTNF3 | GO:0045721 | negative regulation of gluconeogenesis | 20952387 |
| Tgene | C1QTNF3 | GO:0050715 | positive regulation of cytokine secretion | 18421280 |
| Tgene | C1QTNF3 | GO:0050728 | negative regulation of inflammatory response | 16213490 |
| Tgene | C1QTNF3 | GO:0070165 | positive regulation of adiponectin secretion | 18421280 |
| Tgene | C1QTNF3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 20739398 |
| Tgene | C1QTNF3 | GO:1900165 | negative regulation of interleukin-6 secretion | 16213490 |
| Tgene | C1QTNF3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 16213490 |
 Fusion gene breakpoints across ADAMTS12 (5'-gene) Fusion gene breakpoints across ADAMTS12 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
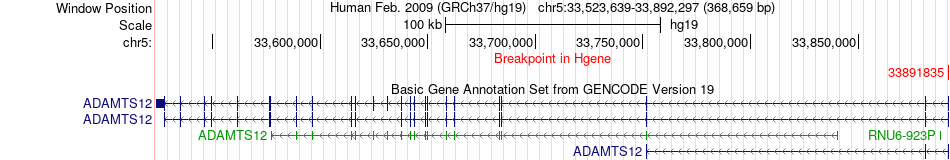 |
 Fusion gene breakpoints across C1QTNF3 (3'-gene) Fusion gene breakpoints across C1QTNF3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
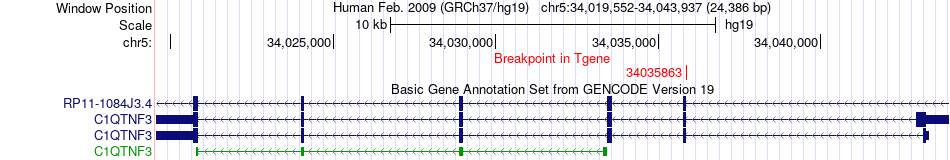 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AN-A0AR-01A | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
Top |
Fusion Gene ORF analysis for ADAMTS12-C1QTNF3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000352040 | ENST00000513065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| 5CDS-intron | ENST00000504830 | ENST00000513065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| 5CDS-intron | ENST00000515401 | ENST00000513065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| In-frame | ENST00000352040 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| In-frame | ENST00000352040 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| In-frame | ENST00000504830 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| In-frame | ENST00000504830 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| In-frame | ENST00000515401 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| In-frame | ENST00000515401 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| intron-3CDS | ENST00000504582 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| intron-3CDS | ENST00000504582 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
| intron-intron | ENST00000504582 | ENST00000513065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000504830 | ADAMTS12 | chr5 | 33891835 | - | ENST00000382065 | C1QTNF3 | chr5 | 34035863 | - | 2255 | 463 | 361 | 1119 | 252 |
| ENST00000504830 | ADAMTS12 | chr5 | 33891835 | - | ENST00000231338 | C1QTNF3 | chr5 | 34035863 | - | 2250 | 463 | 361 | 1119 | 252 |
| ENST00000352040 | ADAMTS12 | chr5 | 33891835 | - | ENST00000382065 | C1QTNF3 | chr5 | 34035863 | - | 2004 | 212 | 110 | 868 | 252 |
| ENST00000352040 | ADAMTS12 | chr5 | 33891835 | - | ENST00000231338 | C1QTNF3 | chr5 | 34035863 | - | 1999 | 212 | 110 | 868 | 252 |
| ENST00000515401 | ADAMTS12 | chr5 | 33891835 | - | ENST00000382065 | C1QTNF3 | chr5 | 34035863 | - | 2082 | 290 | 188 | 946 | 252 |
| ENST00000515401 | ADAMTS12 | chr5 | 33891835 | - | ENST00000231338 | C1QTNF3 | chr5 | 34035863 | - | 2077 | 290 | 188 | 946 | 252 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000504830 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - | 0.001235243 | 0.99876475 |
| ENST00000504830 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - | 0.001241035 | 0.99875903 |
| ENST00000352040 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - | 0.00102778 | 0.9989722 |
| ENST00000352040 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - | 0.001046153 | 0.9989538 |
| ENST00000515401 | ENST00000382065 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - | 0.001018115 | 0.9989819 |
| ENST00000515401 | ENST00000231338 | ADAMTS12 | chr5 | 33891835 | - | C1QTNF3 | chr5 | 34035863 | - | 0.001038027 | 0.9989619 |
Top |
Fusion Genomic Features for ADAMTS12-C1QTNF3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
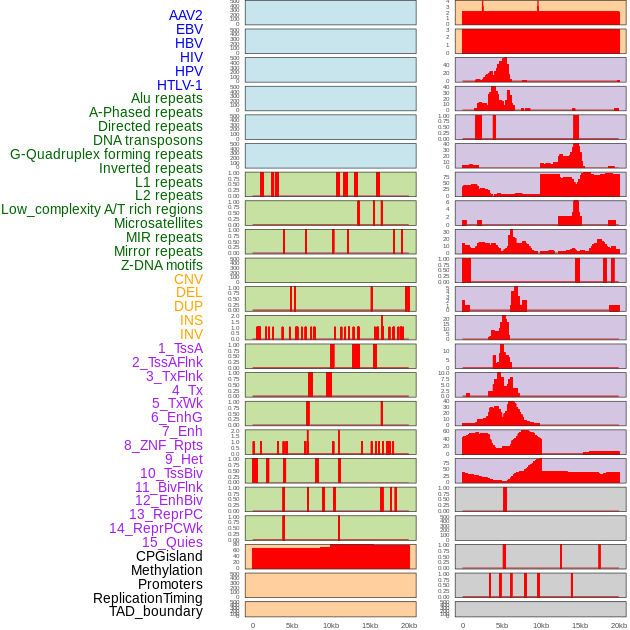 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ADAMTS12-C1QTNF3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:33891835/chr5:34035863) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ADAMTS12 | C1QTNF3 |
| FUNCTION: Metalloprotease that may play a role in the degradation of COMP. Cleaves also alpha-2 macroglobulin and aggregan. Has anti-tumorigenic properties. {ECO:0000269|PubMed:16611630, ECO:0000269|PubMed:17895370, ECO:0000269|PubMed:18485748}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | C1QTNF3 | chr5:33891835 | chr5:34035863 | ENST00000231338 | 0 | 6 | 113_246 | 28 | 247.0 | Domain | C1q | |
| Tgene | C1QTNF3 | chr5:33891835 | chr5:34035863 | ENST00000231338 | 0 | 6 | 51_113 | 28 | 247.0 | Domain | Note=Collagen-like | |
| Tgene | C1QTNF3 | chr5:33891835 | chr5:34035863 | ENST00000382065 | 0 | 6 | 113_246 | 101 | 320.0 | Domain | C1q |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 302_305 | 42 | 1510.0 | Compositional bias | Note=Poly-Glu |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 597_700 | 42 | 1510.0 | Compositional bias | Note=Cys-rich |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 302_305 | 42 | 1595.0 | Compositional bias | Note=Poly-Glu |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 597_700 | 42 | 1595.0 | Compositional bias | Note=Cys-rich |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 1313_1366 | 42 | 1510.0 | Domain | TSP type-1 5 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 1368_1422 | 42 | 1510.0 | Domain | TSP type-1 6 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 1423_1471 | 42 | 1510.0 | Domain | TSP type-1 7 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 1472_1532 | 42 | 1510.0 | Domain | TSP type-1 8 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 1535_1575 | 42 | 1510.0 | Domain | PLAC |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 246_456 | 42 | 1510.0 | Domain | Peptidase M12B |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 465_544 | 42 | 1510.0 | Domain | Note=Disintegrin |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 542_597 | 42 | 1510.0 | Domain | TSP type-1 1 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 823_883 | 42 | 1510.0 | Domain | TSP type-1 2 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 887_943 | 42 | 1510.0 | Domain | TSP type-1 3 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 944_997 | 42 | 1510.0 | Domain | TSP type-1 4 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 1313_1366 | 42 | 1595.0 | Domain | TSP type-1 5 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 1368_1422 | 42 | 1595.0 | Domain | TSP type-1 6 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 1423_1471 | 42 | 1595.0 | Domain | TSP type-1 7 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 1472_1532 | 42 | 1595.0 | Domain | TSP type-1 8 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 1535_1575 | 42 | 1595.0 | Domain | PLAC |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 246_456 | 42 | 1595.0 | Domain | Peptidase M12B |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 465_544 | 42 | 1595.0 | Domain | Note=Disintegrin |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 542_597 | 42 | 1595.0 | Domain | TSP type-1 1 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 823_883 | 42 | 1595.0 | Domain | TSP type-1 2 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 887_943 | 42 | 1595.0 | Domain | TSP type-1 3 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 944_997 | 42 | 1595.0 | Domain | TSP type-1 4 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 208_213 | 42 | 1510.0 | Motif | Cysteine switch |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 208_213 | 42 | 1595.0 | Motif | Cysteine switch |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 701_827 | 42 | 1510.0 | Region | Note=Spacer 1 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000352040 | - | 1 | 22 | 997_1316 | 42 | 1510.0 | Region | Note=Spacer 2 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 701_827 | 42 | 1595.0 | Region | Note=Spacer 1 |
| Hgene | ADAMTS12 | chr5:33891835 | chr5:34035863 | ENST00000504830 | - | 1 | 24 | 997_1316 | 42 | 1595.0 | Region | Note=Spacer 2 |
| Tgene | C1QTNF3 | chr5:33891835 | chr5:34035863 | ENST00000382065 | 0 | 6 | 51_113 | 101 | 320.0 | Domain | Note=Collagen-like |
Top |
Fusion Gene Sequence for ADAMTS12-C1QTNF3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2041_2041_1_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000352040_C1QTNF3_chr5_34035863_ENST00000231338_length(transcript)=1999nt_BP=212nt TCCTGCTGGGCCACAGGCGAGCGCTTTATTTCTGGAGCTGAGGGCTAAAACTTTTTTGACTTTTCTTCTCCTCAACATCTGAATCATGCC ATGTGCCCAGAGGAGCTGGCTTGCAAACCTTTCCGTGGTGGCTCAGCTCCTTAACTTTGGGGCGCTTTGCTATGGGAGACAGCCTCAGCC AGGCCCGGTTCGCTTCCCGGACAGGAGGCAAGTCTCCACAAACCGGAGGACTACCCCCAGACTGCAGTAAGTGTTGTCATGGAGACTACA GCTTTCGAGGCTACCAAGGCCCCCCTGGGCCACCGGGCCCTCCTGGCATTCCAGGAAACCATGGAAACAATGGCAACAATGGAGCCACTG GTCATGAAGGAGCCAAAGGTGAGAAGGGCGACAAAGGTGACCTGGGGCCTCGAGGGGAGCGGGGGCAGCATGGCCCCAAAGGAGAGAAGG GCTACCCGGGGATTCCACCAGAACTTCAGATTGCATTCATGGCTTCTCTGGCAACCCACTTCAGCAATCAGAACAGTGGGATTATCTTCA GCAGTGTTGAGACCAACATTGGAAACTTCTTTGATGTCATGACTGGTAGATTTGGGGCCCCAGTATCAGGTGTGTATTTCTTCACCTTCA GCATGATGAAGCATGAGGATGTTGAGGAAGTGTATGTGTACCTTATGCACAATGGCAACACAGTCTTCAGCATGTACAGCTATGAAATGA AGGGCAAATCAGATACATCCAGCAATCATGCTGTGCTGAAGCTAGCCAAAGGGGATGAGGTTTGGCTGCGAATGGGCAATGGCGCTCTCC ATGGGGACCACCAACGCTTCTCCACCTTTGCAGGATTCCTGCTCTTTGAAACTAAGTAAATATATGACTAGAATAGCTCCACTTTGGGGA AGACTTGTAGCTGAGCTGATTTGTTACGATCTGAGGAACATTAAAGTTGAGGGTTTTACATTGCTGTATTCAAAAAATTATTGGTTGCAA TGTTGTTCACGCTACAGGTACACCAATAATGTTGGACAATTCAGGGGCTCAGAAGAATCAACCACAAAATAGTCTTCTCAGATGACCTTG ACTAATATACTCAGCATCTTTATCACTCTTTCCTTGGCACCTAAAAGATAATTCTCCTCTGACGCAGGTTGGAAATATTTTTTTCTATCA CAGAAGTCATTTGCAAAGAATTTTGACTACTCTGCTTTTAATTTAATACCAGTTTTCAGGAACCCCTGAAGTTTTAAGTTCATTATTCTT TATAACATTTGAGAGAATCGGATGTAGTGATATGACAGGGCTGGGGCAAGAACAGGGGCACTAGCTGCCTTATTAGCTAATTTAGTGCCC TCCGTGTTCAGCTTAGCCTTTGACCCTTTCCTTTTGATCCACAAAATACATTAAAACTCTGAATTCACATACAATGCTATTTTAAAGTCA ATAGATTTTAGCTATAAAGTGCTTGACCAGTAATGTGGTTGTAATTTTGTGTATGTTCCCCCACATCGCCCCCAACTTCGGATGTGGGGT CAGGAGGTTGAGGTTCACTATTAACAAATGTCATAAATATCTCATAGAGGTACAGTGCCAATAGATATTCAAATGTTGCATGTTGACCAG AGGGATTTTATATCTGAAGAACATACACTATTAATAAATACCTTAGAGAAAGATTTTGACCTGGCTTTAGATAAAACTGTGGCAAGAAAA ATGTAATGAGCAATATATGGAAATAAACACACCTTTGTTAAAGATACTTTCTAAACTTGTGTTTAATAAACTTTAATAGTCATAGAATTG TAAATCACTATGGTTAACAGAAAGTGAAAATATTTTCATGCAGATGATGTGAACAGCCATGTGAATAGGTGACTTGGGCACACAGCAGGG TCATATGACTTCAGAAAACTTCGCTTTTCAGTTATTCCATTGTTATAATGTCAACCCTTTAAGACATTGATGTTTAGAGGGCTCACAAAT >2041_2041_1_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000352040_C1QTNF3_chr5_34035863_ENST00000231338_length(amino acids)=252AA_BP=34 MQTFPWWLSSLTLGRFAMGDSLSQARFASRTGGKSPQTGGLPPDCSKCCHGDYSFRGYQGPPGPPGPPGIPGNHGNNGNNGATGHEGAKG EKGDKGDLGPRGERGQHGPKGEKGYPGIPPELQIAFMASLATHFSNQNSGIIFSSVETNIGNFFDVMTGRFGAPVSGVYFFTFSMMKHED -------------------------------------------------------------- >2041_2041_2_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000352040_C1QTNF3_chr5_34035863_ENST00000382065_length(transcript)=2004nt_BP=212nt TCCTGCTGGGCCACAGGCGAGCGCTTTATTTCTGGAGCTGAGGGCTAAAACTTTTTTGACTTTTCTTCTCCTCAACATCTGAATCATGCC ATGTGCCCAGAGGAGCTGGCTTGCAAACCTTTCCGTGGTGGCTCAGCTCCTTAACTTTGGGGCGCTTTGCTATGGGAGACAGCCTCAGCC AGGCCCGGTTCGCTTCCCGGACAGGAGGCAAGTCTCCACAAACCGGAGGACTACCCCCAGACTGCAGTAAGTGTTGTCATGGAGACTACA GCTTTCGAGGCTACCAAGGCCCCCCTGGGCCACCGGGCCCTCCTGGCATTCCAGGAAACCATGGAAACAATGGCAACAATGGAGCCACTG GTCATGAAGGAGCCAAAGGTGAGAAGGGCGACAAAGGTGACCTGGGGCCTCGAGGGGAGCGGGGGCAGCATGGCCCCAAAGGAGAGAAGG GCTACCCGGGGATTCCACCAGAACTTCAGATTGCATTCATGGCTTCTCTGGCAACCCACTTCAGCAATCAGAACAGTGGGATTATCTTCA GCAGTGTTGAGACCAACATTGGAAACTTCTTTGATGTCATGACTGGTAGATTTGGGGCCCCAGTATCAGGTGTGTATTTCTTCACCTTCA GCATGATGAAGCATGAGGATGTTGAGGAAGTGTATGTGTACCTTATGCACAATGGCAACACAGTCTTCAGCATGTACAGCTATGAAATGA AGGGCAAATCAGATACATCCAGCAATCATGCTGTGCTGAAGCTAGCCAAAGGGGATGAGGTTTGGCTGCGAATGGGCAATGGCGCTCTCC ATGGGGACCACCAACGCTTCTCCACCTTTGCAGGATTCCTGCTCTTTGAAACTAAGTAAATATATGACTAGAATAGCTCCACTTTGGGGA AGACTTGTAGCTGAGCTGATTTGTTACGATCTGAGGAACATTAAAGTTGAGGGTTTTACATTGCTGTATTCAAAAAATTATTGGTTGCAA TGTTGTTCACGCTACAGGTACACCAATAATGTTGGACAATTCAGGGGCTCAGAAGAATCAACCACAAAATAGTCTTCTCAGATGACCTTG ACTAATATACTCAGCATCTTTATCACTCTTTCCTTGGCACCTAAAAGATAATTCTCCTCTGACGCAGGTTGGAAATATTTTTTTCTATCA CAGAAGTCATTTGCAAAGAATTTTGACTACTCTGCTTTTAATTTAATACCAGTTTTCAGGAACCCCTGAAGTTTTAAGTTCATTATTCTT TATAACATTTGAGAGAATCGGATGTAGTGATATGACAGGGCTGGGGCAAGAACAGGGGCACTAGCTGCCTTATTAGCTAATTTAGTGCCC TCCGTGTTCAGCTTAGCCTTTGACCCTTTCCTTTTGATCCACAAAATACATTAAAACTCTGAATTCACATACAATGCTATTTTAAAGTCA ATAGATTTTAGCTATAAAGTGCTTGACCAGTAATGTGGTTGTAATTTTGTGTATGTTCCCCCACATCGCCCCCAACTTCGGATGTGGGGT CAGGAGGTTGAGGTTCACTATTAACAAATGTCATAAATATCTCATAGAGGTACAGTGCCAATAGATATTCAAATGTTGCATGTTGACCAG AGGGATTTTATATCTGAAGAACATACACTATTAATAAATACCTTAGAGAAAGATTTTGACCTGGCTTTAGATAAAACTGTGGCAAGAAAA ATGTAATGAGCAATATATGGAAATAAACACACCTTTGTTAAAGATACTTTCTAAACTTGTGTTTAATAAACTTTAATAGTCATAGAATTG TAAATCACTATGGTTAACAGAAAGTGAAAATATTTTCATGCAGATGATGTGAACAGCCATGTGAATAGGTGACTTGGGCACACAGCAGGG TCATATGACTTCAGAAAACTTCGCTTTTCAGTTATTCCATTGTTATAATGTCAACCCTTTAAGACATTGATGTTTAGAGGGCTCACAAAT >2041_2041_2_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000352040_C1QTNF3_chr5_34035863_ENST00000382065_length(amino acids)=252AA_BP=34 MQTFPWWLSSLTLGRFAMGDSLSQARFASRTGGKSPQTGGLPPDCSKCCHGDYSFRGYQGPPGPPGPPGIPGNHGNNGNNGATGHEGAKG EKGDKGDLGPRGERGQHGPKGEKGYPGIPPELQIAFMASLATHFSNQNSGIIFSSVETNIGNFFDVMTGRFGAPVSGVYFFTFSMMKHED -------------------------------------------------------------- >2041_2041_3_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000504830_C1QTNF3_chr5_34035863_ENST00000231338_length(transcript)=2250nt_BP=463nt GAGCGGGCGGGCTGCGAGGCCGCGGGGCATGCGGGAGGCGGAGGGGTGGGGACCGGGGTGGGCTGCGCCCTCCCGCTCTGGCCCATTCCA CACCCCGCCGAAAGCGGACACTGTCAGCTGAATCACTCCCCTTTTAGGAGGAGGGAGGGGAAAAAGGTGTCTAGCTCCTTTCTGCTTAAA AAAGCACAGGGAGATCGCGGGCAGCTTTGCAGTCGCTGCCTTCTCGCGCCTGACCATGCACCCCTGCATCTTCCTGCTGGGCCACAGGCG AGCGCTTTATTTCTGGAGCTGAGGGCTAAAACTTTTTTGACTTTTCTTCTCCTCAACATCTGAATCATGCCATGTGCCCAGAGGAGCTGG CTTGCAAACCTTTCCGTGGTGGCTCAGCTCCTTAACTTTGGGGCGCTTTGCTATGGGAGACAGCCTCAGCCAGGCCCGGTTCGCTTCCCG GACAGGAGGCAAGTCTCCACAAACCGGAGGACTACCCCCAGACTGCAGTAAGTGTTGTCATGGAGACTACAGCTTTCGAGGCTACCAAGG CCCCCCTGGGCCACCGGGCCCTCCTGGCATTCCAGGAAACCATGGAAACAATGGCAACAATGGAGCCACTGGTCATGAAGGAGCCAAAGG TGAGAAGGGCGACAAAGGTGACCTGGGGCCTCGAGGGGAGCGGGGGCAGCATGGCCCCAAAGGAGAGAAGGGCTACCCGGGGATTCCACC AGAACTTCAGATTGCATTCATGGCTTCTCTGGCAACCCACTTCAGCAATCAGAACAGTGGGATTATCTTCAGCAGTGTTGAGACCAACAT TGGAAACTTCTTTGATGTCATGACTGGTAGATTTGGGGCCCCAGTATCAGGTGTGTATTTCTTCACCTTCAGCATGATGAAGCATGAGGA TGTTGAGGAAGTGTATGTGTACCTTATGCACAATGGCAACACAGTCTTCAGCATGTACAGCTATGAAATGAAGGGCAAATCAGATACATC CAGCAATCATGCTGTGCTGAAGCTAGCCAAAGGGGATGAGGTTTGGCTGCGAATGGGCAATGGCGCTCTCCATGGGGACCACCAACGCTT CTCCACCTTTGCAGGATTCCTGCTCTTTGAAACTAAGTAAATATATGACTAGAATAGCTCCACTTTGGGGAAGACTTGTAGCTGAGCTGA TTTGTTACGATCTGAGGAACATTAAAGTTGAGGGTTTTACATTGCTGTATTCAAAAAATTATTGGTTGCAATGTTGTTCACGCTACAGGT ACACCAATAATGTTGGACAATTCAGGGGCTCAGAAGAATCAACCACAAAATAGTCTTCTCAGATGACCTTGACTAATATACTCAGCATCT TTATCACTCTTTCCTTGGCACCTAAAAGATAATTCTCCTCTGACGCAGGTTGGAAATATTTTTTTCTATCACAGAAGTCATTTGCAAAGA ATTTTGACTACTCTGCTTTTAATTTAATACCAGTTTTCAGGAACCCCTGAAGTTTTAAGTTCATTATTCTTTATAACATTTGAGAGAATC GGATGTAGTGATATGACAGGGCTGGGGCAAGAACAGGGGCACTAGCTGCCTTATTAGCTAATTTAGTGCCCTCCGTGTTCAGCTTAGCCT TTGACCCTTTCCTTTTGATCCACAAAATACATTAAAACTCTGAATTCACATACAATGCTATTTTAAAGTCAATAGATTTTAGCTATAAAG TGCTTGACCAGTAATGTGGTTGTAATTTTGTGTATGTTCCCCCACATCGCCCCCAACTTCGGATGTGGGGTCAGGAGGTTGAGGTTCACT ATTAACAAATGTCATAAATATCTCATAGAGGTACAGTGCCAATAGATATTCAAATGTTGCATGTTGACCAGAGGGATTTTATATCTGAAG AACATACACTATTAATAAATACCTTAGAGAAAGATTTTGACCTGGCTTTAGATAAAACTGTGGCAAGAAAAATGTAATGAGCAATATATG GAAATAAACACACCTTTGTTAAAGATACTTTCTAAACTTGTGTTTAATAAACTTTAATAGTCATAGAATTGTAAATCACTATGGTTAACA GAAAGTGAAAATATTTTCATGCAGATGATGTGAACAGCCATGTGAATAGGTGACTTGGGCACACAGCAGGGTCATATGACTTCAGAAAAC TTCGCTTTTCAGTTATTCCATTGTTATAATGTCAACCCTTTAAGACATTGATGTTTAGAGGGCTCACAAATAAAATCTGAATACCTGTAA >2041_2041_3_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000504830_C1QTNF3_chr5_34035863_ENST00000231338_length(amino acids)=252AA_BP=34 MQTFPWWLSSLTLGRFAMGDSLSQARFASRTGGKSPQTGGLPPDCSKCCHGDYSFRGYQGPPGPPGPPGIPGNHGNNGNNGATGHEGAKG EKGDKGDLGPRGERGQHGPKGEKGYPGIPPELQIAFMASLATHFSNQNSGIIFSSVETNIGNFFDVMTGRFGAPVSGVYFFTFSMMKHED -------------------------------------------------------------- >2041_2041_4_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000504830_C1QTNF3_chr5_34035863_ENST00000382065_length(transcript)=2255nt_BP=463nt GAGCGGGCGGGCTGCGAGGCCGCGGGGCATGCGGGAGGCGGAGGGGTGGGGACCGGGGTGGGCTGCGCCCTCCCGCTCTGGCCCATTCCA CACCCCGCCGAAAGCGGACACTGTCAGCTGAATCACTCCCCTTTTAGGAGGAGGGAGGGGAAAAAGGTGTCTAGCTCCTTTCTGCTTAAA AAAGCACAGGGAGATCGCGGGCAGCTTTGCAGTCGCTGCCTTCTCGCGCCTGACCATGCACCCCTGCATCTTCCTGCTGGGCCACAGGCG AGCGCTTTATTTCTGGAGCTGAGGGCTAAAACTTTTTTGACTTTTCTTCTCCTCAACATCTGAATCATGCCATGTGCCCAGAGGAGCTGG CTTGCAAACCTTTCCGTGGTGGCTCAGCTCCTTAACTTTGGGGCGCTTTGCTATGGGAGACAGCCTCAGCCAGGCCCGGTTCGCTTCCCG GACAGGAGGCAAGTCTCCACAAACCGGAGGACTACCCCCAGACTGCAGTAAGTGTTGTCATGGAGACTACAGCTTTCGAGGCTACCAAGG CCCCCCTGGGCCACCGGGCCCTCCTGGCATTCCAGGAAACCATGGAAACAATGGCAACAATGGAGCCACTGGTCATGAAGGAGCCAAAGG TGAGAAGGGCGACAAAGGTGACCTGGGGCCTCGAGGGGAGCGGGGGCAGCATGGCCCCAAAGGAGAGAAGGGCTACCCGGGGATTCCACC AGAACTTCAGATTGCATTCATGGCTTCTCTGGCAACCCACTTCAGCAATCAGAACAGTGGGATTATCTTCAGCAGTGTTGAGACCAACAT TGGAAACTTCTTTGATGTCATGACTGGTAGATTTGGGGCCCCAGTATCAGGTGTGTATTTCTTCACCTTCAGCATGATGAAGCATGAGGA TGTTGAGGAAGTGTATGTGTACCTTATGCACAATGGCAACACAGTCTTCAGCATGTACAGCTATGAAATGAAGGGCAAATCAGATACATC CAGCAATCATGCTGTGCTGAAGCTAGCCAAAGGGGATGAGGTTTGGCTGCGAATGGGCAATGGCGCTCTCCATGGGGACCACCAACGCTT CTCCACCTTTGCAGGATTCCTGCTCTTTGAAACTAAGTAAATATATGACTAGAATAGCTCCACTTTGGGGAAGACTTGTAGCTGAGCTGA TTTGTTACGATCTGAGGAACATTAAAGTTGAGGGTTTTACATTGCTGTATTCAAAAAATTATTGGTTGCAATGTTGTTCACGCTACAGGT ACACCAATAATGTTGGACAATTCAGGGGCTCAGAAGAATCAACCACAAAATAGTCTTCTCAGATGACCTTGACTAATATACTCAGCATCT TTATCACTCTTTCCTTGGCACCTAAAAGATAATTCTCCTCTGACGCAGGTTGGAAATATTTTTTTCTATCACAGAAGTCATTTGCAAAGA ATTTTGACTACTCTGCTTTTAATTTAATACCAGTTTTCAGGAACCCCTGAAGTTTTAAGTTCATTATTCTTTATAACATTTGAGAGAATC GGATGTAGTGATATGACAGGGCTGGGGCAAGAACAGGGGCACTAGCTGCCTTATTAGCTAATTTAGTGCCCTCCGTGTTCAGCTTAGCCT TTGACCCTTTCCTTTTGATCCACAAAATACATTAAAACTCTGAATTCACATACAATGCTATTTTAAAGTCAATAGATTTTAGCTATAAAG TGCTTGACCAGTAATGTGGTTGTAATTTTGTGTATGTTCCCCCACATCGCCCCCAACTTCGGATGTGGGGTCAGGAGGTTGAGGTTCACT ATTAACAAATGTCATAAATATCTCATAGAGGTACAGTGCCAATAGATATTCAAATGTTGCATGTTGACCAGAGGGATTTTATATCTGAAG AACATACACTATTAATAAATACCTTAGAGAAAGATTTTGACCTGGCTTTAGATAAAACTGTGGCAAGAAAAATGTAATGAGCAATATATG GAAATAAACACACCTTTGTTAAAGATACTTTCTAAACTTGTGTTTAATAAACTTTAATAGTCATAGAATTGTAAATCACTATGGTTAACA GAAAGTGAAAATATTTTCATGCAGATGATGTGAACAGCCATGTGAATAGGTGACTTGGGCACACAGCAGGGTCATATGACTTCAGAAAAC TTCGCTTTTCAGTTATTCCATTGTTATAATGTCAACCCTTTAAGACATTGATGTTTAGAGGGCTCACAAATAAAATCTGAATACCTGTAA >2041_2041_4_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000504830_C1QTNF3_chr5_34035863_ENST00000382065_length(amino acids)=252AA_BP=34 MQTFPWWLSSLTLGRFAMGDSLSQARFASRTGGKSPQTGGLPPDCSKCCHGDYSFRGYQGPPGPPGPPGIPGNHGNNGNNGATGHEGAKG EKGDKGDLGPRGERGQHGPKGEKGYPGIPPELQIAFMASLATHFSNQNSGIIFSSVETNIGNFFDVMTGRFGAPVSGVYFFTFSMMKHED -------------------------------------------------------------- >2041_2041_5_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000515401_C1QTNF3_chr5_34035863_ENST00000231338_length(transcript)=2077nt_BP=290nt GCTTAAAAAAGCACAGGGAGATCGCGGGCAGCTTTGCAGTCGCTGCCTTCTCGCGCCTGACCATGCACCCCTGCATCTTCCTGCTGGGCC ACAGGCGAGCGCTTTATTTCTGGAGCTGAGGGCTAAAACTTTTTTGACTTTTCTTCTCCTCAACATCTGAATCATGCCATGTGCCCAGAG GAGCTGGCTTGCAAACCTTTCCGTGGTGGCTCAGCTCCTTAACTTTGGGGCGCTTTGCTATGGGAGACAGCCTCAGCCAGGCCCGGTTCG CTTCCCGGACAGGAGGCAAGTCTCCACAAACCGGAGGACTACCCCCAGACTGCAGTAAGTGTTGTCATGGAGACTACAGCTTTCGAGGCT ACCAAGGCCCCCCTGGGCCACCGGGCCCTCCTGGCATTCCAGGAAACCATGGAAACAATGGCAACAATGGAGCCACTGGTCATGAAGGAG CCAAAGGTGAGAAGGGCGACAAAGGTGACCTGGGGCCTCGAGGGGAGCGGGGGCAGCATGGCCCCAAAGGAGAGAAGGGCTACCCGGGGA TTCCACCAGAACTTCAGATTGCATTCATGGCTTCTCTGGCAACCCACTTCAGCAATCAGAACAGTGGGATTATCTTCAGCAGTGTTGAGA CCAACATTGGAAACTTCTTTGATGTCATGACTGGTAGATTTGGGGCCCCAGTATCAGGTGTGTATTTCTTCACCTTCAGCATGATGAAGC ATGAGGATGTTGAGGAAGTGTATGTGTACCTTATGCACAATGGCAACACAGTCTTCAGCATGTACAGCTATGAAATGAAGGGCAAATCAG ATACATCCAGCAATCATGCTGTGCTGAAGCTAGCCAAAGGGGATGAGGTTTGGCTGCGAATGGGCAATGGCGCTCTCCATGGGGACCACC AACGCTTCTCCACCTTTGCAGGATTCCTGCTCTTTGAAACTAAGTAAATATATGACTAGAATAGCTCCACTTTGGGGAAGACTTGTAGCT GAGCTGATTTGTTACGATCTGAGGAACATTAAAGTTGAGGGTTTTACATTGCTGTATTCAAAAAATTATTGGTTGCAATGTTGTTCACGC TACAGGTACACCAATAATGTTGGACAATTCAGGGGCTCAGAAGAATCAACCACAAAATAGTCTTCTCAGATGACCTTGACTAATATACTC AGCATCTTTATCACTCTTTCCTTGGCACCTAAAAGATAATTCTCCTCTGACGCAGGTTGGAAATATTTTTTTCTATCACAGAAGTCATTT GCAAAGAATTTTGACTACTCTGCTTTTAATTTAATACCAGTTTTCAGGAACCCCTGAAGTTTTAAGTTCATTATTCTTTATAACATTTGA GAGAATCGGATGTAGTGATATGACAGGGCTGGGGCAAGAACAGGGGCACTAGCTGCCTTATTAGCTAATTTAGTGCCCTCCGTGTTCAGC TTAGCCTTTGACCCTTTCCTTTTGATCCACAAAATACATTAAAACTCTGAATTCACATACAATGCTATTTTAAAGTCAATAGATTTTAGC TATAAAGTGCTTGACCAGTAATGTGGTTGTAATTTTGTGTATGTTCCCCCACATCGCCCCCAACTTCGGATGTGGGGTCAGGAGGTTGAG GTTCACTATTAACAAATGTCATAAATATCTCATAGAGGTACAGTGCCAATAGATATTCAAATGTTGCATGTTGACCAGAGGGATTTTATA TCTGAAGAACATACACTATTAATAAATACCTTAGAGAAAGATTTTGACCTGGCTTTAGATAAAACTGTGGCAAGAAAAATGTAATGAGCA ATATATGGAAATAAACACACCTTTGTTAAAGATACTTTCTAAACTTGTGTTTAATAAACTTTAATAGTCATAGAATTGTAAATCACTATG GTTAACAGAAAGTGAAAATATTTTCATGCAGATGATGTGAACAGCCATGTGAATAGGTGACTTGGGCACACAGCAGGGTCATATGACTTC AGAAAACTTCGCTTTTCAGTTATTCCATTGTTATAATGTCAACCCTTTAAGACATTGATGTTTAGAGGGCTCACAAATAAAATCTGAATA >2041_2041_5_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000515401_C1QTNF3_chr5_34035863_ENST00000231338_length(amino acids)=252AA_BP=34 MQTFPWWLSSLTLGRFAMGDSLSQARFASRTGGKSPQTGGLPPDCSKCCHGDYSFRGYQGPPGPPGPPGIPGNHGNNGNNGATGHEGAKG EKGDKGDLGPRGERGQHGPKGEKGYPGIPPELQIAFMASLATHFSNQNSGIIFSSVETNIGNFFDVMTGRFGAPVSGVYFFTFSMMKHED -------------------------------------------------------------- >2041_2041_6_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000515401_C1QTNF3_chr5_34035863_ENST00000382065_length(transcript)=2082nt_BP=290nt GCTTAAAAAAGCACAGGGAGATCGCGGGCAGCTTTGCAGTCGCTGCCTTCTCGCGCCTGACCATGCACCCCTGCATCTTCCTGCTGGGCC ACAGGCGAGCGCTTTATTTCTGGAGCTGAGGGCTAAAACTTTTTTGACTTTTCTTCTCCTCAACATCTGAATCATGCCATGTGCCCAGAG GAGCTGGCTTGCAAACCTTTCCGTGGTGGCTCAGCTCCTTAACTTTGGGGCGCTTTGCTATGGGAGACAGCCTCAGCCAGGCCCGGTTCG CTTCCCGGACAGGAGGCAAGTCTCCACAAACCGGAGGACTACCCCCAGACTGCAGTAAGTGTTGTCATGGAGACTACAGCTTTCGAGGCT ACCAAGGCCCCCCTGGGCCACCGGGCCCTCCTGGCATTCCAGGAAACCATGGAAACAATGGCAACAATGGAGCCACTGGTCATGAAGGAG CCAAAGGTGAGAAGGGCGACAAAGGTGACCTGGGGCCTCGAGGGGAGCGGGGGCAGCATGGCCCCAAAGGAGAGAAGGGCTACCCGGGGA TTCCACCAGAACTTCAGATTGCATTCATGGCTTCTCTGGCAACCCACTTCAGCAATCAGAACAGTGGGATTATCTTCAGCAGTGTTGAGA CCAACATTGGAAACTTCTTTGATGTCATGACTGGTAGATTTGGGGCCCCAGTATCAGGTGTGTATTTCTTCACCTTCAGCATGATGAAGC ATGAGGATGTTGAGGAAGTGTATGTGTACCTTATGCACAATGGCAACACAGTCTTCAGCATGTACAGCTATGAAATGAAGGGCAAATCAG ATACATCCAGCAATCATGCTGTGCTGAAGCTAGCCAAAGGGGATGAGGTTTGGCTGCGAATGGGCAATGGCGCTCTCCATGGGGACCACC AACGCTTCTCCACCTTTGCAGGATTCCTGCTCTTTGAAACTAAGTAAATATATGACTAGAATAGCTCCACTTTGGGGAAGACTTGTAGCT GAGCTGATTTGTTACGATCTGAGGAACATTAAAGTTGAGGGTTTTACATTGCTGTATTCAAAAAATTATTGGTTGCAATGTTGTTCACGC TACAGGTACACCAATAATGTTGGACAATTCAGGGGCTCAGAAGAATCAACCACAAAATAGTCTTCTCAGATGACCTTGACTAATATACTC AGCATCTTTATCACTCTTTCCTTGGCACCTAAAAGATAATTCTCCTCTGACGCAGGTTGGAAATATTTTTTTCTATCACAGAAGTCATTT GCAAAGAATTTTGACTACTCTGCTTTTAATTTAATACCAGTTTTCAGGAACCCCTGAAGTTTTAAGTTCATTATTCTTTATAACATTTGA GAGAATCGGATGTAGTGATATGACAGGGCTGGGGCAAGAACAGGGGCACTAGCTGCCTTATTAGCTAATTTAGTGCCCTCCGTGTTCAGC TTAGCCTTTGACCCTTTCCTTTTGATCCACAAAATACATTAAAACTCTGAATTCACATACAATGCTATTTTAAAGTCAATAGATTTTAGC TATAAAGTGCTTGACCAGTAATGTGGTTGTAATTTTGTGTATGTTCCCCCACATCGCCCCCAACTTCGGATGTGGGGTCAGGAGGTTGAG GTTCACTATTAACAAATGTCATAAATATCTCATAGAGGTACAGTGCCAATAGATATTCAAATGTTGCATGTTGACCAGAGGGATTTTATA TCTGAAGAACATACACTATTAATAAATACCTTAGAGAAAGATTTTGACCTGGCTTTAGATAAAACTGTGGCAAGAAAAATGTAATGAGCA ATATATGGAAATAAACACACCTTTGTTAAAGATACTTTCTAAACTTGTGTTTAATAAACTTTAATAGTCATAGAATTGTAAATCACTATG GTTAACAGAAAGTGAAAATATTTTCATGCAGATGATGTGAACAGCCATGTGAATAGGTGACTTGGGCACACAGCAGGGTCATATGACTTC AGAAAACTTCGCTTTTCAGTTATTCCATTGTTATAATGTCAACCCTTTAAGACATTGATGTTTAGAGGGCTCACAAATAAAATCTGAATA >2041_2041_6_ADAMTS12-C1QTNF3_ADAMTS12_chr5_33891835_ENST00000515401_C1QTNF3_chr5_34035863_ENST00000382065_length(amino acids)=252AA_BP=34 MQTFPWWLSSLTLGRFAMGDSLSQARFASRTGGKSPQTGGLPPDCSKCCHGDYSFRGYQGPPGPPGPPGIPGNHGNNGNNGATGHEGAKG EKGDKGDLGPRGERGQHGPKGEKGYPGIPPELQIAFMASLATHFSNQNSGIIFSSVETNIGNFFDVMTGRFGAPVSGVYFFTFSMMKHED -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ADAMTS12-C1QTNF3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ADAMTS12-C1QTNF3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ADAMTS12-C1QTNF3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
