
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CTPS2-AKAP8 (FusionGDB2 ID:20415) |
Fusion Gene Summary for CTPS2-AKAP8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CTPS2-AKAP8 | Fusion gene ID: 20415 | Hgene | Tgene | Gene symbol | CTPS2 | AKAP8 | Gene ID | 56474 | 10270 |
| Gene name | CTP synthase 2 | A-kinase anchoring protein 8 | |
| Synonyms | - | AKAP 95|AKAP-8|AKAP-95|AKAP95 | |
| Cytomap | Xp22.2 | 19p13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | CTP synthase 2CTP synthase IICTP synthetase type 2UTP-ammonia ligase 2cytidine 5'-triphosphate synthetase 2 | A-kinase anchor protein 8A kinase (PRKA) anchor protein 8A-kinase anchor protein, 95kDa | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9ULX6 | |
| Ensembl transtripts involved in fusion gene | ENST00000359276, ENST00000380241, ENST00000443824, ENST00000483053, | ENST00000269701, | |
| Fusion gene scores | * DoF score | 8 X 8 X 6=384 | 4 X 5 X 4=80 |
| # samples | 11 | 7 | |
| ** MAII score | log2(11/384*10)=-1.8036027871965 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/80*10)=-0.192645077942396 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CTPS2 [Title/Abstract] AND AKAP8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CTPS2(16711263)-AKAP8(15480990), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CTPS2 (5'-gene) Fusion gene breakpoints across CTPS2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
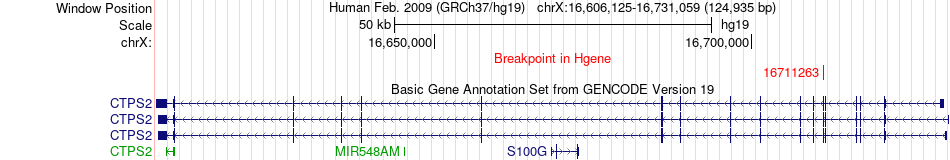 |
 Fusion gene breakpoints across AKAP8 (3'-gene) Fusion gene breakpoints across AKAP8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
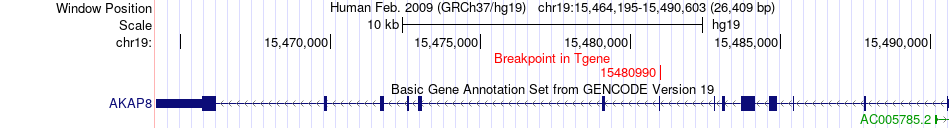 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-LN-A7HY | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - |
Top |
Fusion Gene ORF analysis for CTPS2-AKAP8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000359276 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - |
| In-frame | ENST00000380241 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - |
| In-frame | ENST00000443824 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - |
| intron-3CDS | ENST00000483053 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000443824 | CTPS2 | chrX | 16711263 | - | ENST00000269701 | AKAP8 | chr19 | 15480990 | - | 3954 | 1383 | 738 | 2423 | 561 |
| ENST00000359276 | CTPS2 | chrX | 16711263 | - | ENST00000269701 | AKAP8 | chr19 | 15480990 | - | 3567 | 996 | 351 | 2036 | 561 |
| ENST00000380241 | CTPS2 | chrX | 16711263 | - | ENST00000269701 | AKAP8 | chr19 | 15480990 | - | 3464 | 893 | 116 | 1933 | 605 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000443824 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - | 0.000531853 | 0.99946815 |
| ENST00000359276 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - | 0.000596947 | 0.999403 |
| ENST00000380241 | ENST00000269701 | CTPS2 | chrX | 16711263 | - | AKAP8 | chr19 | 15480990 | - | 0.000572882 | 0.9994272 |
Top |
Fusion Genomic Features for CTPS2-AKAP8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
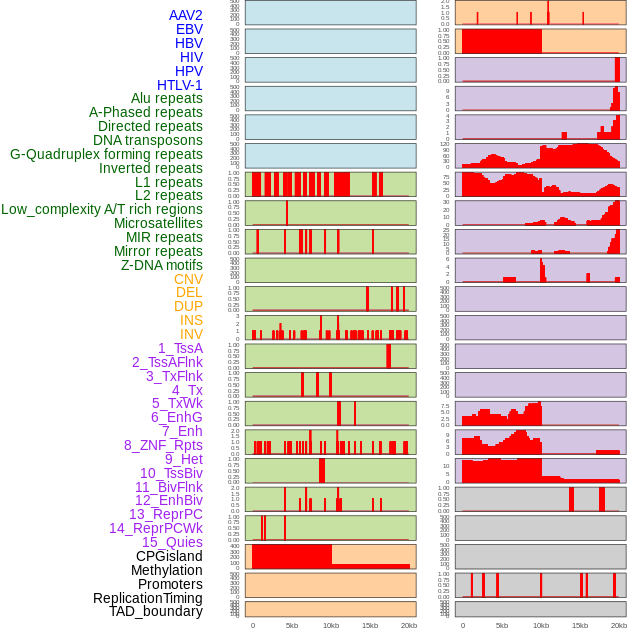 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for CTPS2-AKAP8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:16711263/chr19:15480990) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | AKAP8 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Could play a role in constitutive transport element (CTE)-mediated gene expression by association with DHX9. Increases CTE-dependent nuclear unspliced mRNA export (PubMed:10748171, PubMed:11402034). Proposed to target PRKACA to the nucleus but does not seem to be implicated in the binding of regulatory subunit II of PKA (PubMed:10761695, PubMed:11884601). May be involved in nuclear envelope breakdown and chromatin condensation. May be involved in anchoring nuclear membranes to chromatin in interphase and in releasing membranes from chromating at mitosis (PubMed:11034899). May regulate the initiation phase of DNA replication when associated with TMPO isoform Beta (PubMed:12538639). Required for cell cycle G2/M transition and histone deacetylation during mitosis. In mitotic cells recruits HDAC3 to the vicinity of chromatin leading to deacetylation and subsequent phosphorylation at 'Ser-10' of histone H3; in this function seems to act redundantly with AKAP8 (PubMed:16980585). May be involved in regulation of pre-mRNA splicing (PubMed:17594903). {ECO:0000269|PubMed:10748171, ECO:0000269|PubMed:11034899, ECO:0000269|PubMed:11402034, ECO:0000269|PubMed:11884601, ECO:0000269|PubMed:12538639, ECO:0000269|PubMed:16980585, ECO:0000305|PubMed:10761695}.; FUNCTION: (Microbial infection) In case of EBV infection, may target PRKACA to EBNA-LP-containing nuclear sites to modulate transcription from specific promoters. {ECO:0000269|PubMed:11884601}.; FUNCTION: (Microbial infection) Can synergize with DHX9 to activate the CTE-mediated gene expression of type D retroviruses. {ECO:0000269|PubMed:11402034}.; FUNCTION: (Microbial infection) In case of HIV-1 infection, involved in the DHX9-promoted annealing of host tRNA(Lys3) to viral genomic RNA as a primer in reverse transcription; in vitro negatively regulates DHX9 annealing activity. {ECO:0000269|PubMed:25034436}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 387_450 | 346 | 693.0 | Region | Involved in chromatin-binding | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 525_569 | 346 | 693.0 | Region | Involved in condensin complex recruitment | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 572_589 | 346 | 693.0 | Region | RII-binding | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 392_414 | 346 | 693.0 | Zinc finger | C2H2 AKAP95-type 1 | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 481_504 | 346 | 693.0 | Zinc finger | C2H2 AKAP95-type 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CTPS2 | chrX:16711263 | chr19:15480990 | ENST00000359276 | - | 6 | 19 | 300_554 | 213 | 939.6666666666666 | Domain | Glutamine amidotransferase type-1 |
| Hgene | CTPS2 | chrX:16711263 | chr19:15480990 | ENST00000380241 | - | 6 | 19 | 300_554 | 213 | 974.0 | Domain | Glutamine amidotransferase type-1 |
| Hgene | CTPS2 | chrX:16711263 | chr19:15480990 | ENST00000443824 | - | 6 | 19 | 300_554 | 213 | 911.6666666666666 | Domain | Glutamine amidotransferase type-1 |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 107_118 | 346 | 693.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 289_306 | 346 | 693.0 | Motif | Bipartite nuclear localization signal |
Top |
Fusion Gene Sequence for CTPS2-AKAP8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >20415_20415_1_CTPS2-AKAP8_CTPS2_chrX_16711263_ENST00000359276_AKAP8_chr19_15480990_ENST00000269701_length(transcript)=3567nt_BP=996nt GGGTGACGGGAGCTGCCTGTGCTGGAGGAATCACTTTTTAGGCGCTTGTTTTGGACCATTGCACAAACCCGGGTGCAAACCCCAAGCTCA CCAGCGTGAGTGAGCTGGGCCAGCAGCAGGGAGGAGAGGGGAAGGTGGGCGAGGAGGGCGCCGCGCACCCCGAGGCCCGTGTGGGCGGTG GGAAGATCCCGGGGGCGGCTTTGGACAGCCCCGGCAGCGACCCCTTCCCCAGCCCGACAGGTGAGCGCCAGGCCAGCCGCGGGGTGGAGC CCGCCGTGCCCACCGGCCACCCTCCCCGGTGCTACCACCACCGCGCAGATTATATCTGGGTGTTGGCACCCAGCCACTATTCTGCCAATG AAGTACATCCTGGTCACGGGTGGGGTCATCTCAGGCATTGGTAAAGGGATCATTGCCAGCAGCATTGGAACGATTCTAAAATCATGTGGA CTCCGAGTTACTGCCATAAAAATCGACCCCTATATTAACATCGATGCTGGCACTTTTTCACCTTATGAACACGGTGAAGTCTTCGTCTTA AATGATGGTGGAGAAGTTGATTTAGACCTTGGAAATTATGAAAGATTTTTGGATATTAATCTTTATAAAGACAACAATATCACCACGGGG AAGATATATCAGCATGTGATCAATAAAGAGAGGCGTGGTGATTACCTGGGGAAAACAGTGCAAGTTGTCCCTCACATTACTGATGCTGTC CAGGAGTGGGTTATGAATCAAGCCAAGGTGCCGGTGGATGGTAATAAGGAAGAGCCCCAAATATGCGTTATTGAGCTGGGAGGCACCATT GGAGACATCGAAGGAATGCCGTTTGTGGAGGCGTTTAGACAATTCCAGTTTAAGGCGAAAAGAGAGAATTTCTGTAATATCCACGTTAGC CTTGTCCCACAGCTCAGTGCTACCGGAGAACAAAAAACCAAACCCACCCAAAACAGCGTCCGCGCACTGAGGGGTTTAGGCCTGTCTCCA GATCTGGAGGATGAACTCTGCGACTCTGGGAGGCAAAGAGGAGAGAAGGAGGACGAGGACGAGGATGTGAAGAAGAGAAGGGAAAAGCAA AGGAGAAGAGACAGGACGCGGGACCGTGCAGCCGACAGAATTCAGTTTGCCTGTTCTGTATGCAAGTTCCGTAGCTTTGATGACGAAGAG ATCCAGAAGCATCTGCAAAGCAAATTTCACAAAGAGACCCTGCGGTTCATAAGCACCAAGCTGCCCGACAAGACCGTGGAGTTCCTCCAG GAATACATTGTAAACAGAAATAAGAAAATTGAGAAGCGGCGTCAGGAATTGATGGAGAAAGAAACCGCAAAACCAAAACCAGATCCTTTC AAAGGGATTGGCCAGGAGCACTTCTTCAAGAAGATCGAGGCTGCTCACTGCCTGGCCTGCGACATGCTAATTCCTGCACAGCCGCAGCTC CTCCAGCGGCACCTGCACTCCGTGGACCACAATCACAACCGCAGGTTGGCTGCTGAACAGTTCAAGAAAACCAGTCTCCATGTGGCTAAG AGTGTTTTGAACAACAGACATATAGTGAAGATGCTGGAAAAATACCTCAAGGGTGAGGACCCTTTCACCAGTGAAACTGTTGATCCAGAA ATGGAAGGAGATGACAATTTAGGAGGTGAGGATAAGAAAGAGACACCTGAGGAGGTGGCCGCGGACGTCTTAGCAGAGGTGATTACAGCA GCAGTGAGGGCCGTAGATGGGGAAGGAGCGCCCGCTCCAGAGAGCAGCGGGGAGCCGGCTGAGGACGAAGGCCCCACGGACACAGCGGAG GCCGGTAGTGATCCTCAAGCCGAACAGCTGCTGGAAGAGCAGGTGCCCTGTGGAACGGCACATGAGAAGGGCGTCCCCAAGGCCAGAAGT GAGGCTGCAGAGGCTGGAAATGGCGCCGAGACAATGGCAGCAGAGGCAGAAAGTGCCCAAACCAGAGTTGCTCCTGCCCCAGCTGCCGCG GATGCTGAAGTGGAACAAACTGATGCAGAGTCTAAAGACGCTGTTCCCACAGAATGATGCTCATTTCCCTGTTCCAGGGAAGGCGTTGGG ATGATGGATGCGTTGGTCTTTCTCCCTTGGTTTGTAAGCAGTACAAGGGCGTGTGCTCCCAGAATATGCTGTAATCTAATTTTGGTGAAG AGACCCAGCGTTTCCTCCTGAGCAGTGCCTCTCACGGCTTGTCTCATGCAGTCGTGTGGCTTCTTGCCCAGGTTTCAAAGCTGAAGTACA TTGTCCTTAGCGGCTGTAACATGTCTCTTGACAGTAGTGCACTTGGAATAATAAAGGTTGGGTGATTATATCTTGATGATACATTACTTG TTCAATACAGCCACTGATGGAATGCTTCCTTTTTTATTTTTTTCCTTAATTTTTTTTTTTATTTGGTTGGGAACAGCTGAATACTAGGAA TATATCTTGCTCTATAGAGGATTTTTTTTTGTATGTTTCAAGCTTCAGCCTTTAACCTATACCTTTGTAGTGCACCATATGGTGTGTGAC TTTCACAGGACTTCGCAGCACCTGGTTCACATGTGGCACTGACCGCGTCACATCCACGCACTCCCAAAGGCCAGAAGTATCTGACCGACC TACGCCACTGGAAACACACCCACCGCAACCTCAAGAACCAGACTGTGCAGAGGGCATTGCGTCCCAATCTTTAGTCCTTGCTGAATCAGT TCTCTAATATTTTACCTCATTTGTGTTCCACCTCTAGATTACTTCAGGTTTTTTTCCTTTAAAATTAGTTACTACCACTCAAATGTATTT ACAAAGAGAATTTGGCCAGGCACGGTGATGCATACCTATAATCCCAGCACTTCGGGAGGCCGTGGTGAGAGGATAGCTTAAGCCCAGGAG TTCAAGACCAACCTGGACAACATAGCAAGACCCCATCTCTTAAAAAAAAAGGAAAGAAAACTTGATGTGATTGCCATAGGTGGAATAATC CAACATAAATTGCCATAGATAGAAGGTATCTGTAATATATATATATATATATAAAATGAAATATATGTTTCATTTTAGAGAAATAACTAT TACTTTAGATCTTTCCAAATCTGAGAAAGGGAGGCTAGCATGTGTTCAAGGTTAGCACGCAACAGAATTTCCTAAAATCAGAAGAATTGG AAGATCCTCCCCTTTTGAAATGGCCCTGCTGTGTCAGTTTCCCTGTGGCCTTTTGAACTGTACATCTCACATGTTGGGAAACGCTGGCCA CTGGGAAATCATTAGAAAGGAGGCTGTAGAATATTTGCCGAGCCTCTACTGTATACCAGGGGCTAACTCACCAAGCACATTCTAGGAATT GGGCCCTGCTCATGAGGAGCCTTAGTGGAGATTCCAGGTGAATATTTATGAAAAAGTCAACATTAGAACTGAAAATGGAAATAAACTGCT TGAAAAGACGATGGTGCCTCTGGGTCATTTCTGCTCAGCATCCGTTTGTGTTGTGTAGATGCGTCTTAATCGCAGGGTGAGCATGCCTTT >20415_20415_1_CTPS2-AKAP8_CTPS2_chrX_16711263_ENST00000359276_AKAP8_chr19_15480990_ENST00000269701_length(amino acids)=561AA_BP=205 MPMKYILVTGGVISGIGKGIIASSIGTILKSCGLRVTAIKIDPYINIDAGTFSPYEHGEVFVLNDGGEVDLDLGNYERFLDINLYKDNNI TTGKIYQHVINKERRGDYLGKTVQVVPHITDAVQEWVMNQAKVPVDGNKEEPQICVIELGGTIGDIEGMPFVEAFRQFQFKAKRENFCNI HVSLVPQLSATGEQKTKPTQNSVRALRGLGLSPDLEDELCDSGRQRGEKEDEDEDVKKRREKQRRRDRTRDRAADRIQFACSVCKFRSFD DEEIQKHLQSKFHKETLRFISTKLPDKTVEFLQEYIVNRNKKIEKRRQELMEKETAKPKPDPFKGIGQEHFFKKIEAAHCLACDMLIPAQ PQLLQRHLHSVDHNHNRRLAAEQFKKTSLHVAKSVLNNRHIVKMLEKYLKGEDPFTSETVDPEMEGDDNLGGEDKKETPEEVAADVLAEV ITAAVRAVDGEGAPAPESSGEPAEDEGPTDTAEAGSDPQAEQLLEEQVPCGTAHEKGVPKARSEAAEAGNGAETMAAEAESAQTRVAPAP -------------------------------------------------------------- >20415_20415_2_CTPS2-AKAP8_CTPS2_chrX_16711263_ENST00000380241_AKAP8_chr19_15480990_ENST00000269701_length(transcript)=3464nt_BP=893nt AAAGGCAGGCATTCTCTTTCCTTTTCCTTCTCTCCTGAGCGCTCCTGCAGTTCCTGGGGCGTAGTAGGGGATCCACAAGCGTTTGTGACC AGTGAAGTTCTTTACAAGGGTGAGATCTGCACGGGAGGACCCGAGCGAGGGTCTCGGCTTGCCAGGAAGCCGGGGTTCCCCGGGAAGCGT GGAGTTCACCCGCGCACTCGAAGTGCCTTTGCAAAATTATATCTGGGTGTTGGCACCCAGCCACTATTCTGCCAATGAAGTACATCCTGG TCACGGGTGGGGTCATCTCAGGCATTGGTAAAGGGATCATTGCCAGCAGCATTGGAACGATTCTAAAATCATGTGGACTCCGAGTTACTG CCATAAAAATCGACCCCTATATTAACATCGATGCTGGCACTTTTTCACCTTATGAACACGGTGAAGTCTTCGTCTTAAATGATGGTGGAG AAGTTGATTTAGACCTTGGAAATTATGAAAGATTTTTGGATATTAATCTTTATAAAGACAACAATATCACCACGGGGAAGATATATCAGC ATGTGATCAATAAAGAGAGGCGTGGTGATTACCTGGGGAAAACAGTGCAAGTTGTCCCTCACATTACTGATGCTGTCCAGGAGTGGGTTA TGAATCAAGCCAAGGTGCCGGTGGATGGTAATAAGGAAGAGCCCCAAATATGCGTTATTGAGCTGGGAGGCACCATTGGAGACATCGAAG GAATGCCGTTTGTGGAGGCGTTTAGACAATTCCAGTTTAAGGCGAAAAGAGAGAATTTCTGTAATATCCACGTTAGCCTTGTCCCACAGC TCAGTGCTACCGGAGAACAAAAAACCAAACCCACCCAAAACAGCGTCCGCGCACTGAGGGGTTTAGGCCTGTCTCCAGATCTGGAGGATG AACTCTGCGACTCTGGGAGGCAAAGAGGAGAGAAGGAGGACGAGGACGAGGATGTGAAGAAGAGAAGGGAAAAGCAAAGGAGAAGAGACA GGACGCGGGACCGTGCAGCCGACAGAATTCAGTTTGCCTGTTCTGTATGCAAGTTCCGTAGCTTTGATGACGAAGAGATCCAGAAGCATC TGCAAAGCAAATTTCACAAAGAGACCCTGCGGTTCATAAGCACCAAGCTGCCCGACAAGACCGTGGAGTTCCTCCAGGAATACATTGTAA ACAGAAATAAGAAAATTGAGAAGCGGCGTCAGGAATTGATGGAGAAAGAAACCGCAAAACCAAAACCAGATCCTTTCAAAGGGATTGGCC AGGAGCACTTCTTCAAGAAGATCGAGGCTGCTCACTGCCTGGCCTGCGACATGCTAATTCCTGCACAGCCGCAGCTCCTCCAGCGGCACC TGCACTCCGTGGACCACAATCACAACCGCAGGTTGGCTGCTGAACAGTTCAAGAAAACCAGTCTCCATGTGGCTAAGAGTGTTTTGAACA ACAGACATATAGTGAAGATGCTGGAAAAATACCTCAAGGGTGAGGACCCTTTCACCAGTGAAACTGTTGATCCAGAAATGGAAGGAGATG ACAATTTAGGAGGTGAGGATAAGAAAGAGACACCTGAGGAGGTGGCCGCGGACGTCTTAGCAGAGGTGATTACAGCAGCAGTGAGGGCCG TAGATGGGGAAGGAGCGCCCGCTCCAGAGAGCAGCGGGGAGCCGGCTGAGGACGAAGGCCCCACGGACACAGCGGAGGCCGGTAGTGATC CTCAAGCCGAACAGCTGCTGGAAGAGCAGGTGCCCTGTGGAACGGCACATGAGAAGGGCGTCCCCAAGGCCAGAAGTGAGGCTGCAGAGG CTGGAAATGGCGCCGAGACAATGGCAGCAGAGGCAGAAAGTGCCCAAACCAGAGTTGCTCCTGCCCCAGCTGCCGCGGATGCTGAAGTGG AACAAACTGATGCAGAGTCTAAAGACGCTGTTCCCACAGAATGATGCTCATTTCCCTGTTCCAGGGAAGGCGTTGGGATGATGGATGCGT TGGTCTTTCTCCCTTGGTTTGTAAGCAGTACAAGGGCGTGTGCTCCCAGAATATGCTGTAATCTAATTTTGGTGAAGAGACCCAGCGTTT CCTCCTGAGCAGTGCCTCTCACGGCTTGTCTCATGCAGTCGTGTGGCTTCTTGCCCAGGTTTCAAAGCTGAAGTACATTGTCCTTAGCGG CTGTAACATGTCTCTTGACAGTAGTGCACTTGGAATAATAAAGGTTGGGTGATTATATCTTGATGATACATTACTTGTTCAATACAGCCA CTGATGGAATGCTTCCTTTTTTATTTTTTTCCTTAATTTTTTTTTTTATTTGGTTGGGAACAGCTGAATACTAGGAATATATCTTGCTCT ATAGAGGATTTTTTTTTGTATGTTTCAAGCTTCAGCCTTTAACCTATACCTTTGTAGTGCACCATATGGTGTGTGACTTTCACAGGACTT CGCAGCACCTGGTTCACATGTGGCACTGACCGCGTCACATCCACGCACTCCCAAAGGCCAGAAGTATCTGACCGACCTACGCCACTGGAA ACACACCCACCGCAACCTCAAGAACCAGACTGTGCAGAGGGCATTGCGTCCCAATCTTTAGTCCTTGCTGAATCAGTTCTCTAATATTTT ACCTCATTTGTGTTCCACCTCTAGATTACTTCAGGTTTTTTTCCTTTAAAATTAGTTACTACCACTCAAATGTATTTACAAAGAGAATTT GGCCAGGCACGGTGATGCATACCTATAATCCCAGCACTTCGGGAGGCCGTGGTGAGAGGATAGCTTAAGCCCAGGAGTTCAAGACCAACC TGGACAACATAGCAAGACCCCATCTCTTAAAAAAAAAGGAAAGAAAACTTGATGTGATTGCCATAGGTGGAATAATCCAACATAAATTGC CATAGATAGAAGGTATCTGTAATATATATATATATATATAAAATGAAATATATGTTTCATTTTAGAGAAATAACTATTACTTTAGATCTT TCCAAATCTGAGAAAGGGAGGCTAGCATGTGTTCAAGGTTAGCACGCAACAGAATTTCCTAAAATCAGAAGAATTGGAAGATCCTCCCCT TTTGAAATGGCCCTGCTGTGTCAGTTTCCCTGTGGCCTTTTGAACTGTACATCTCACATGTTGGGAAACGCTGGCCACTGGGAAATCATT AGAAAGGAGGCTGTAGAATATTTGCCGAGCCTCTACTGTATACCAGGGGCTAACTCACCAAGCACATTCTAGGAATTGGGCCCTGCTCAT GAGGAGCCTTAGTGGAGATTCCAGGTGAATATTTATGAAAAAGTCAACATTAGAACTGAAAATGGAAATAAACTGCTTGAAAAGACGATG GTGCCTCTGGGTCATTTCTGCTCAGCATCCGTTTGTGTTGTGTAGATGCGTCTTAATCGCAGGGTGAGCATGCCTTTTAAGCTTGTTCTC >20415_20415_2_CTPS2-AKAP8_CTPS2_chrX_16711263_ENST00000380241_AKAP8_chr19_15480990_ENST00000269701_length(amino acids)=605AA_BP=249 MHGRTRARVSACQEAGVPREAWSSPAHSKCLCKIISGCWHPATILPMKYILVTGGVISGIGKGIIASSIGTILKSCGLRVTAIKIDPYIN IDAGTFSPYEHGEVFVLNDGGEVDLDLGNYERFLDINLYKDNNITTGKIYQHVINKERRGDYLGKTVQVVPHITDAVQEWVMNQAKVPVD GNKEEPQICVIELGGTIGDIEGMPFVEAFRQFQFKAKRENFCNIHVSLVPQLSATGEQKTKPTQNSVRALRGLGLSPDLEDELCDSGRQR GEKEDEDEDVKKRREKQRRRDRTRDRAADRIQFACSVCKFRSFDDEEIQKHLQSKFHKETLRFISTKLPDKTVEFLQEYIVNRNKKIEKR RQELMEKETAKPKPDPFKGIGQEHFFKKIEAAHCLACDMLIPAQPQLLQRHLHSVDHNHNRRLAAEQFKKTSLHVAKSVLNNRHIVKMLE KYLKGEDPFTSETVDPEMEGDDNLGGEDKKETPEEVAADVLAEVITAAVRAVDGEGAPAPESSGEPAEDEGPTDTAEAGSDPQAEQLLEE -------------------------------------------------------------- >20415_20415_3_CTPS2-AKAP8_CTPS2_chrX_16711263_ENST00000443824_AKAP8_chr19_15480990_ENST00000269701_length(transcript)=3954nt_BP=1383nt TCCTTTGTCCGCTCTCTGATGGCGCCGGCTCCCTGCCCAGCGCTGAGTCGGGTCCGGCCGCCAGCCCCGCGCTCGCAGACCTCGGCTGCC GGGTGTGGCGCGGGGACTGGGGAACGCTGGCCCGTGCCCAGTGCGGTTGGAGCCTGTCCCGCGCGTCCCCGGGACGCGCTTCTTCCCGCC TCCGCCCGCGCCAGCGCCCGCACCCGGATCCCCACTTCTCCCGGCCCTCGGGAGCCAGGAGAGCCCTGAGATGGCGTTGGCGGAAGTGAA GCCCCGAGCGGGGGTCTGCGAGGCTGGTGATCAGCGCCGGTAACATGGCCTTTCTGTCCTCTCCCCGGTCCCAGTGCACCCCTTAAACAA CGACCCCCGCGTTTTCCCCGTACTAGATGGTTAGGGCGCATAGTGCCGAACTACGCTGCTGCTACAGAATAGCTTTTTTGGGGGCAACAT AAAAAAGAATTGTATGTATGGTTTATATACAACAAAATGTCCCATTTTCAGTGTGCAAATCGACGGGATTTGACAAATTATACACTCCTG TAACCATTATCCCAAAGTGAACATTGACCATTTCCTTCACCCTGCAAAGTTCCCTGGTACCTCCTTTCTGTCAATCCACCCCAGGCCCCG CACTCAACCCCGGTTCTGATTGTTATCATAGCTTAGCTTTGACTGTTCTAGAACTTCATAGAACAAGATCATCAGATTATATCTGGGTGT TGGCACCCAGCCACTATTCTGCCAATGAAGTACATCCTGGTCACGGGTGGGGTCATCTCAGGCATTGGTAAAGGGATCATTGCCAGCAGC ATTGGAACGATTCTAAAATCATGTGGACTCCGAGTTACTGCCATAAAAATCGACCCCTATATTAACATCGATGCTGGCACTTTTTCACCT TATGAACACGGTGAAGTCTTCGTCTTAAATGATGGTGGAGAAGTTGATTTAGACCTTGGAAATTATGAAAGATTTTTGGATATTAATCTT TATAAAGACAACAATATCACCACGGGGAAGATATATCAGCATGTGATCAATAAAGAGAGGCGTGGTGATTACCTGGGGAAAACAGTGCAA GTTGTCCCTCACATTACTGATGCTGTCCAGGAGTGGGTTATGAATCAAGCCAAGGTGCCGGTGGATGGTAATAAGGAAGAGCCCCAAATA TGCGTTATTGAGCTGGGAGGCACCATTGGAGACATCGAAGGAATGCCGTTTGTGGAGGCGTTTAGACAATTCCAGTTTAAGGCGAAAAGA GAGAATTTCTGTAATATCCACGTTAGCCTTGTCCCACAGCTCAGTGCTACCGGAGAACAAAAAACCAAACCCACCCAAAACAGCGTCCGC GCACTGAGGGGTTTAGGCCTGTCTCCAGATCTGGAGGATGAACTCTGCGACTCTGGGAGGCAAAGAGGAGAGAAGGAGGACGAGGACGAG GATGTGAAGAAGAGAAGGGAAAAGCAAAGGAGAAGAGACAGGACGCGGGACCGTGCAGCCGACAGAATTCAGTTTGCCTGTTCTGTATGC AAGTTCCGTAGCTTTGATGACGAAGAGATCCAGAAGCATCTGCAAAGCAAATTTCACAAAGAGACCCTGCGGTTCATAAGCACCAAGCTG CCCGACAAGACCGTGGAGTTCCTCCAGGAATACATTGTAAACAGAAATAAGAAAATTGAGAAGCGGCGTCAGGAATTGATGGAGAAAGAA ACCGCAAAACCAAAACCAGATCCTTTCAAAGGGATTGGCCAGGAGCACTTCTTCAAGAAGATCGAGGCTGCTCACTGCCTGGCCTGCGAC ATGCTAATTCCTGCACAGCCGCAGCTCCTCCAGCGGCACCTGCACTCCGTGGACCACAATCACAACCGCAGGTTGGCTGCTGAACAGTTC AAGAAAACCAGTCTCCATGTGGCTAAGAGTGTTTTGAACAACAGACATATAGTGAAGATGCTGGAAAAATACCTCAAGGGTGAGGACCCT TTCACCAGTGAAACTGTTGATCCAGAAATGGAAGGAGATGACAATTTAGGAGGTGAGGATAAGAAAGAGACACCTGAGGAGGTGGCCGCG GACGTCTTAGCAGAGGTGATTACAGCAGCAGTGAGGGCCGTAGATGGGGAAGGAGCGCCCGCTCCAGAGAGCAGCGGGGAGCCGGCTGAG GACGAAGGCCCCACGGACACAGCGGAGGCCGGTAGTGATCCTCAAGCCGAACAGCTGCTGGAAGAGCAGGTGCCCTGTGGAACGGCACAT GAGAAGGGCGTCCCCAAGGCCAGAAGTGAGGCTGCAGAGGCTGGAAATGGCGCCGAGACAATGGCAGCAGAGGCAGAAAGTGCCCAAACC AGAGTTGCTCCTGCCCCAGCTGCCGCGGATGCTGAAGTGGAACAAACTGATGCAGAGTCTAAAGACGCTGTTCCCACAGAATGATGCTCA TTTCCCTGTTCCAGGGAAGGCGTTGGGATGATGGATGCGTTGGTCTTTCTCCCTTGGTTTGTAAGCAGTACAAGGGCGTGTGCTCCCAGA ATATGCTGTAATCTAATTTTGGTGAAGAGACCCAGCGTTTCCTCCTGAGCAGTGCCTCTCACGGCTTGTCTCATGCAGTCGTGTGGCTTC TTGCCCAGGTTTCAAAGCTGAAGTACATTGTCCTTAGCGGCTGTAACATGTCTCTTGACAGTAGTGCACTTGGAATAATAAAGGTTGGGT GATTATATCTTGATGATACATTACTTGTTCAATACAGCCACTGATGGAATGCTTCCTTTTTTATTTTTTTCCTTAATTTTTTTTTTTATT TGGTTGGGAACAGCTGAATACTAGGAATATATCTTGCTCTATAGAGGATTTTTTTTTGTATGTTTCAAGCTTCAGCCTTTAACCTATACC TTTGTAGTGCACCATATGGTGTGTGACTTTCACAGGACTTCGCAGCACCTGGTTCACATGTGGCACTGACCGCGTCACATCCACGCACTC CCAAAGGCCAGAAGTATCTGACCGACCTACGCCACTGGAAACACACCCACCGCAACCTCAAGAACCAGACTGTGCAGAGGGCATTGCGTC CCAATCTTTAGTCCTTGCTGAATCAGTTCTCTAATATTTTACCTCATTTGTGTTCCACCTCTAGATTACTTCAGGTTTTTTTCCTTTAAA ATTAGTTACTACCACTCAAATGTATTTACAAAGAGAATTTGGCCAGGCACGGTGATGCATACCTATAATCCCAGCACTTCGGGAGGCCGT GGTGAGAGGATAGCTTAAGCCCAGGAGTTCAAGACCAACCTGGACAACATAGCAAGACCCCATCTCTTAAAAAAAAAGGAAAGAAAACTT GATGTGATTGCCATAGGTGGAATAATCCAACATAAATTGCCATAGATAGAAGGTATCTGTAATATATATATATATATATAAAATGAAATA TATGTTTCATTTTAGAGAAATAACTATTACTTTAGATCTTTCCAAATCTGAGAAAGGGAGGCTAGCATGTGTTCAAGGTTAGCACGCAAC AGAATTTCCTAAAATCAGAAGAATTGGAAGATCCTCCCCTTTTGAAATGGCCCTGCTGTGTCAGTTTCCCTGTGGCCTTTTGAACTGTAC ATCTCACATGTTGGGAAACGCTGGCCACTGGGAAATCATTAGAAAGGAGGCTGTAGAATATTTGCCGAGCCTCTACTGTATACCAGGGGC TAACTCACCAAGCACATTCTAGGAATTGGGCCCTGCTCATGAGGAGCCTTAGTGGAGATTCCAGGTGAATATTTATGAAAAAGTCAACAT TAGAACTGAAAATGGAAATAAACTGCTTGAAAAGACGATGGTGCCTCTGGGTCATTTCTGCTCAGCATCCGTTTGTGTTGTGTAGATGCG >20415_20415_3_CTPS2-AKAP8_CTPS2_chrX_16711263_ENST00000443824_AKAP8_chr19_15480990_ENST00000269701_length(amino acids)=561AA_BP=205 MPMKYILVTGGVISGIGKGIIASSIGTILKSCGLRVTAIKIDPYINIDAGTFSPYEHGEVFVLNDGGEVDLDLGNYERFLDINLYKDNNI TTGKIYQHVINKERRGDYLGKTVQVVPHITDAVQEWVMNQAKVPVDGNKEEPQICVIELGGTIGDIEGMPFVEAFRQFQFKAKRENFCNI HVSLVPQLSATGEQKTKPTQNSVRALRGLGLSPDLEDELCDSGRQRGEKEDEDEDVKKRREKQRRRDRTRDRAADRIQFACSVCKFRSFD DEEIQKHLQSKFHKETLRFISTKLPDKTVEFLQEYIVNRNKKIEKRRQELMEKETAKPKPDPFKGIGQEHFFKKIEAAHCLACDMLIPAQ PQLLQRHLHSVDHNHNRRLAAEQFKKTSLHVAKSVLNNRHIVKMLEKYLKGEDPFTSETVDPEMEGDDNLGGEDKKETPEEVAADVLAEV ITAAVRAVDGEGAPAPESSGEPAEDEGPTDTAEAGSDPQAEQLLEEQVPCGTAHEKGVPKARSEAAEAGNGAETMAAEAESAQTRVAPAP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CTPS2-AKAP8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 109_201 | 346.0 | 693.0 | DDX5 | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 1_210 | 346.0 | 693.0 | DPY30 | |
| Tgene | AKAP8 | chrX:16711263 | chr19:15480990 | ENST00000269701 | 6 | 14 | 1_195 | 346.0 | 693.0 | MCM2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CTPS2-AKAP8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CTPS2-AKAP8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
